The biotin protein ligase from S. aureus has been overexpressed in E. coli, purified, crystallized by the hanging-drop vapour-diffusion method and analysed using X-ray diffraction.
Keywords: biotin protein ligase, Staphylococcus aureus
Abstract
Biotin protein ligase from Staphylococcus aureus catalyses the biotinylation of acetyl-CoA carboxylase and pyruvate carboxylase. Recombinant biotin protein ligase from S. aureus has been cloned, expressed and purified. Crystals were grown using the hanging-drop vapour-diffusion method using PEG 8000 as the precipitant at 295 K. X-ray diffraction data were collected to 2.3 Å resolution from crystals using synchrotron X-ray radiation at 100 K. The diffraction was consistent with the tetragonal space group P42212, with unit-cell parameters a = b = 93.665, c = 131.95.
1. Introduction
Staphylococcus aureus is a clinically important opportunistic pathogen that has acquired resistance to almost all antibiotics on the market, including penicillin, methicillin and vancomycin (Gomes et al., 2006 ▶). There is a desperate need to develop new therapeutic agents to target this pathogen. These agents may be a novel class of compounds that target differing pathways to those already on the market. Biotin protein ligase (BPL) is one such target (Payne et al., 2007 ▶) as it is an essential enzyme (Cronan & Wallace, 1995 ▶) that is responsible for the post-translational attachment of biotin to biotin-dependent enzymes. Biotinylation occurs in a two-step reaction in which BPL produces the reaction intermediate biotinyl-5′-AMP from biotin and ATP before transferring the biotin moiety onto a specific lysine residue in the active site of the biotin-dependent enzyme (Lane et al., 1964 ▶). S. aureus expresses two biotin-requiring enzymes, namely acetyl-CoA carboxylase and pyruvate carboxylase, that participate in important metabolic pathways. Acetyl-CoA carboxylase catalyses the production of malonyl-CoA from acetyl-CoA, which is the first committed step in fatty-acid biosynthesis (Wakil et al., 1983 ▶). Pyruvate carboxylase is a gluconeogenic enzyme that catalyses the conversion of pyruvate to oxaloacetate (Wallace et al., 1998 ▶; Attwood & Keech, 1984 ▶). The structure of S. aureus pyruvate carboxylase has recently been determined (Xiang & Tong, 2008 ▶). Whilst the biotin enzymes have been identified as targets for new antibiotics (Forsyth et al., 2002 ▶), exploiting BPL in the same manner has yet to be reported.
To date, structures of BPL from the Gram-negative bacterium Escherichia coli (EcBPL; Wood et al., 2006 ▶; Weaver et al., 2001 ▶; Wilson et al., 1992 ▶) and the archeon Pyrococcus horikoshii (PhBPL; Bagautdinov et al., 2005 ▶, 2008 ▶) have been well characterized. The coordinates for the BPL structures from Mycobacterium tuberculosis (PDB code 2cgh), Aquifex aeolicus (PDB code 2eay) and Methanococcus jannaschii (PDB code 2ej9) have been deposited in the PDB, but no analyses of these structures have been published. All BPL structures solved thus far have a conserved catalytic core composed of two domains with structural homology to SH2 and SH3 domains. Residues within the SH2-like domain are required for binding biotin, whereas the C-terminal SH3-like domain is required for nucleotide binding and forms a cap over the active site upon ligand binding (Wilson et al., 1992 ▶; Beckett, 2005 ▶; Chapman-Smith et al., 2001 ▶). The microbial BPLs can be divided into two separate classes. The class I BPLs are simple two-domain structures considered to be the minimal catalytic unit. The BPLs from P. horikoshii, My. tuberculosis, A. aeolicus and Me. jannaschii all belong to this class. Interestingly, PhBPL is a constitutive homodimer, although the biological significance of this is not understood. The class II BPLs are more complex and contain an additional N-terminal domain that is required for DNA binding. EcBPL, commonly known as the biotin-inducible repressor protein (BirA), has been extensively investigated by genetic, mutational and structural studies. BirA is monomeric in the absence of ligand, but can be induced to form a homodimer upon binding of biotin or biotinyl-5′-AMP. The orientation of the N-terminal domains in the homodimer facilitates binding to a specific DNA sequence that is present in the operator region of the biotin-biosynthetic operon, where it functions as a transcriptional repressor. Thus, BirA is a bifunctional protein that regulates both the synthesis and utilization of biotin. Importantly, the PhBPL and BirA dimers are structurally distinct and employ different surfaces for dimerization. From DNA-sequence analysis, the BPL from S. aureus (SaBPL) is proposed to contain an N-terminal DNA-binding domain and to belong to class II. However, SaBPL has a higher sequence identity to PhBPL (29% compared with 22% for EcBPL). Here, we present the expression, purification and crystallization of a BPL from a Gram-positive bacterium. Detailed knowledge of the SaBPL structure may facilitate the discovery and development of new therapeutic agents.
