The crystal structure of the core-binding domain of bacteriophage lambda integrase has been determined at 2.0 Å resolution.
Keywords: bacteriophage lambda integrase, binding-coupled folding
Abstract
Bacteriophage lambda integrase catalyzes site-specific DNA recombination. A helical bundle domain in the enzyme, called the core-binding domain (IntCB), promotes the catalysis of an intermediate DNA-cleavage reaction that is critical for recombination and is not well folded in solution in the absence of DNA. To gain structural insights into the mechanism behind the accessory role of this domain in catalysis, an attempt was made to crystallize an IntCB–DNA complex, but crystals of free IntCB were fortuitously obtained. The three-dimensional structure of DNA-free IntCB was solved at 2.0 Å resolution by molecular replacement using as the search model the previously available DNA-bound 2.8 Å structure of the IntCB domain in a larger construct of lambda integrase. The crystal structure of DNA-free IntCB resembles the DNA-bound structure of IntCB, but exhibits subtle differences in the DNA-binding face and lacks electron density for ten residues in the C-terminus that form a portion of a linker connecting IntCB to the C-terminal catalytic domain of the enzyme. Thus, this work reveals the domain in the absence of DNA and allows comparison with the DNA-bound form of this catalytically activating domain.
1. Introduction
Bacteriophage lambda integrase (λ-Int), a site-specific DNA recombinase encoded by bacteriophage lambda, catalyzes genetic recombination between the virus and the host bacterium to enable the lytic and the lysogenic phases of the viral life cycle (Campbell, 1962 ▶). Recombination is carried out by four molecules of λ-Int acting on conserved genomic sequences, called core sites, and proceeds via a Holliday-junction intermediate formed by a series of DNA-cleavage, strand-exchange and ligation reactions (for reviews, see Sadowski, 1993 ▶; Radman-Livaja et al., 2006 ▶; Landy, 1989 ▶). Enzyme activity at the core site is mediated by two of the three domains of λ-Int: the central DNA-binding domain, commonly known as the core-binding domain (IntCB), and the C-terminal catalytic domain (IntCat), which enables the DNA-cleavage reaction through a conserved tyrosine nucleophile (Tirumalai et al., 1997 ▶, 1998 ▶). The crystal structures of DNA-bound partial and full-length constructs of λ-Int provide critical knowledge about the organization of the active site and reveal details of interactions of the enzyme with DNA, as well as with the other enzyme molecules in the dimeric or tetrameric assemblies (Aihara et al., 2003 ▶; Biswas et al., 2005 ▶). These crystal structures show that IntCB and IntCat bind to opposite faces of the recognition sequence by adopting a clamp-like architecture with an extended linker connecting the two domains. Experiments in our laboratory with isolated protein constructs of IntCB and IntCat showed that IntCB contributes to an increase in DNA cleavage by IntCat even when they are not covalently linked together (Subramaniam et al., 2007 ▶). We proposed that IntCB might render the substrate DNA more suitable for cleavage by IntCat by inducing structural changes in the DNA.
Characterization of IntCB–DNA interactions in solution using biophysical techniques revealed that IntCB indeed induces structural changes in the DNA and concomitantly undergoes a transition from a molten globule-like structure to a well folded structure upon binding DNA (Kamadurai & Foster, 2007 ▶). In order to characterize the IntCB-mediated DNA structural changes in more detail, we attempted to crystallize an IntCB–cognate DNA complex; fortuitously, we observed diffraction-quality crystals of IntCB that were free of DNA under certain crystallization conditions, although the presence of DNA in the mother liquor strongly promoted crystallization. The crystal structure of DNA-free IntCB was solved at 2.0 Å resolution by molecular replacement and was found to resemble the DNA-bound conformation observed in the other lower resolution (3.0, 2.8 Å) crystal structures of complete and partial constructs of lambda integrase (Aihara et al., 2003 ▶; Biswas et al., 2005 ▶), but with subtle changes in the DNA-binding face and in the C-terminus.
