Crystallization of the prefoldin β subunit Yke2 is reported. This protein is a novel and potentially important target for anti-cancer therapeutics.
Keywords: prefoldin, β subunit, Gim complex, Yke2
Abstract
The Gim complex (GimC) from Saccharomyces cerevisiae is a heterohexameric protein complex, also known as prefoldin (PFD), which binds and stabilizes unfolded target polypeptides and subsequently delivers them to chaperonins for completion of folding. In this study, the crystallization and preliminary X-ray analysis of one of the β subunits of the Gim complex (Yke2) from S. cerevisiae are described. The purified protein was crystallized by the vapour-diffusion method, producing two types of crystals that belonged to the orthorhombic space group C222 or the primitive monoclinic space group P21. The unit-cell parameters for the C-centred orthorhombic crystal were a = 48.2, b = 168.86, c = 131.81 Å and the unit-cell parameters for the primitive monoclinic crystal were a = 47.83, b = 134.90, c = 81.50 Å, β = 100.71°. The Yke2 crystals diffracted to 4.2 and 3.1 Å resolution, respectively, on a rotating-anode generator under cryoconditions. This is the first report concerning the crystallization of a β subunit of a eukaryotic prefoldin.
1. Introduction
Microtubules are hollow cylinders that are part of the cytoskeleton of eukaryotic cells. They play an important role in numerous cellular processes such as chromosome segregation in mitosis and meiosis, organelle positioning and intracellular transport.
The walls of the microtubule cylinder are formed by α- and β-tubulin subunits, with γ-tubulin being involved in microtubule nucleation (Mitchison & Kirschner, 1984 ▶; Stearns et al., 1991 ▶).
The Gim complex, also known as prefoldin, is a heterohexameric molecular chaperone that is present in both eukaryotes and archaea. Chaperones are crucial for maintaining the native protein conformation and preventing nonspecific aggregation. The Gim-complex chaperone specifically promotes the formation of properly folded and functional α- and γ-tubulin (Geissler et al., 1998 ▶; Hansen et al., 1999 ▶).
The yeast Gim complex is assembled from six different proteins (Geissler et al., 1998 ▶; Siegers et al., 1999 ▶; Vainberg et al., 1998 ▶), whereas the archaeal homologue is a hexamer built from only two subunit proteins, α and β (Leroux et al., 1999 ▶; Siegert et al., 2000 ▶). Prefoldin subunits can be subdivided into two classes, namely α and β subunits. Archaea possess one member of each class (PFDα and PFDβ), whereas eukaryotes have two different but related subunits of the α class and four subunits of the β class (Leroux et al., 1999 ▶).
Deletion of the GIM genes causes microtubule defects and decreased levels of α-, β- and γ-tubulin. The Gim proteins are phylogenetically conserved proteins, to the point that the mouse and human homologues function in yeast.
Tubulin-disrupting agents are presently used as anti-cancer drugs against lung cancer, the most lethal cancer in the western world. However, β-tubulin mutations detected in non-small-cell lung-cancer tumours cause chemoresistance (Monzó et al., 1999 ▶). Therefore, structural investigation of the Gim complex might be of potential interest as an alternative target for the future design of new tubulin-disrupting therapeutic agents.
While the structure of the archaeal prefoldin is known, that of the eukaryotic prefoldin remains unsolved. Thus, we have undertaken the resolution of the structure of this complex and of its components. Here, we report crystallization and diffraction data for Yke2, a PFDβ subunit.
2. Materials and methods
2.1. Vector construction and cloning
The cDNA segment encoding full-length Yke2 was amplified by PCR from S. cerevisiae genomic DNA using the primer pair (5′-GCGcatatgATGTCTGAATTAGGTGCCAAA-3′ and 5′-CGCggtaccTTACCTTCCTGGGCCAGTGG-3′), where lower-case letters indicate nucleotides that were introduced to generate restriction-enzyme sites. The amplified fragments were cloned into the NdeI–KpnI sites of the pOPTH vector, kindly provided by Dr Olga Perisic (LMB, Cambridge, England), for expression in Escherichia coli. The recombinant protein includes a six-histidine tag at its N-terminus, immediately followed by the complete Yke2 sequence.