2. Methods and results
2.1. DNA manipulations
The gene for S. aureus BPL was obtained by genomic PCR using an isolate of S. aureus. Oligonucleotides were designed using the DNA sequence deposited in GenBank (accession No. NP_371980; gi:15924446; genomic sequence of methicillin-resistant S. aureus; Kuroda et al., 2001 ▶). To facilitate subsequent cloning, a PciI restriction site was engineered into the 5′ primer (B138/38, 5′-ACATGTCAAAATATAGTCAAGATGTACTTCAATTACTC) and BamHI and HindIII restriction sites were engineered into the 3′ primer (B139/40, 5′-GGATCCAAGCTTAAAAATCTATATCTGCACTAATAAAACG). PCR was performed under standard conditions (Ling et al., 1991 ▶) using either Vent DNA polymerase (New England Biolabs) for genomic PCR or Dynazyme EXT DNA polymerase (New England Biolabs) to link single adenosine bases onto the 5′ ends of the fragment for TA cloning. Plasmid pGEM(S. aureus BPL) was generated by ligating the resulting 1 kbp fragment from genomic PCR into pGEM-T Easy vector (Promega). To facilitate the purification of SaBPL by metal-chelating chromatography, a hexahistidine sequence was engineered onto the C-terminus of the protein. This was performed using PCR with oligonucleotides B138/38 and B140/55 (5′-AAGCTTAATGATGATGATGATGATGACCAAAATCTATATCTGCACTAATTAAACG) and pGEM(S. aureus BPL) as a template. The PCR fragment was cloned into pGEM-T Easy, yielding pGEM(S. aureus BPL-H6). For overexpression of recombinant S. aureus BPL-H6 in E. coli, the 1 kbp fragment liberated from pGEM(S. aureus BPL-H6) upon digestion with PciI and HindIII was ligated into NcoI- and HindIII-treated pET16b (Novagen). The resulting plasmid, pET(S. aureus BPL-H6), was transformed into E. coli BL21 (DE3) for recombinant protein expression. All constructs were confirmed by DNA sequencing. The isolated DNA sequence was confirmed to be a functional BPL using a complementation assay with the temperature-sensitive birA − mutant E. coli strain CY218 (Chapman-Smith et al., 1994 ▶).
2.2. Recombinant protein expression and purification
E. coli BL21 (DE3) cells harbouring pET(S. aureus BPL-H6) were grown at 310 K in 2YT medium supplemented with 200 µg ml−1 ampicillin to an OD600 of 0.6. Recombinant protein expression was induced with 1 mM IPTG for 4 h at 303 K. The cells were harvested by centrifugation at 2968g for 20 min at 277 K before resuspension in buffer A (20 mM Tris–HCl, 0.5 M NaCl pH 7.9, 20 mM imidazole) with 1 mM PMSF. Cells were lysed by sonication and French press. Cellular debris was removed by centrifugation at 2968g for 20 min. SaBPL-H6 was then purified using immobilized metal-affinity chromatography (IMAC). The filtered supernatant was applied onto a 5 ml His-Trap HP column (GE Healthcare) equilibrated with buffer A. The column was washed with ten column volumes of buffer A followed by three column volumes of buffer A containing 50 mM imidazole before SaBPL-H6 was eluted with buffer A containing 100 mM imidazole. Fractions containing SaBPL-H6 were determined by SDS–PAGE and Western blotting probed with Ni–NTA alkaline phosphatase. These fractions were pooled, concentrated and buffer-exchanged into buffer B (50 mM sodium phosphate, 5% glycerol, 1 mM EDTA and 1 mM DTT) using an Amicon Ultra Centrifugal Filter Device (Millipore).