2. Materials and methods
The IntCB construct used in this study contains residues 75–176 of the full-length 356-residue lambda integrase and includes a starting methionine. It was obtained by deletion mutagenesis of an IntCB construct encoding residues 63–176 (Tirumalai et al., 1998 ▶). As a ligand, a 15-base-pair cognate DNA duplex, 5′-d(GCT CAA GTT AGT ACG)-3′, and its complement were used. Oligonucleotide preparation, protein expression and purification were performed using methods described previously (Subramaniam et al., 2003 ▶, 2007 ▶; Kamadurai et al., 2003 ▶). The IntCB–DNA complex was assembled in 25 mM Tris pH 8.5 and 10 mM sodium chloride (assembly buffer) at a final protein concentration of 0.6 mM with a 10% stoichiometric excess of DNA or at an equimolar concentration of 1.75 mM. In all crystallization trials, 1 µl stock solution was mixed with 1 µl reservoir solution at room temperature and the crystallization trays were incubated at 277 K. Initial crystals were obtained via the sitting-drop method with reservoir solution condition No. B5 from the Crystal Screen HT kit (Hampton Research) containing 0.1 M Tris pH 8.5, 0.2 M lithium sulfate and 30% PEG 4000. Subsequent trials were carried out with and without the DNA ligand with the same reservoir solution composition but prepared in-house. Crystal constituents were identified by dissolving the crystal in the assembly buffer followed by analysis using Coomassie Blue-stained SDS–PAGE and ethidium bromide-stained DNA agarose gels. To derivatize with platinum, a saturated aqueous solution of K2PtCl4 was diluted with the pre-equilibrated reservoir solution and a crystal was soaked in this K2PtCl4-containing reservoir solution for 5 min prior to data collection.
The crystals were mounted on nylon loops (Hampton Research) and were flash-frozen either in a nitrogen–helium stream at 93 K or in liquid nitrogen, with the reservoir solution serving as the cryoprotectant. Data were collected using a Rigaku rotating-anode generator equipped with an R-AXIS IV++ detector. The data were scaled and averaged using CrystalClear (Rigaku Inc.) and then imported into the CCP4 software suite (Collaborative Computational Project, Number 4, 1994 ▶) for subsequent analysis.
For molecular replacement, the coordinates corresponding to IntCB in PDB entry 1p7d (chain A, residues 74–176; Aihara et al., 2003 ▶) were used as the search model and the search was implemented using the automated search feature in Phaser (McCoy, 2007 ▶). The initial structure from molecular replacement was then refined using REFMAC (Murshudov et al., 1997 ▶) followed by visual inspection and structure manipulation using Coot (Emsley & Cowtan, 2004 ▶). The stereochemical quality of the final structure, listed in Table 1 ▶, was assessed using PROCHECK (Laskowski et al., 1993 ▶).
Table 1. Diffraction data and crystallographic refinement statistics.
Values in parentheses are for the highest resolution shell.
| Data collection | |
| Space group | P41212 |
| Unit-cell parameters (, ) | a = b = 46.69, c = 111.53, = = = 90 |
| Resolution range () | 43.072.00 (2.072.00) |
| Total reflections | 57414 (3708) |
| Unique reflections | 8503 (641) |
| R merge (%) | 3.5 (17.4) |
| Completeness (%) | 95.4 (74.5) |
| I/(I) | 34.2 (7.9) |
| Refinement | |
| R cryst (%) | 22.8 |
| R free (%) | 28.4 |
| R.m.s. deviations from ideal geometry | |
| Bond lengths () | 0.02 |
| Bond angles () | 1.6 |
| Ramachandran plot statistics | |
| Residues in most favored regions (%) | 95.2 |
| Residues in additional allowed regions (%) | 4.8 |
| Residues in disallowed regions (%) | 0 |
| Average B value, all atoms (2) | 23.91 |
3. Results and discussion
Large single crystals were obtained in crystallization wells containing an IntCB–DNA mixture after one week of incubation at 277 K. Even though the starting material contained both IntCB and the DNA ligand, analysis of these crystals on SDS–PAGE and DNA agarose gels revealed only the presence of IntCB, indicating that IntCB crystallized without the DNA. Crystal structures of DNA-bound IntCB in larger constructs of lambda integrase (Aihara et al., 2003 ▶; Biswas et al., 2005 ▶) show that IntCB–DNA interactions are mediated predominantly by electrostatic interactions involving the phosphate backbone of the DNA ligand. Consistent with this observation, binding studies showed that IntCB–DNA interactions are weakened upon an increase in ionic strength (Kamadurai, 2007 ▶). The absence of DNA in these IntCB crystals could be attributed to weakened protein–DNA interactions arising from the moderately high ionic strength of the crystallization condition. Nevertheless, the presence of the DNA oligonucleotide strongly promoted protein crystallization, as omission of the DNA ligand significantly reduced the yield of diffraction-quality crystals.
