Abstract
We used targeted gene disruption in mice to ablate nonmuscle myosin heavy chain B (NMHC-B), one of the two isoforms of nonmuscle myosin II present in all vertebrate cells. Approximately 65% of the NMHC-B−/− embryos died prior to birth, and those that were born suffered from congestive heart failure and died during the first day. No abnormalities were detected in NMHC-B+/− mice. The absence of NMHC-B resulted in a significant increase in the transverse diameters of the cardiac myocytes from 7.8 ± 1.8 μm (right ventricle) and 7.8 ± 1.3 μm (left ventricle) in NMHC-B+/+ and B+/− mice to 14.7 ± 1.1 μm and 13.8 ± 2.3 μm, respectively, in NMHC-B−/− mice (in both cases, P < 0.001). The increase in size of the cardiac myocytes was seen as early as embryonic day 12.5 (4.5 ± 0.2 μm for NMHC-B+/+ and B+/− vs. 7.2 ± 0.6 μm for NMHC-B−/− mice (P < 0.01)). Six of seven NMHC-B−/− newborn mice analyzed by serial sectioning also showed structural cardiac defects, including a ventricular septal defect, an aortic root that either straddled the defect or originated from the right ventricle, and muscular obstruction to right ventricular outflow. Some of the hearts of NMHC-B−/− mice showed evidence for up-regulation of NMHC-A protein. These studies suggest that nonmuscle myosin II-B is required for normal cardiac myocyte development and that its absence results in structural defects resembling, in part, two common human congenital heart diseases, tetralogy of Fallot and double outlet right ventricle.
In vertebrates, most tissues contain two different isoforms of nonmuscle myosin II, II-A and II-B, which are composed of a pair of heavy chains (200 kDa) and two pairs of light chains (20 and 17 kDa) (reviewed in refs. 1 and 2). In humans, the two genes encoding nonmuscle myosin heavy chains (NMHCs) are located on different chromosomes (NMHC-A on 22q11.2 and NMHC-B on 17p13) (3, 4) and, in a variety of species, these genes are expressed in a tissue-specific (5–7) and differentiation-dependent (8–10) manner. Vertebrate neuronal cells are enriched in NMHC-B (7, 10–12), whereas splenic tissue and intestinal epithelial cells are enriched in NMHC-A (7, 13). Nonmuscle myosin II has been shown to play a role in cytokinesis in Dictyostelium (14), in cell shape changes during Drosophila development (15), and in determining cell polarity in Caenorhabditis elegans (16).
The two nonmuscle myosin II isoforms present in vertebrates differ in the rate at which they hydrolyze ATP and propel actin filaments in the in vitro motility assay (17). While there is some overlap in their intracellular distribution, NMHC-B has been shown to be localized near the plasma membrane of some cells, whereas NMHC-A is found in stress fibers, suggesting that each isoform might have a specific function (12, 17, 18). However, a number of cells, including rat basophil leukemic cells (19) and human platelets (13, 18), contain only myosin II-A, whereas a monkey kidney epithelial cell line (COS-7 cells) contains only myosin II-B. Furthermore, cardiac myocytes from newborn mice (8) and primary cultures of embryonic chicken cardiac myocytes were found to contain only NMHC-B and not NMHC-A (20, 21). Thus, it was of interest to study what effect deletion of the NMHC-B gene product would have upon cardiac development and whether ablation of NMHC-B could be compensated for by NMHC-A in tissues that expressed both isoforms.
MATERIALS AND METHODS
Construction of the Targeting Vector.
A 6.5-kb genomic fragment encompassing exon 2 (351 bp) was cloned from a 129/Sv mouse genomic library (Stratagene). Mutagenesis was performed by digesting the SpeI subclone at a unique BamHI site and inserting a 1.8-kb PGK-Neo transcriptional unit into exon 2, just after the initiating ATG (Fig. 1A). The final construct contained the neomycin-resistance fragment in the 3′-5′ orientation. The nucleotide sequences at the exon 2–PGK–Neo boundaries were confirmed by restriction mapping and sequencing.
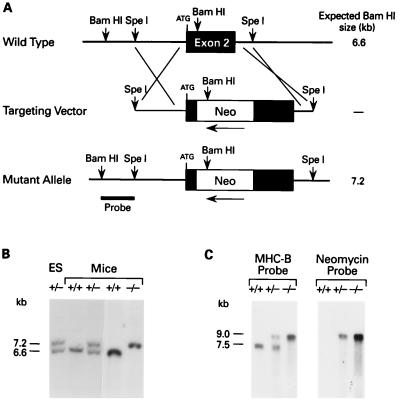
Figure 1.
