NCDV VP8*64–224 was expressed in E. coli, purified and crystallized in the presence of a sialic acid derivative. X-ray diffraction data were obtained to a resolution of 2.0 Å and the crystallographic structure was determined by molecular replacement.
Keywords: bovine rotavirus, carbohydrate-recognizing domain
Abstract
The infectivity of rotavirus is dramatically enhanced by proteolytic cleavage of its outer layer VP4 spike protein into two functional domains, VP8* and VP5*. The carbohydrate-recognizing domain VP8* is proposed to bind sialic acid-containing host cell-surface glycans and this is followed by a series of subsequent virus–cell interactions. Live attenuated human and bovine rotavirus vaccine candidates for the prevention of gastroenteritis have been derived from bovine rotavirus strain NCDV. The NCDV VP8*64–224 was overexpressed, purified to homogeneity and crystallized in the presence of an N-acetylneuraminic acid derivative. X-ray diffraction data were collected to a resolution of 2.0 Å and the crystallographic structure of NCDV VP8*64–224 was determined by molecular replacement.
1. Introduction
Rotaviruses are recognized as the leading pathogens associated with severe diarrhoea in children and young domestic animals and are responsible for more than 30% of all diarrhoea-caused hospitalizations and an estimated 600 000 infant deaths each year (Parashar et al., 2006 ▶). Rotavirus-induced neonatal diarrhoea in calves occurs in many countries regardless of technology, management or sanitary procedures. Furthermore, economic loss in beef and dairy farms has driven significant efforts towards the development of vaccines and chemotherapeutic agents to control bovine neonatal diarrhoea. The Nebraska Calf Diarrhoea Virus (NCDV) strain of rotavirus has been used for cattle vaccination (Lu et al., 1994 ▶). The live oral human rotavirus vaccine candidate RIT 4237 has been derived from NCDV (Vesikari et al., 1984 ▶). Mature rotaviruses are non-enveloped and consist of a triple-layered particle containing a genome of 11 double-stranded RNA segments. The infectivity of some animal rotavirus strains is decreased by sialidase treatment of host cells. These ‘sialidase-sensitive’ strains contrast with human strains, which appear to be ‘sialidase-insensitive’. It is noteworthy that sialidase treatment, for example with Vibrio cholerae sialidase, removes terminal sialic acids but not those that are located internally within oligosaccharide chains, such as those that can act as viral receptors on host-cell surfaces. Thus, sialidase-insensitive rotaviruses may still utilize sialic acid as part of their carbohydrate-recognition motif. Certainly, human rotavirus strains KUN and MO interact with the ganglioside GM1 that contains an internal sialic acid during infection of MA104 monkey kidney cells (Guo et al., 1999 ▶). However, whether or not this interaction involves the sialic acid moiety is unknown. Thus, the strict requirement of cell-surface sialic acid for efficient rotavirus infectivity has yet to be ascertained. Models for initial rotavirus–cell interactions suggest that sialic acid-containing receptors might be recognized as common receptors for both sialidase-sensitive and sialidase-insensitive rotavirus strains, along with integrins and heat-shock proteins (Graham et al., 2003 ▶; Lopez & Arias, 2004 ▶). Both the outer layer viral spike protein (VP4) and the surface-coat protein (VP7) are involved in virus–host cell interactions at the early infection stage (Graham et al., 2003 ▶; Delorme et al., 2001 ▶). Rotavirus infectivity is markedly increased by trypsin treatment (Crawford et al., 2001 ▶), which cleaves VP4 into functional domains VP8* and VP5* which remain associated with the virion (Arias et al., 1996 ▶). The carbohydrate-recognizing domain VP8* has essential functions in receptor binding and cell attachment and has been suggested to bind sialic acid-containing host cell-surface glycans in some strains (Fiore et al., 1991 ▶; Dormitzer, Sun, Blixt et al., 2002 ▶). The crystallographic structures of the VP8* domains of sialidase-sensitive rhesus rotavirus (RRV; Dormitzer, Sun, Wagner et al., 2002 ▶) and porcine rotavirus strain CRW-8 (Scott et al., 2005 ▶; Blanchard et al., 2007 ▶) in complex with the sialic acid derivative methyl α-d-N-acetylneuraminide (Neu5Acα2Me) have provided some insight into the nature of sialic acid recognition by these different rotavirus strains. A site on the VP8* surface is consistent between RRV and CRW-8 in binding the sialic acid monosaccharide (Dormitzer, Sun, Wagner et al., 2002 ▶; Blanchard et al., 2007 ▶), but whether there is just one, or more than one, ligand-interaction site during engagement with larger oligosaccharide receptors remains to be determined. Furthermore, the ability of