IntCB crystals belonged to space group P41212 and in general had good diffraction qualities, with the highest resolution in the region of 2.0 Å. A K2PtCl4-soaked crystal showed exceptionally good diffraction data [R merge, I/σ(I)] and completeness and hence the data set collected on this crystal was used for structure determination, although no density was observed for the heavy metal. Matthews coefficient analysis with the mass of IntCB and the observed unit-cell parameters (a = b = 46.69, c = 111.53 Å) showed that the asymmetric unit consists of only one copy of IntCB. We were able to find a unique molecular-replacement solution using the IntCB portion of the crystal structure of a DNA-bound partial λ-Int construct that contains both IntCB and IntCat (PDB code 1p7d; Aihara et al., 2003 ▶) as the search model. The IntCB structure was refined to a final R cryst of 22.8% and an R free of 28.4%. The difference in these values is moderately higher than that expected at this resolution; however, comparison of the model against a composite OMIT map confirmed the accuracy of the model and additional refinement to improve statistics did not result in significant changes in the model. Refinement and stereochemical statistics of the final structure are included in Table 1 ▶.
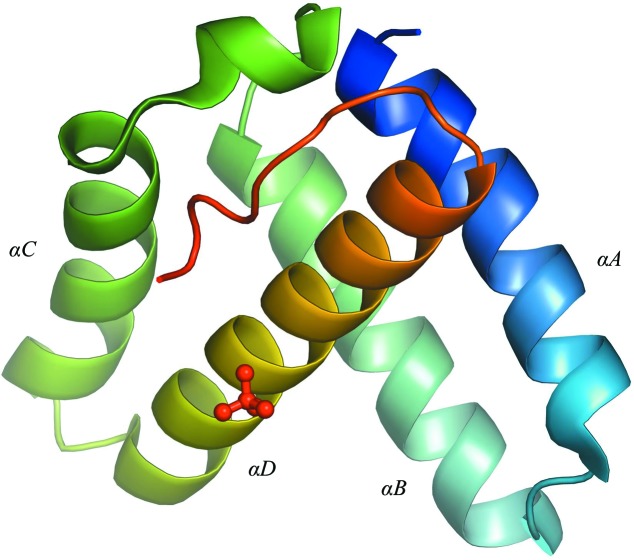
The refined structure of DNA-free IntCB reveals a four-helix bundle architecture defined by the helices A–D (Fig. 1 ▶). The crystal-packing interactions involved the fringes of the helices, including the helix–turn–helix DNA-binding motifs constituted by helices A and B and by helices C and D, and occlude the space for DNA, which may also explain why DNA failed to cocrystallize. The arrangement of IntCB in the crystal is unlike the arrangement in the functionally important tetramer assembly (Biswas et al., 2005 ▶), suggesting that the interprotomer interactions observed in the tetrameric assembly were not critical for stabilizing the IntCB structure in the crystal.
Figure 1.
Cartoon representation of the crystal structure of DNA-free IntCB. The structure is shown in a rainbow color gradient from the N-terminus (blue) to the C-terminus (red). The lone sulfate ion is shown as a ball-and-stick model. IntCB folds into a four-helix bundle (helices A–D) with a flexible linker region at the C-terminus. The IntCB construct used in this study contains residues 75–176 of the full-length lambda integrase and a starting methionine (residue 74), but electron density was only observed between residues 74 and 166. The B factors are nearly uniform through out the protein, averaging 24 ± 5 Å2.
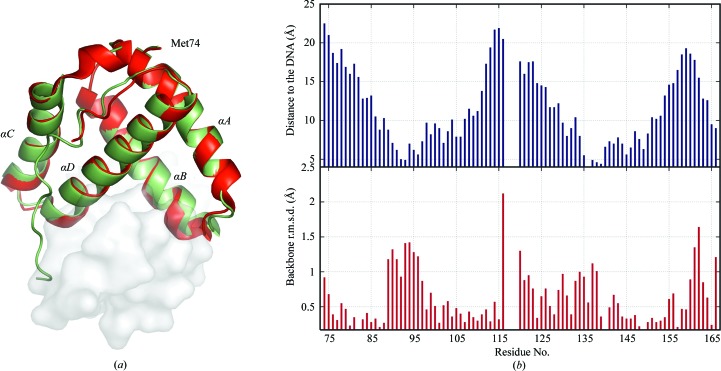
The final structure of DNA-free IntCB resembles the DNA-bound structure (Aihara et al., 2003 ▶) with an average backbone r.m.s.d. of 0.6 Å (Fig. 2 ▶ a). Modest but noticeable structural differences between the two structures are observed in regions that are proximal to the DNA (Fig. 2 ▶ b). A sulfate ion bound to Arg152 that mimics the interactions of the residue with the phosphate backbone in the DNA-bound structure (Aihara et al., 2003 ▶) was observed in the structure, as well as 98 ordered water molecules. No electron density was observed for ten C-terminal residues (167–176) that form a part of the flexible linker region connecting IntCB to the C-terminal catalytic domain of lambda integrase (Aihara et al., 2003 ▶). Electron densities were also lacking for some of the surface-exposed side chains, particularly those of Lys93, Lys95 and Arg109 that interact with the phosphate backbone, suggesting disorder in the absence of DNA. However, the side chain of Asn99, which makes a base-specific contact with DNA (Aihara et al., 2003 ▶), is ordered and is observed to interact with the side chain of Lys103. Similarly, interaction with the sulfate ion and crystal-packing interactions appeared to have contributed to the ordering of the side chains of Arg152 and residues 116–120, respectively.
