Mcm10 is a highly conserved nuclear protein that plays a key role in the initiation and elongation processes of DNA replication by providing a physical link between the Mcm2–7 complex and DNA polymerases. In this study, the central domain of human Mcm10 was crystallized using the hanging-drop vapour-diffusion method in the presence of PEG 3350.
Keywords: Mcm10, zinc-binding domain, DNA replication
Abstract
The initiation of eukaryotic DNA replication requires the tightly controlled assembly of a set of replication factors. Mcm10 is a highly conserved nuclear protein that plays a key role in the initiation and elongation processes of DNA replication by providing a physical link between the Mcm2–7 complex and DNA polymerases. The central domain, which contains the CCCH zinc-binding motif, is most conserved within Mcm10 and binds to DNA and several proteins, including proliferative cell nuclear antigen. In this study, the central domain of human Mcm10 was crystallized using the hanging-drop vapour-diffusion method in the presence of PEG 3350. An X-ray diffraction data set was collected to a resolution of 2.6 Å on a synchrotron beamline. The crystals formed belonged to space group R3, with unit-cell parameters a = b = 99.5, c = 133.0 Å. According to Matthews coefficient calculations, the crystals were predicted to contain six MCM10 central domain molecules in the asymmetric unit.
1. Introduction
In eukaryotic cells, DNA replication is initiated from multiple replication origins that are distributed on each chromosome. The activation of these replication origins requires the controlled assembly of a number of replication-initiator proteins to form a replication fork during the cell cycle. Initially, the pre-replicative complex (pre-RC) is formed by the stepwise assembly of the origin-recognition complex (ORC), cell-division cycle 6 (Cdc6), chromosome licensing and DNA-replication factor 1 (Cdt1) and the minichromosome maintenance 2–7 (Mcm2–7) complex onto chromatin (Bell & Dutta, 2002 ▶). The phosphorylation of Mcm2–7 by kinases and the association of additional replication factors to the pre-RC complex transforms the pre-RC complex to the pre-initiation complex (pre-IC) and ultimately into the functional replication forks that unwind the DNA and synthesize new DNA strands (Mendez & Stillman, 2003 ▶; Blow & Dutta, 2005 ▶). It is believed that DNA unwinding is achieved by the cell-division cycle 45 (Cdc45)/Mcm2–7/go ichinisan (GINS) complex (Moyer et al., 2006 ▶). Mcm10 is a conserved nuclear DNA-binding protein that is essential for loading Cdc45 onto chromatin and forming a Cdc45/Mcm2–7/GINS helicase (Wohlschlegel et al., 2002 ▶; Cook et al., 2003 ▶; Gregan et al., 2003 ▶; Sawyer et al., 2004 ▶). In addition, Mcm10 is required to maintain DNA polymerase α (Pol α) primase on chromatin during the replication-elongation step (Ricke & Bielinsky, 2004 ▶; Yang et al., 2005 ▶).
Mcm10 interacts with the components of the pre-RC and replication forks and presumably regulates both replication initiation and elongation by mediating the interactions between the proteins found in pre-RC and other replication factors. Mcm10 interacts with ORC, Mcm2–7, Cdc45, Pol δ and Pol ∊ (Homesley et al., 2000 ▶; Izumi et al., 2000 ▶; Kawasaki et al., 2000 ▶). In budding yeast, the Mcm10 homologue participates in activation of the pre-RC complex by recruiting dumbbell-forming 4 (Dbf4)/cell-division cycle 7 (Cdc7) kinase and stimulating the phosphorylation of the Mcm2–7 complex (Lee et al., 2003 ▶). In yeast and humans, Mcm10 regulates the stability of the catalytic subunit of Pol α through the formation of a complex and recruits Pol α to the replication origins (Ricke & Bielinsky, 2004 ▶; Chattopadhyay & Bielinsky, 2007 ▶). In addition, di-ubiquitinated Mcm10 interacts with proliferative cell nuclear antigen (PCNA), which is essential for replication and cell growth in budding yeast (Das-Bradoo et al., 2006 ▶). Therefore, MCM10 might play a key role in lagging-strand synthesis by facilitating RNA/DNA synthesis by Pol α and recruiting PCNA.