different strains to demonstrate specificity toward particular carbohydrates is of interest, with ganglioside-binding specificity reported as being distinct between sialidase-sensitive and sialidase-insensitive strains (Delorme et al., 2001 ▶). There are also differences within sialidase-sensitive strains; for example, RRV VP8* exhibits a tenfold lower affinity for the sialic acid N-glycolylneuraminic acid (Neu5Gc) compared with N-acetylneuraminic acid (Neu5Ac); the latter has a K d of 1.2 mM (Dormitzer, Sun, Blixt et al., 2002 ▶). In contrast, porcine OSU (which has a VP8* with 97% sequence identity to that of CRW-8) preferably binds gangliosides containing Neu5Gc over Neu5Ac (Rolsma et al., 1998 ▶). The sialidase-sensitive bovine NCDV and monkey SA11 recognize the terminal sialic acid-containing Neu5Gc-GM3 ganglioside, but interestingly neither bound to Neu5Ac-GM3 (Delorme et al., 2001 ▶). This apparent preference for N-glycolyl (NHGc) over N-acetyl (NHAc) groups contrasts with that shown by RRV (Dormitzer, Sun, Blixt et al., 2002 ▶).
As infection is triggered by rotavirus binding to host cells, it follows that suppression of this binding by competition for interaction with viral receptors will reduce virus infectivity. As the VP8* domain is involved in virus–cell interactions associated with sialic acid-containing carbohydrates, investigation into the molecular basis of the VP8*–sialic acid interaction in different rotavirus strains will further the understanding of early rotavirus-infection events and contribute to the development of rotavirus-specific therapeutics. We initiated a structural investigation to characterize the host-specificity of sialic acid recognition by the sialidase-sensitive bovine rotavirus NCDV, aiming in the first instance to explore the potential for VP8* interaction with N-acetyl derivatives. Here, we present the first report of the expression, purification and crystallization of bovine NCDV VP8*64–224 together with the preliminary details of X-ray crystallographic structure determination by molecular replacement.
2. Experimental procedures and results
2.1. Expression and purification of NCDV VP8*64–224
The NCDV VP8*64–224 construct was provided by Dr Gavan Holloway (University of Melbourne). The amino-acid sequence of NCDV VP8*64–224 is identical to that published (accession No. AB119636; Entrez Database, NCBI, NIH) except for the substitution of threonine by isoleucine at position 208. Escherichia coli strain BL21 (DE3) was transformed with pGEX-NCDV-VP8* and cells were grown at 310 K in Luria–Bertani medium supplemented with 150 µg ml−1 ampicillin. After reaching an OD600 of 0.6, the cultures were incubated at 298 K for 1 h and then induced with 1 mM isopropyl β-d-1-thiogalactopyranoside (BioVectra DCL). Cells were harvested after a 4 h induction by centrifugation at 6000g for 15 min. The supernatant was discarded and the cell pellets were frozen. Frozen cell pellets were thawed in PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4 pH 7.3) supplemented with 0.5 mM phenylmethylsulfonyl fluoride (PMSF; Roche Diagnostics). The cells were lysed with 3 mg ml−1 lysozyme supplemented with 1%(w/v) Triton X-100 and 20 µg ml−1 DNaseI. After 30 min centrifugation at 20 000g, cell debris was removed and the supernatant was passed over a glutathione Sepharose column (Amersham-Pharmacia Biotech) and washed with TNC buffer (20 mM Tris pH 8.0, 100 mM NaCl, 1 mM CaCl2). Proteolytic digestion was performed using 10 µg ml−1 TPCK-treated trypsin (Worthington Biochemical) for 2 h at room temperature. After digestion, a benzamidine Sepharose (Amersham-Pharmacia Biotech) column pre-equilibrated with TNC was connected to the glutathione Sepharose column. The N-terminal GST tag was cleaved from the protein with trypsin, leaving a remnant Gly-Ser peptide sequence, and the protein was eluted with a high-salt buffer (20 mM sodium phosphate pH 7.5, 1 M NaCl); the eluant was supplemented with 1 mM PMSF and 2.5 mM benzamidine. After dialyzing with TNE (20 mM Tris–HCl, 100 mM NaCl, 1 mM EDTA pH 8.0), the protein was further purified via size-exclusion chromatography using Sephacryl S100 resin (Sigma) pre-equilibrated with TNE. Purified VP8* protein was analysed for homogeneity by SDS–PAGE analysis (Fig. 1 ▶) and dynamic light-scattering (DLS) [CoolBatch + 90T instrument (Precision Detectors), 293 K, protein concentration of 20 mg ml−1, 20 repeats]. The DLS indicated one protein species with a hydrodynamic radius (R h) of 1.42 nm, corresponding to monomeric NCDV VP8*64–224, which is consistent with our previous DLS results for VP8* from rotavirus strains human Wa and CRW-8 (Kraschnefski et al., 2005 ▶; Scott et al., 2005 ▶). The polydispersity is 16.5% and falls in the range 15–30% that is indicative of good homogeneity.