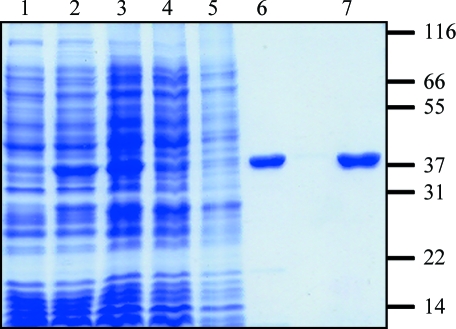
Analysis of the IMAC-purified protein by SDS–PAGE showed that the sample was not homogeneous. Ion-exchange chromatography has previously been employed for purification of E. coli and yeast BPL (Polyak et al., 1999 ▶; Chapman-Smith et al., 2001 ▶). Therefore, the IMAC-purified protein was applied onto a 15 ml SP Sepharose Fast Flow (GE Healthcare) column equilibrated in buffer B. The sample was then fractionated using a 0–400 mM NaCl gradient over 40 min, with SaBPL-H6 eluting at 200 mM NaCl. Fractions containing SaBPL-H6 were confirmed using an in vitro biotinylation assay (Chapman-Smith et al., 1999 ▶). SDS–PAGE revealed the protein to be homogenous (Fig. 1 ▶). Mass spectrometry measured the protein mass as 37 891.3 (the predicted M r was 37 893.4). Following concentration, SaBPL-H6 was stored at 1–2 mg ml−1 in storage buffer (50 mM Tris–HCl pH 7.5, 0.5 mM EDTA pH 8.0, 250 mM NaCl) at 193 K. The enzyme retained catalytic activity for at least six months.
Figure 1.
Purification of recombinant SaBPL-H6. The purification strategy was monitored by SDS–PAGE on a 12% polyacrylamide gel. The His6-tagged protein migrated at a relative mobility of ∼38 kDa. Lane 1 contains crude cell lysate before recombinant protein expression, lane 2 contains crude lysate after induction with 1 mM IPTG and lane 3 contains 50 µg of the soluble fraction after cell lysis. SaBPL-H6 was then purified by IMAC chromatography (lane 4, 50 µg unbound protein; lane 5, 50 µg of the wash fraction; lane 6, 5 µg eluted protein) and S-Sepharose ion-exchange chromoatography (lane 7, 5 µg of SaBPL-H6 from pooled fractions containing BPL activity). Molecular-weight standards (labelled in kDa) are shown on the right.
2.3. Crystallization
SaBPL-H6 was concentrated to 12.5 mg ml−1 using a Vivaspin 10 kDa molecular-weight cutoff concentrator (Sartorius Stedim Biotech, Victoria, Australia) and buffer-exchanged into 20 mM Tris–HCl pH 7.5. Initial crystallization conditions were found using the Sigma protein–protein interaction screen (Sigma–Aldrich, NSW, Australia). In brief, 500 µl of each reagent from the 48 pre-composed Sigma protein–protein interaction screen conditions were dispensed into Limbro tissue-culture plates. 1 µl protein-solution drops were mixed with 1 µl reservoir solution and equilibrated over the reservoir at 293 K on Hampton siliconized 22 mm glass slides using the hanging-drop vapour-diffusion method. Crystals were observed in two conditions: 20% PEG 3350, 0.1 M MOPS pH 7.5, 0.1 M MgCl and 12% PEG 8000, 0.1 M Tris–HCl pH 8.0 (condition Nos. 11 and 34 respectively). The first condition produced thin plate-shaped crystals (0.05 × 0.05 × 0.01 mm) that bundled together, while the second condition produced rectangular brick-like single crystals (approximate dimensions 0.2 × 0.15 × 0.1 mm). Both crystal forms gave rise to X-ray diffraction that was consistent with protein crystals. Crystallization optimization was performed by screening PEG molecular weights (2000–120 000) and concentrations (6–22%), the pH of the buffer (7–9.5) and the type of buffer (including HEPES, MES and MOPS) used for each condition and varying the protein concentration (10–40 mg ml−1). The optimum crystals grew using a 500 µl reservoir containing 8% PEG 8000 and 0.1 M Tris–HCl pH 8.0. 1 µl protein solution (15 mg ml−1) was mixed with 1 µl reservoir solution and equilibrated at either 277 or 293 K using the hanging-drop vapour-diffusion method. Crystals appeared within 24 h at both temperatures, with dimensions of approximately 0.3 × 0.2 × 0.1 mm. A single crystal was picked up using a Hampton silicon loop and streaked through cryoprotectant solutions containing a 1:4, a 1:3 and then a 1:2 ratio of 100% glycerol:reservoir condition and flash-cooled at 100 K. X-ray diffraction data were collected on the high-throughput protein crystallography beamline at the Australian Synchrotron using a MAR 165 CCD detector. 180 diffraction images were recorded. The oscillation angle for each frame was 0.5° and the exposure time was 20 s. The diffraction data were integrated using MOSFLM (Leslie, 1999 ▶) and the intensities were merged and scaled using SCALA (Collaborative Computational Project, Number 4, 1994 ▶). Wilson scaling was applied using TRUNCATE (Collaborative Computational Project, Number 4, 1994 ▶). Molecular replacement is under way.