2.2. Protein expression and purification
The pOPTH-Yke2 vector was used to transform E. coli strain BL21 (DE3) for expression of the protein. Transformants were grown to mid-log phase in LB medium at 310 K. Protein expression was induced by adding isopropyl β-d-1-thiogalactopyranoside to the cells to a final concentration of 0.5 mM and the cultures were maintained for an additional 3 h. Cells were harvested by centrifugation at 4000g and the cell pellets were resuspended in 40 ml lysis buffer [50 mM phosphate buffer pH 7.2, 0.1 M NaCl, 5 mM 2-mercaptoethanol and protease-inhibitor cocktail (Complete EDTA-free; Roche, Mannheim, Germany)]. The cells were lysed by ultrasound sonication (15 min; 9.9 s on, 5.0 s off; 40% power applied with a 12T probe in a Vibracell 75042 sonicator; Biobloc, Strasbourg, France). Cellular debris was removed by centrifugation at 140 000g in a 45Ti rotor and the supernatant was loaded onto a 5 ml HiTrap Chelating HP Column (GE Healthcare Biosciences AB, Uppsala, Sweden). The column was prepared by charging with 2.5 ml 100 mM NiSO4, followed by equilibration with lysis buffer. After loading, the column was washed with ten column volumes of wash buffer (50 mM phosphate buffer pH 7.2, 0.1 M NaCl, 5 mM 2-mercaptoethanol, 10 mM imidazole) and the protein was eluted using a gradient from wash buffer to elution buffer (50 mM phosphate buffer pH 7.2, 0.1 M NaCl, 5 mM 2-mercaptoethanol, 500 mM imidazole). A flow rate of 5 ml min−1 was used for elution and the proportion of elution buffer was increased by 6.2% per minute.
Further purification was attained by gel filtration on a Superdex 75 16/60 preparation-grade column equilibrated in 20 mM bis-tris, 100 mM NaCl, 1 mM 2-mercaptoethanol pH 7.0. Most of the protein eluted as a monomer, as calculated from the elution volumes of protein standards. Dynamic light scattering was performed using a Protein Solutions DynaPro (Wyatt Technology) instrument at a protein concentration of 1 mg ml−1 using a cuvette of 3 mm path length. A hydrodynamic radius of 1.7 nm and an estimated polydispersity of 18.3% were obtained. Purified protein was concentrated to 24 mg ml−1 in 20 mM bis-tris pH 7.0, 100 mM NaCl, 1 mM 2-mercaptoethanol, plunged into liquid nitrogen and stored in small aliquots at 193 K until use in crystallization experiments. The identity of the purified protein was confirmed by mass spectrometry.
2.3. Crystallization
Initial crystallization trials were performed with JBScreen HTS I (Jena Bioscience GmbH, Jena, Germany) using the hanging-drop vapour-diffusion method. An initial crystallization condition consisting of 30%(w/v) PEG 3000, 100 mM Tris–HCl pH 8.5 and 0.1 M lithium sulfate was modified to give optimally sized crystals of the Yke2 protein. The best conditions were found using 2 µl drops (1:1 ratio of protein and reservoir solutions) with 24 mg ml−1 protein solution and 26%(w/v) PEG 3000, 100 mM bis-tris pH 7.0 and 0.1 M lithium sulfate for the orthorhombic crystals or 23%(w/v) PEG 400, 100 mM Na MES pH 6.0 and 0.1 M sodium acetate for the monoclinic crystals. Crystals grew in 24–48 h.
2.4. X-ray data collection and processing
Yke2 crystals were flash-cooled in mother liquor supplemented with 16%(v/v) glycerol. Diffraction data were collected using a rotating-anode generator under cryoconditions. Data were indexed, integrated, scaled and merged with MOSFLM and SCALA from the CCP4 program suite (Collaborative Computational Project, Number 4, 1994 ▶).
3. Results and discussion
The Yke2 crystals belonged to either the orthorhombic system with space group C222 or the primitive monoclinic system with space group P21 (Fig. 1 ▶). Diffraction data extended to 4.2 and 3.1 Å resolution (Fig. 2 ▶), respectively. Crystal and data-processing statistics are summarized in Table 1 ▶.
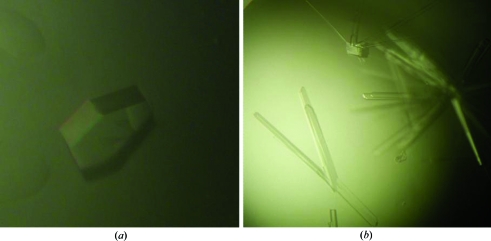
Figure 1.
Crystals of Yke2 obtained by the hanging-drop vapour-diffusion technique using (a) 26%(w/v) PEG 3000, 100 mM bis-tris pH 7.0 and 0.1 M lithium sulfate and (b) 23%(w/v) PEG 400, 100 mM Na MES pH 6.0 and 0.1 M sodium acetate. The crystals shown have approximate dimensions of 0.35 × 0.30 × 0.30 mm (a) and 0.70 × 0.06 × 0.06 mm (b).
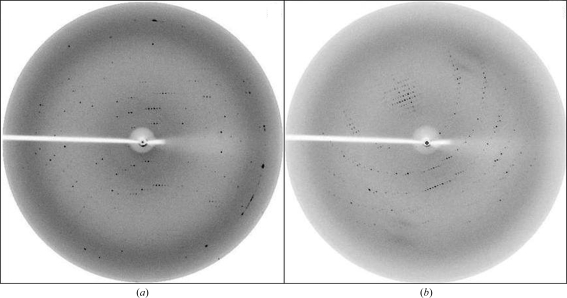
Figure 2.