Figure 1.
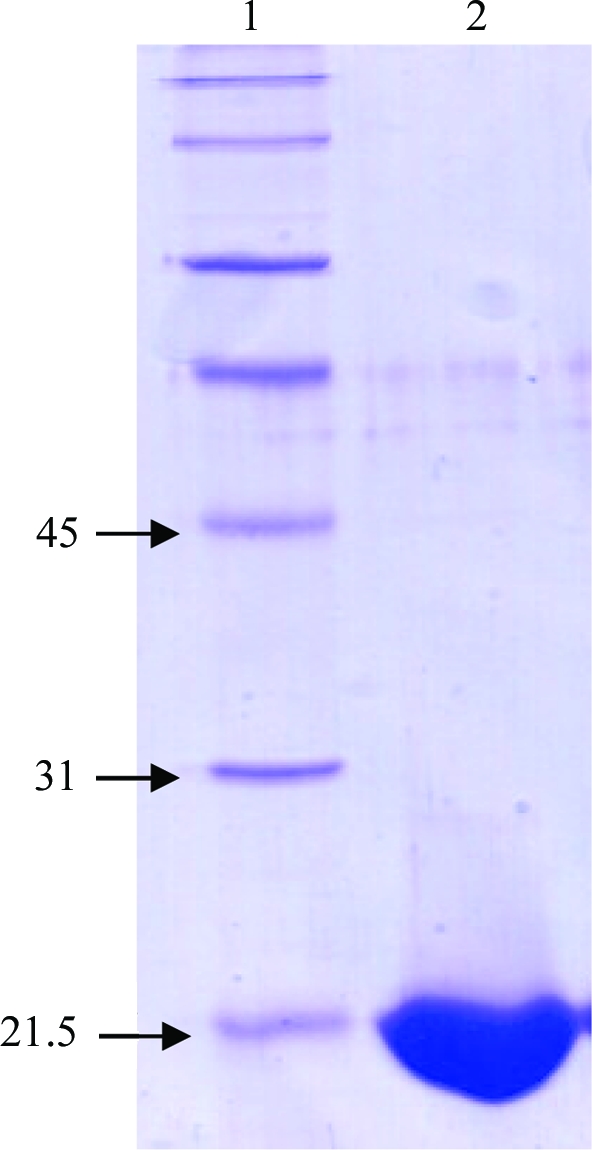
SDS–PAGE (12%, stained with Coomassie Blue) analysis of NCDV VP8*64–224. Lane 1, ladder of molecular-weight standards. Lane 2, band (slightly below 21.5 kDa) representing NCDV VP8*64–224 protein.