Although Mcm10 is a highly conserved protein from yeast to humans, its sequence conservation is limited to the central 200 amino acids (Fig. 1 ▶ a). The conserved central domain contains an oligonucleotide/oligosaccharide-binding (OB) fold and an invariant CCCH zinc-binding motif (Figs. 1 ▶ a and 1 ▶ b). Within the OB fold, the PCNA-binding motif mediates the interaction with PCNA, whereas the heat-shock protein 10 (Hsp10)-like motif provides part of the interaction site for the catalytic subunit of Pol α (Fien et al., 2004 ▶; Ricke & Bielinsky, 2004 ▶, 2006 ▶; Das-Bradoo et al., 2006 ▶). The low sequence similarity of Mcm10 to other proteins limits our understanding of the biochemical and biological role of this important protein in replication initiation and elongation. A recent electron-microscopy study reported that human Mcm10 forms a hexameric ring structure (Okorokov et al., 2007 ▶). However, in order to clearly understand the regulatory role of Mcm10 in DNA replication, it is essential to determine the high-resolution structure of Mcm10. This paper reports the purification and preliminary X-ray analysis of the conserved domain of human Mcm10 that contains the binding motifs for zinc ion and other replication factors.
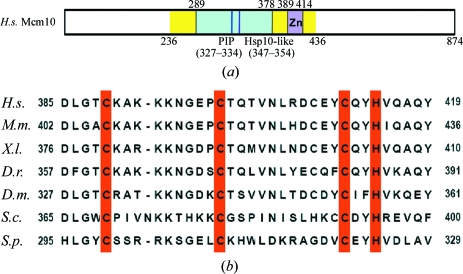
Figure 1.
(a) Schematic diagram of the primary structure of hMcm10. The conserved region used in crystallization analysis (residues 236–436) is coloured yellow and the zinc-binding motif is marked by a magenta box. The OB-fold region is shown in cyan and the PCNA-interacting protein (PIP) motif and Hsp10-like motif are shown as blue and purple lines, respectively. (b) Sequence-comparison analysis of the zinc-binding motif of Mcm10. The Mcm10 proteins from human (H.s.; gi:55958695), mouse (M.m.; gi:123232611), frog (X.l.; gi:11276023), zebra fish (D.r.; gi:55962511), fly (D.m.; gi:22946978), budding yeast (S.c.; gi:6322041) and fission yeast (S.p., gi:12229739) were used for analysis and the conserved zinc-binding residues are marked in red.
2. Experimental procedures
2.1. Expression and purification
The gene encoding the conserved domain (residues 236–436) of human Mcm10 (hMcm10; GenBank entry BC101727.1) was amplified from the cDNA library of the human 293 kidney cell through the polymerase chain reaction (PCR). The amplified gene was cloned into the pET28a (Novagene) expression vector using NheI and SalI restriction sites. This vector was then transformed into Escherichia coli strain BL21 (DE3) for recombinant protein expression. The cells were grown in Luria broth (LB) medium containing 50 µg ml−1 kanamycin at 310 K until the OD600 reached approximately 0.6. Expression of the hMcm10 central domain was induced by the addition of IPTG to a final concentration of 0.4 mM. At the same time, the cells were supplemented with a final concentration of 0.2 mM ZnSO4. Recombinant protein expression was induced for 8 h at 298 K. The cells from 1 l culture were first pelleted and then resuspended in 40 ml cold lysis buffer containing 400 mM NaCl, 25 mM Na2HPO4 pH 7.4, 10 mM β-mercaptoethanol and lysed by sonication. The supernatant containing the His-tagged hMcm10 central domain was collected by centrifugation. All of the purification steps were carried out at 277 K.