3. Results and discussion
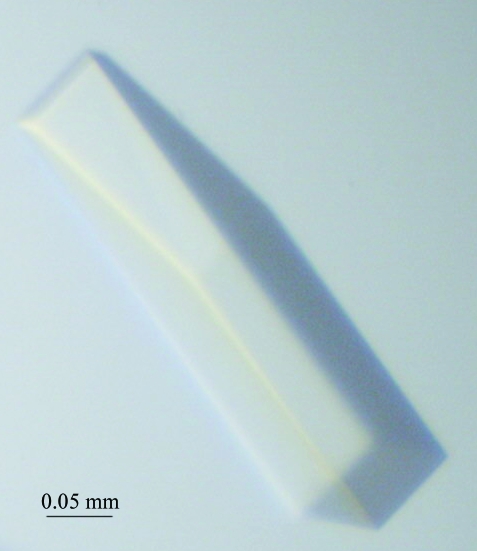
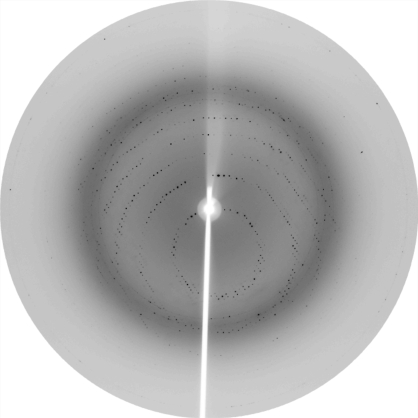
The gene for BPL from S. aureus was isolated and cloned into an expression system for recombinant expression in E. coli. Using two chromatography steps, the protein was purified to homogeneity for crystallization. The purification procedure was optimized to facilitate rapid isolation of the enzyme. This allowed the protein to be rapidly purified from cell lysates in 1 d, thus minimizing storage and freeze–thawing of the enzyme. Using this homogeneous protein, crystals of SaBPL-H6 were grown and X-ray diffraction data were collected. Crystals appeared within 24 h, with some crystals growing as large as 0.3 × 0.2 × 0.1 mm (Fig. 2 ▶). A crystal grown at 293 K degraded within 24 h, while crystals grown at 277 K were stable for up to one week. The crystals produced at 277 K were used for diffraction data collection (Fig. 3 ▶). The diffraction data were consistent with space group P422. Systematic absences suggested that the crystals belonged to space group P42212. Based upon the Matthews coefficient, V M = 2.16 Å3 Da−1, which is consistent with approximately 40% solvent content, we anticipate that there will be one molecule in the asymmetric unit. Diffraction data are summarized in Table 1 ▶.
Figure 2.
Crystal of biotin protein ligase from S. aureus of approximately 0.3 × 0.2 × 0.1 mm in size.
Figure 3.
X-ray diffraction from a native S. aureus BPL crystal. The edge of the detector corresponds to 2.16 Å resolution.
Table 1. Crystallographic data and refinement statistics for SaBPL.
Values in parentheses are for the highest resolution bin.
| Wavelength (Å) | 0.95367 |
| Resolution range (Å) | 29–2.33 (2.33–2.4) |
| Total observations | 543518 (15545) |
| Unique reflections | 30148 (2076) |
| Completeness (%) | 100 (99.6) |
| Redundancy | 7.4 (7.1) |
| Mean I/σ(I) | 8.9 (2.3) |
| Unit-cell parameters (Å, °) | a = 93.665, b = 93.665, c = 131.95, α = β = γ = 90 |
| Rp.i.m.† (%) | 5.0 (44.9) |
R p.i.m. is the precision-indicating (multiplicity-weighted) R merge (Diederichs & Karplus, 1997 ▶).