Typical diffraction patterns of Yke2 crystals with a 0.5° rotation oscillation. (a) Orthorhombic system, space group C222, corresponding to the crystal shown in Fig. 1 ▶(a). The edge of the detector corresponds to a resolution of 3.0 Å. (b) Primitive monoclinic system, space group P21, corresponding to the crystal shown in Fig. 1 ▶(b). The edge of the detector corresponds to a resolution of 2.7 Å.
Table 1. Crystal parameters and data-processing statistics.
Values in parentheses are for the highest resolution shell.
| Space group | C222 | P21 |
|---|---|---|
| Unit-cell parameters (Å, °) | a = 48.2, b = 168.86, c = 131.81 | a = 47.83, b = 134.90, c = 81.50, β = 100.71 |
| Resolution limits (Å) | 84.51–4.20 (4.43–4.2) | 80.06–3.1 (3.27–3.1) |
| Total No. of frames (Δϕ = 0.5°) | 252 | 307 |
| Mosaicity (°) | 1.0 | 1.0 |
| Total No. of observations | 14206 (2087) | 58721 (8489) |
| Unique reflections | 4054 (582) | 18282 (2677) |
| Multiplicity | 3.5 (3.6) | 3.2 (3.2) |
| Rmeas† | 0.163 (0.546) | 0.141 (0.546) |
| Rp.i.m.‡ | 0.085 (0.284) | 0.071 (0.288) |
| 〈I/σ(I)〉 | 4.1 (2.4) | 7.3 (2.5) |
| Completeness (%) | 97.2 (96.6) | 98.9 (100.0) |
R
meas = 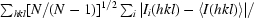
 , where Ii(hkl) are the observed intensities, 〈I(hkl)〉 are the average intensities and N is the multiplicity of reflection hkl.
, where Ii(hkl) are the observed intensities, 〈I(hkl)〉 are the average intensities and N is the multiplicity of reflection hkl.
R
p.i.m. = 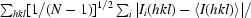
 , where Ii(hkl) are the observed intensities, 〈I(hkl)〉 are the average intensities and N is the multiplicity of reflection hkl.
, where Ii(hkl) are the observed intensities, 〈I(hkl)〉 are the average intensities and N is the multiplicity of reflection hkl.
Using the known molecular weight of the monomer (14.11 kDa) and assuming the presence of four molecules in the asymmetric unit for the orthorhombic crystal, a Matthews coefficient of 2.38 Å3 Da−1 was calculated. Molecular replacement was carried out with MOLREP using a prefoldin β subunit from the X-ray structure of Methanothermobacter thermoautotrophicus prefoldin (PDB code 1fxk; Siegert et al., 2000 ▶) as a search model. MOLREP converged to a possible solution with four molecules in the asymmetric unit.
Acknowledgments
This work was supported by the Spanish National Cancer Research Centre (CNIO) and was partially supported by the EU Grant 3D Repertoire (contract LSHG-CT-2005-512028) and by Grant SAF2006-10269 from the Ministerio de Ciencia y Tecnología, Spain.
References
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Geissler, S., Siegers, K. & Schiebel, E. (1998). EMBO J.17, 952–966. [DOI] [PMC free article] [PubMed]
- Hansen, W. J., Cowan, N. J. & Welch, W. J. (1999). J. Cell Biol.145, 265–277. [DOI] [PMC free article] [PubMed]
- Leroux, M. R., Fändrich, M., Klunker, D., Siegers, K., Lupas, A. N., Brown, J. R., Schiebel, E., Dobson, C. M. & Hartl, F. U. (1999). EMBO J.18, 6730–6743. [DOI] [PMC free article] [PubMed]
- Mitchison, T. & Kirschner, M. (1984). Nature (London), 312, 232–237. [DOI] [PubMed]
- Monzó, M., Rosell, R., Sánchez, J. J., Lee, J. S., O’Brate, A., González-Larriba, J. L., Alberola, V., Lorenzo, J. C., Núñez, L., Ro, J. Y. & Martín, C. (1999). J. Clin. Oncol.17, 1786–1793. [DOI] [PubMed]
- Siegers, K., Waldmann, T., Leroux, M. R., Grein, K., Shevchenko, A., Schiebel, E. & Hartl, F. U. (1999). EMBO J.18, 75–84. [DOI] [PMC free article] [PubMed]
- Siegert, R., Leroux, M. R., Scheufler, C., Hartl, F. U. & Moarefi, I. (2000). Cell, 103, 621–632. [DOI] [PubMed]
- Stearns, T., Evans, L. & Kirschner, M. (1991). Cell, 65, 825–836. [DOI] [PubMed]
- Vainberg, I. E., Lewis, S. A., Rommelaere, H., Ampe, C., Vandekerckhove, J., Klein, H. L. & Cowan, N. J. (1998). Cell, 93, 863–873. [DOI] [PubMed]