2.2. Crystallization and X-ray diffraction data collection
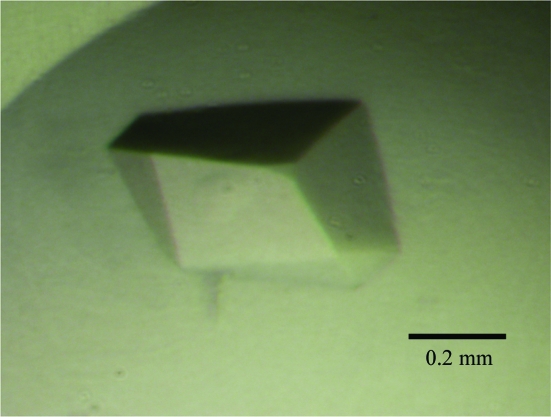
NCDV VP8*64–224 was concentrated to 20 mg ml−1 in TNE and mixed with an in-house-synthesized Neu5Acα2Me derivative to give a protein:ligand ratio of 1:50, which our experience with CRW-8 VP8* has shown to favour complex formation (Scott et al., 2005 ▶). Crystallization trials were carried out with Crystal Screen kits (Hampton Research) at 303 K using the hanging-drop vapour-diffusion method and drops comprising 1 µl protein–ligand mixture and 1 µl reservoir solution. Crystals formed in 3–5 d using reservoir solution of Crystal Screen I condition No. 35 (without further optimization), comprising 1.6 M sodium/potassium phosphate, 0.1 M HEPES pH 7.5, and grew to dimensions of 0.3 × 0.4 × 0.15 mm (Fig. 2 ▶). X-ray diffraction data were collected at room temperature (295 K) on a Bruker SMART 6000 diffractometer equipped with a SMART 6000 CCD detector and a MacScience M06XCE rotating-anode generator (Cu Kα, λ = 1.5418 Å). The crystal-to-detector distance was set at 55 mm and 600 frames were collected from one crystal (no significant decay was exhibited) with an oscillation angle of 0.3° and an exposure time of 60 s. X-ray diffraction data were collected to 2.0 Å resolution (Table 1 ▶) from a capillary-mounted crystal grown in the presence of ligand (Fig. 2 ▶). Data were processed with SAINT (Bruker AXS, Madison, Wisconsin, USA) and scaled and merged using SCALA (CCP4 suite; Collaborative Computational Project, Number 4, 1994 ▶). The crystal exhibited a trigonal system, with unit-cell parameters a = b = 56.28, c = 103.89 Å and crystal class P3 or P32. A Matthews coefficient (Matthews, 1968 ▶) V M of 2.64 Å3 Da−1 and an associated solvent content of 53.4% were calculated for two molecules in the asymmetric unit in the lower symmetry class or for one molecule in the asymmetric unit in the higher symmetry class. A homology model of NCDV VP8*64–224 was built using MODELLER (Sali & Blundell, 1993 ▶) based upon amino-acid sequence alignment and the crystal structure coordinates of porcine rotavirus CRW-8 VP8* (74% amino-acid sequence identity; PDB code 2i2s; Blanchard et al., 2007 ▶). Molecular replacement was performed using AMoRe (Navaza, 1994 ▶). A solution was determined in space group P3121, giving the anticipated single molecule in the asymmetric unit. The rotation function calculated at 15.0 to 3.0 Å resolution gave a highest rotation peak with an R factor of 56.4% and a correlation coefficient of 32.3%, exhibiting some distinction from the next highest peak, which had an R factor of 56.9% and a correlation coefficient of 30.3%. The translation-function calculation gave a highest peak correlating to the top rotation-function solution, with an R factor of 42.9% and a correlation coefficient of 60.3%, which are clearly distinct from those of the next highest peak, which had an R factor of 48.2% and a correlation coefficient of 51.9%. Rigid-body refinement was applied to this molecule and an R factor of 39.1% and a correlation coefficient of 71.8% were obtained. Initial refinement (ten cycles) of the structure gave an R factor of 28.4% and an R free of 30.5%; further model building and refinement are in progress.
Figure 2.
Crystal of NCDV VP8*64–224 in complex with the sialic acid derivative. The resulting X-ray diffraction data (Table 1 ▶) enabled structure determination. The scale bar represents 0.2 mm.
Table 1. X-ray diffraction data statistics.
Values in parentheses are for the highest resolution shell.
| Resolution (Å) | 48.74–1.99 (2.10–1.99) |
| Total No. of observations | 81100 (4173) |
| Total No. of unique observations | 13539 (1847) |
| Redundancy | 5.99 (2.26) |
| Completeness (%) | 99.2 (95.1) |
| Mean I/σ(I) | 9.0 (2.9) |
| Rmerge† (%) | 6.4 (26.3) |
| Crystal system, space group | Trigonal, P3121 |
| Unit-cell parameters (Å, °) | a = b = 56.28, c = 103.89, α = β = 90, γ = 120 |
R
merge = 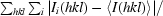
 .