In order to isolate the central domain of hMcm10, the supernatant was loaded onto an Ni–NTA His-bind column (Amersham Pharmacia) equilibrated with lysis buffer. The bound hMcm10 protein was eluted from the column using 300 mM imidazole along with 300 mM NaCl, 25 mM Na2HPO4 pH 7.4, 5 mM β-mercaptoethanol. The eluted protein was dialyzed against 25 mM Tris–HCl pH 8.0, 80 mM NaCl and 5 mM DTT to remove imidazole. The N-terminal (6×His) tag of the central domain of hMcm10 protein was removed by incubating the protein with thrombin (5 units of thrombin per milligram of hMcm10) for 18 h. Subsequently, the hMcm10 protein (MW = 23 137 Da), which contains an additional N-terminal six residues (GSHMAS), was loaded onto a MonoS HR 16/10 column equilibrated with 25 mM Tris–HCl pH 7.5 and 5 mM DTT and eluted with an NaCl gradient using buffer containing 25 mM Tris–HCl pH 7.5, 1 M NaCl and 5 mM DTT. The protein was further purified using a Superdex 75 16/60 gel-filtration column equilibrated with 20 mM Tris–HCl pH 7.4, 250 mM NaCl and 5 mM DTT. The eluted hMcm10 protein was finally concentrated to 15 mg ml−1 using an Amicon concentrator.
2.2. Crystallization and data collection
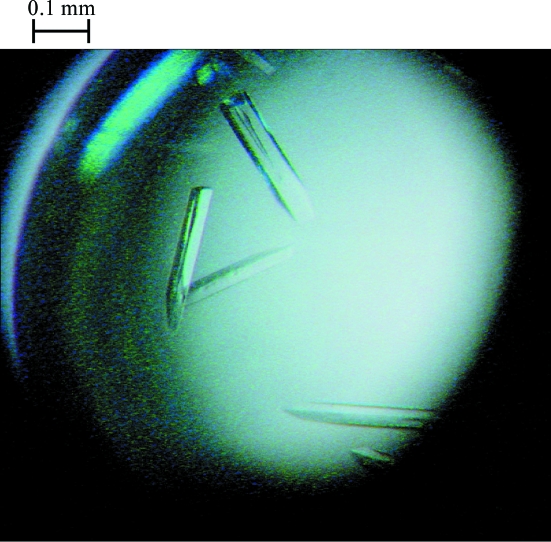
Crystals of the hMcm10 central domain were grown at 301 K by the hanging-drop vapour-diffusion method. The best crystals were obtained by mixing 2 µl of a 15 mg ml−1 solution of the hMcm10 central domain in 20 mM Tris–HCl pH 7.4, 250 mM NaCl and 5 mM DTT with an equal volume of the buffer used in the crystallization well (19% PEG 3350, 0.34 M Li2SO4 and 0.1 M Tris–HCl pH 8.5) and then equilibrating the mixture against a 1 ml crystallization well. The crystals grew over the course of a week and reached maximum dimensions of approximately 0.4 × 0.05 × 0.05 mm (Fig. 2 ▶).
Figure 2.
Crystals of the hMcm10 conserved domain grown using 19% PEG 3350, 0.34 M Li2SO4 and 0.1 M Tris–HCl pH 8.5. Their approximate dimensions are 0.4 × 0.05 × 0.05 mm.
The crystals were transferred into a cryoprotectant consisting of reservoir solution with 30%(w/v) sucrose and were flash-frozen using liquid nitrogen. The data set for the hMcm10 central domain was collected on beamline 4A (MXW) of the Pohang Accelerator Laboratory (Pohang, South Korea) and diffraction data were obtained to a resolution of 2.6 Å. The data were processed using the HKL-2000 program suite (Otwinowski & Minor, 1997 ▶). The crystal belonged to the rhombohedral space group R3, with unit-cell parameters a = b = 99.5, c = 133.0 Å. Assuming six molecules per asymmetric unit, the Matthews coefficient (V M) was calculated to be 2.83 Å3 Da−1, which corresponds to a solvent content of approximately 56.5% (Matthews, 1968 ▶). Table 1 ▶ gives a summary of the crystallographic data.
Table 1. Data-collection and processing statistics.
Values in parentheses are for the highest resolution shell.
| X-ray source | Beamline 4A, PAL |
| Detector type | ADSC Quantum 210 CCD |
| Space group | R3 |
| Temperature (K) | 103 |
| Measurement time (s) | 2 |
| No. of images | 360 |
| Wavelength (Å) | 1.00 |
| Unit-cell parameters (Å) | a = b = 99.5, c = 133.0 |
| Resolution range (Å) | 50–2.6 (2.69–2.6) |
| Redundancy | 7.1 (3.5) |
| Observed reflections [I > 1σ(I)] | 96548 (97390) |
| Unique reflections | 13766 (847) |
| Completeness† (%) | 91.9 (56.3) |
| Rmerge‡ | 0.090 (0.187) |
| 〈I/σ(I)〉 | 34.6 (4.2) |
Because of the incompleteness of the data in the highest resolution shell, we chose to limit the resolution to 2.6 Å.