Figure 2.
Structural differences between free and DNA-bound IntCB. (a) Superimposition of free and DNA-bound crystal structures of IntCB. The structure without DNA is shown in red and with DNA (PDB code 1p7d; Aihara et al., 2003 ▶) in green; the DNA is shown as a semi-transparent surface. Overall, the structures overlay well, but subtle changes are observed in the DNA-binding region; also, a portion of a loop (residues 167–176) is missing in the DNA-free IntCB structure. Unlike the DNA-bound structure, the free structure contains a 310-helix between helices B and C. (b) Correlation between structural differences between free and DNA-bound IntCB (PDB code 1p7d; Aihara et al., 2003 ▶) and proximity to DNA. Top, the closest approach of any backbone atoms in each residue in IntCB to the DNA; r.m.s.d. values and the distances to the DNA were calculated for the amide N, Cα and carbonyl C atoms using MOLMOL (Koradi et al., 1996 ▶). Bottom, the backbone r.m.s.d. between the structures in (a). These values could not be computed for residues 117–119 as the DNA-bound structure lacks electron density in this segment.
In summary, we report the crystal structure of the core-binding domain of the prototypical site-specific DNA recombinase in the absence of DNA. Although the protein is not well folded in solution, by including a DNA ligand in the crystallizing solution high-quality protein-only crystals were observed. Overall, the structure of the free protein resembles the DNA-bound structure, but with subtle differences in the backbone and side chains of residues involved in DNA binding.
Supplementary Material
PDB reference: IntCB, 2oxo, r2oxosf
References
- Aihara, H., Kwon, H. J., Nunes-Duby, S. E., Landy, A. & Ellenberger, T. (2003). Mol. Cell, 12, 187–198. [DOI] [PubMed]
- Biswas, T., Aihara, H., Radman-Livaja, M., Filman, D., Landy, A. & Ellenberger, T. (2005). Nature (London), 435, 1059–1066. [DOI] [PMC free article] [PubMed]
- Campbell, A. (1962). Advances in Genetics, pp. 101–145. New York: Academic Press.
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Kamadurai, H. B. (2007). Thesis. The Ohio State University, USA.
- Kamadurai, H. B. & Foster, M. P. (2007). Biochemistry, 46, 13939–13947. [DOI] [PubMed]
- Kamadurai, H. B., Subramaniam, S., Jones, R. B., Green-Church, K. B. & Foster, M. P. (2003). Protein Sci. 12, 620–626. [DOI] [PMC free article] [PubMed]
- Koradi, R., Billeter, M. & Wuthrich, K. (1996). J. Mol. Graph. 14, 51–55. [DOI] [PubMed]
- Landy, A. (1989). Annu. Rev. Biochem. 58, 913–949. [DOI] [PubMed]
- Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). J. Appl. Cryst. 26, 283–291.
- McCoy, A. J. (2007). Acta Cryst. D63, 32–41. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Radman-Livaja, M., Biswas, T., Ellenberger, T., Landy, A. & Aihara, H. (2006). Curr. Opin. Struct. Biol. 16, 42–50. [DOI] [PMC free article] [PubMed]
- Sadowski, P. D. (1993). FASEB J. 7, 760–767. [DOI] [PubMed]
- Subramaniam, S., Kamadurai, H. B. & Foster, M. P. (2007). J. Mol. Biol. 370, 303–314. [DOI] [PMC free article] [PubMed]
- Subramaniam, S., Tewari, A. K., Nunes-Duby, S. E. & Foster, M. P. (2003). J. Mol. Biol. 329, 423–439. [DOI] [PubMed]
- Tirumalai, R. S., Healey, E. & Landy, A. (1997). Proc. Natl Acad. Sci. USA, 94, 6104–6109. [DOI] [PMC free article] [PubMed]
- Tirumalai, R. S., Kwon, H. J., Cardente, E. H., Ellenberger, T. & Landy, A. (1998). J. Mol. Biol. 279, 513–527. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: IntCB, 2oxo, r2oxosf