Acknowledgments
We wish to thank Dr Renato Morona for access to the S. aureus bacterium for cloning and Julian Adams and the staff at the Protein Crystallography beamline at the Australian Synchrotron for their training and guidance. This work was supported by the Biochemistry Discipline, School of Molecular and Biomedical Sciences, University of Adelaide. SWP, GWB and JCW were awarded a Commercial Development Initiative grant, BioInnovationSA. MCJW acknowledges support from the National Health and Medical Research Council and the Australian Research Council.
References
- Attwood, P. V. & Keech, D. B. (1984). Curr. Top. Cell Regul.23, 1–55. [DOI] [PubMed]
- Bagautdinov, B., Kuroishi, C., Sugahara, M. & Kunishima, N. (2005). J. Mol. Biol.353, 322–333. [DOI] [PubMed]
- Bagautdinov, B., Matsuura, Y., Bagautdinova, S. & Kunishima, N. (2008). J. Biol. Chem. doi: 10.1074/jbc.M709116200. [DOI] [PubMed]
- Beckett, D. (2005). J. Nutr. Biochem.16, 411–415. [DOI] [PubMed]
- Chapman-Smith, A., Morris, T. W., Wallace, J. C. & Cronan, J. E. Jr (1999). J. Biol. Chem.274, 1449–1457. [DOI] [PubMed]
- Chapman-Smith, A., Mulhern, T. D., Whelan, F., Cronan, J. E. Jr & Wallace, J. C. (2001). Protein Sci.10, 2608–2617. [DOI] [PMC free article] [PubMed]
- Chapman-Smith, A., Turner, D. L., Cronan, J. E., Morris, T. W. & Wallace, J. C. (1994). Biochem. J.302, 881–887. [DOI] [PMC free article] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Cronan, J. E. Jr & Wallace, J. C. (1995). FEMS Microbiol. Lett.130, 221–229. [DOI] [PubMed]
- Diederichs, K. & Karplus, P. A. (1997). Nature Struct. Biol.4, 269–275. [DOI] [PubMed]
- Forsyth, R. A. et al. (2002). Mol. Microbiol.43, 1387–1400. [DOI] [PubMed]
- Gomes, A. R., Westh, H. & de Lencastre, H. (2006). Antimicrob. Agents Chemother.50, 3237–3244. [DOI] [PMC free article] [PubMed]
- Kuroda, M. et al. (2001). Lancet, 357, 1225–1240. [DOI] [PubMed]
- Lane, M. D., Rominger, K. L., Young, D. L. & Lynen, F. (1964). J. Biol. Chem.239, 2865–2871. [PubMed]
- Leslie, A. G. W. (1999). Acta Cryst. D55, 1696–1702. [DOI] [PubMed]
- Ling, L. L., Keohavong, P., Dias, C. & Thilly, W. G. (1991). PCR Methods Appl.1, 63–69. [DOI] [PubMed]
- Payne, D. J., Gwynn, M. N., Holmes, D. J. & Pompliano, D. L. (2007). Nature Rev. Drug Discov.6, 29–40. [DOI] [PubMed]
- Polyak, S. W., Chapman-Smith, A., Brautigan, P. & Wallace, J. C. (1999). J. Biol. Chem.274, 32847–32854. [DOI] [PubMed]
- Wakil, S. J., Stoops, J. K. & Joshi, V. C. (1983). Annu. Rev. Biochem.52, 537–579. [DOI] [PubMed]
- Wallace, J. C., Jitrapakdee, S. & Chapman-Smith, A. (1998). Int. J. Biochem. Cell Biol.30, 1–5. [DOI] [PubMed]
- Weaver, L. H., Kwon, K., Beckett, D. & Matthews, B. W. (2001). Proc. Natl Acad. Sci. USA, 98, 6045–6050. [DOI] [PMC free article] [PubMed]
- Wilson, K. P., Shewchuk, L. M., Brennan, R. G., Otsuka, A. J. & Matthews, B. W. (1992). Proc. Natl Acad. Sci. USA, 89, 9257–9261. [DOI] [PMC free article] [PubMed]
- Wood, Z. A., Weaver, L. H., Brown, P. H., Beckett, D. & Matthews, B. W. (2006). J. Mol. Biol.357, 509–523. [DOI] [PubMed]
- Xiang, S. & Tong, L. (2008). Nature Struct. Mol. Biol.15, 295–302. [DOI] [PubMed]