.
Acknowledgments
HB, MJK, BSC and MvI gratefully acknowledge the financial support of the Australian Research Council (ARC). AG thanks the Ecole de Biologie Industrielle (EBI), Paris, France for the award of an internship scholarship undertaken within the group of HB. BSC is the recipient of a Senior Research Fellowship (350253) from the National Health and Medical Research Council of Australia. MvI thanks the ARC for the award of an Australian Federation Fellowship.
References
- Arias, C. F., Romero, P., Alvarez, V. & Lopez, S. (1996). J. Virol.70, 5832–5839. [DOI] [PMC free article] [PubMed]
- Blanchard, H., Yu, X., Coulson, B. S. & von Itzstein, M. (2007). J. Mol. Biol.367, 1215–1226. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Crawford, S. E., Mukherjee, S. K., Estes, M. K., Lawton, J. A., Shaw, A. L., Ramig, R. F. & Prasad, B. V. (2001). J. Virol.75, 6052–6061. [DOI] [PMC free article] [PubMed]
- Delorme, C., Brussow, H., Sidoti, J., Roche, N., Karisson, K. A., Neeser, J. R. & Teneberg, S. (2001). J. Virol.75, 2276–2287. [DOI] [PMC free article] [PubMed]
- Dormitzer, P. R., Sun, Z. Y., Blixt, O., Paulson, J. C., Wagner, G. & Harrison, S. C. (2002). J. Virol.76, 10512–10517. [DOI] [PMC free article] [PubMed]
- Dormitzer, P. R., Sun, Z. Y., Wagner, G. & Harrison, S. C. (2002). EMBO J.21, 885–897. [DOI] [PMC free article] [PubMed]
- Fiore, L., Greenberg, H. B. & Mackow, E. R. (1991). Virology, 181, 553–563. [DOI] [PubMed]
- Graham, K. L., Halasz, P., Tan, Y., Hewish, M. J., Takada, Y., Mackow, E. R., Robinson, M. K. & Coulson, B. S. (2003). J. Virol.77, 9969–9978. [DOI] [PMC free article] [PubMed]
- Guo, C. T., Nakagomi, O., Mochizuki, M., Ishida, H., Kiso, M., Ohta, Y., Suzuki, T., Miyamoto, D., Hidari, K. I. & Suzuki, Y. (1999). J. Biochem. (Tokyo), 126, 683–688. [DOI] [PubMed]
- Kraschnefski, M. J., Scott, S. A., Holloway, G., Coulson, B. S., von Itzstein, M. & Blanchard, H. (2005). Acta Cryst. F61, 989–993. [DOI] [PMC free article] [PubMed]
- Lopez, S. & Arias, C. F. (2004). Trends Microbiol.12, 271–278. [DOI] [PubMed]
- Lu, W., Duhamel, G. E., Benfield, D. A. & Grotelueschen, D. M. (1994). Vet. Microbiol.42, 159–170. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Navaza, J. (1994). Acta Cryst. A50, 157–163.
- Parashar, U. D., Gibson, C. J., Bresse, J. S. & Glass, R. I. (2006). Emerg. Infect. Dis.12, 304–306. [DOI] [PMC free article] [PubMed]
- Rolsma, M. D., Kuhlenschmidt, T. B., Gelberg, H. B. & Kuhlenschmidt, M. S. (1998). J. Virol.72, 9079–9091. [DOI] [PMC free article] [PubMed]
- Sali, A. & Blundell, T. L. (1993). J. Mol. Biol.234, 779–815. [DOI] [PubMed]
- Scott, S. A., Holloway, G., Coulson, B. S., Szyczew, A. J., Kiefel, M. J., von Itzstein, M. & Blanchard, H. (2005). Acta Cryst. F61, 617–620. [DOI] [PMC free article] [PubMed]
- Vesikari, T., Isolauri, E., D’Hondt, E., Delem, A., Andre, F. E. & Zissis, G. (1984). Lancet, 1, 977–981. [DOI] [PubMed]