R
merge = 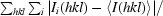
 .
.
Acknowledgments
We thank Heung Soo Lee, Kyung Jin Kim and Kyung Hwa Kim for help with data collection (PAL4A and PAL6C). This work was supported by funds from the National Creative Research Initiatives (Ministry of Science and Technology).
References
- Bell, S. P. & Dutta, A. (2002). Annu. Rev. Biochem.71, 333–374. [DOI] [PubMed]
- Blow, J. J. & Dutta, A. (2005). Nature Rev. Mol. Cell Biol.6, 476–486. [DOI] [PMC free article] [PubMed]
- Chattopadhyay, S. & Bielinsky, A. K. (2007). Mol. Biol. Cell, 18, 4085–4095. [DOI] [PMC free article] [PubMed]
- Cook, C. R., Kung, G., Peterson, F. C., Volkman, B. F. & Lei, M. (2003). J. Biol. Chem.278, 36051–36058. [DOI] [PubMed]
- Das-Bradoo, S., Ricke, R. M. & Bielinsky, A. K. (2006). Mol. Cell. Biol.26, 4806–4817. [DOI] [PMC free article] [PubMed]
- Fien, K., Cho, Y. S., Lee, J. K., Raychaudhuri, S., Tappin, I. & Hurwitz, J. (2004). J. Biol. Chem.279, 16144–16153. [DOI] [PubMed]
- Gregan, J., Lindner, K., Brimage, L., Franklin, R., Namdar, M., Hart, E. A., Aves, S. J. & Kearsey, S. E. (2003). Mol. Biol. Cell, 14, 3876–3887. [DOI] [PMC free article] [PubMed]
- Homesley, L., Lei, M., Kawasaki, Y., Sawyer, S., Christensen, T. & Tye, B. K. (2000). Genes Dev.14, 913–926. [PMC free article] [PubMed]
- Izumi, M., Yanagi, K., Mizuno, T., Yokoi, M., Kawasaki, Y., Moon, K. Y., Hurwitz, J., Yatagai, F. & Hanaoka, F. (2000). Nucleic Acids Res.28, 4769–4777. [DOI] [PMC free article] [PubMed]
- Kawasaki, Y., Hiraga, S. & Sugino, A. (2000). Genes Cells, 5, 975–989. [DOI] [PubMed]
- Lee, J. K., Seo, Y. S. & Hurwitz, J. (2003). Proc. Natl Acad. Sci. USA, 100, 2334–2339. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Mendez, J. & Stillman, B. (2003). Bioassays, 25, 1158–1167. [DOI] [PubMed]
- Moyer, S. E., Lewis, P. W. & Botchan, M. R. (2006). Proc. Natl Acad. Sci. USA, 103, 10236–10241. [DOI] [PMC free article] [PubMed]
- Okorokov, A. L., Waugh, A., Hodgkinson, J., Murthy, A., Hong, H. K., Leo, E., Sherman, M. B., Stoeber, K., Orlova, E. V. & Williams, G. H. (2007). EMBO Rep.8, 925–930. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Ricke, R. M. & Bielinsky, A. K. (2004). Mol. Cell, 16, 173–185. [DOI] [PubMed]
- Ricke, R. M. & Bielinsky, A. K. (2006). J. Biol. Chem.281, 18414–18425. [DOI] [PubMed]
- Sawyer, S. L., Cheng, I. H., Chai, W. & Tye, B. K. (2004). J. Mol. Biol.340, 195–202. [DOI] [PubMed]
- Wohlschlegel, J. A., Dhar, S. K., Prokhorova, T. A., Dutta, A. & Walter, J. C. (2002). Mol. Cell, 9, 233–240. [DOI] [PubMed]
- Yang, X., Gregan, J., Lindner, K., Young, H. & Kearsey, S. E. (2005). BMC Mol. Biol.6, 13. [DOI] [PMC free article] [PubMed]