Targeted disruption of the mouse NMHC-B locus. (A) Genomic map of the mouse NMHC-B locus surrounding exon 2. The locations of the crossover sites are indicated at either end of the targeting vector. Exon 2 contains the initiating ATG, after which a PGK-Neo cassette was inserted in the 3′–5′ direction. The probe used for analysis is indicated on the mutant allele. The fragment size after digestion of genomic DNA with BamHI is indicated on the right. The map is not drawn to scale. (B) Southern analysis of an ES clone (lane 1) and mouse genomic DNA (lanes 2–5). DNA derived from targeted ES cells and mice was digested with BamHI and hybridized with the probe indicated in A. A 6.6-kb fragment indicates a + (wild-type) allele, whereas a 7.2-kb fragment indicates a − (recombinant) allele. (C) Analysis of mouse brain mRNA. A 1.6-kb mouse cDNA probe specific for NMHC-B detected both a 7.5-kb message (+ allele) and a 9.0 kb message (− allele), whereas a 1.0-kb cDNA probe specific for the neomycin cassette detected a 9.0-kb message (− allele) only.
Generation of Transgenic Animals.
J1 embryonic stem (ES) cells (a gift of R. Jaenisch) were grown on mouse embryonic fibroblasts treated with mitomycin C. Leukemia inhibitory factor (500 units/ml) was supplemented in the medium during all passages. A 25-μg sample of linearized targeting vector was electroporated into 2 × 107 cells, which were plated on gelatin-coated 6-cm plates on neomycin-resistant (Neor) embryonic feeder fibroblasts. Selection with G418 was applied 24 h after plating and continued for 7–9 days. Individual Neor colonies were selected and expanded as described (22). Genomic DNA was prepared from individual clones and analyzed by Southern blotting using the probe indicated in Fig. 1A. Five of 341 clones showed evidence for homologous recombination and were used for generation of transgenic mice either by microinjection into embryonic day 3.5 (E3.5) blastocysts or by 8-cell-stage morula aggregation. Chimeric males were scored for germ-line transmission by mating with C57BL/6J females, followed by Southern blotting (see Fig. 1B) and PCR analysis of the progeny.
RNA Analysis.
Whole brains from newborn +/+, +/−, and −/− mice were used to prepare total RNA by using Trizol reagent (Life Technologies) and analyzed by electrophoresis in a formaldehyde/1% agarose gel. After transfer to Nytran (Schleicher & Schuell), the blots were analyzed with a mouse cDNA probe specific for NMHC-B and, after being stripped, by a probe specific for the PGK-Neo cassette (see Fig. 1C).
Immunoblot Analysis.
Samples for immunoblot analysis were prepared as previously described (13, 17) and transferred to Immobilon P membranes (Millipore) after electrophoresis in SDS/6% polyacrylamide gels. Proteins were visualized by using Promega Western Blue stabilized substrate for alkaline phosphatase (brain and kidney) as well as Pierce SuperSignal Substrate for Western blotting (heart and lung). The amount of protein loaded was determined by Pierce protein assays and Coomassie blue staining. Blots were analyzed by using polyclonal antibodies specific for NMHC-B and NMHC-A as described (13).
Histologic and Electron Microscopic Preparations.
Fresh tissues were fixed with 10% formalin, embedded in paraffin, sectioned, and stained with hematoxylin and eosin. To evaluate the degree of cardiac hypertrophy, the transverse diameters of the ventricular myocytes were measured in the histologic sections. A high-resolution TV camera and monitor were attached to a light microscope and the magnification was calibrated with a stage micrometer (Zeiss). The transverse diameters of the myocytes were measured (n = 50 for each ventricle) at the levels of the nuclei to the nearest 0.1 μm. The mean ± SD was calculated for each individual animal and for each group of animals. The data obtained were analyzed by Student’s two-tailed t test. Myocyte disarray was evaluated as absent, mild, moderate, or severe in 20 randomly selected microscopic fields viewed at ×100. To evaluate the presence of nonmuscle myosin II-B, paraffin sections of formalin-fixed hearts from E18.5 embryos and newborn +/+, +/−, and −/− mice were immunostained, using the same antibody used for immunoblotting and a goat anti-rabbit IgG conjugated with fluorescein isothiocyanate as the secondary antibody. The sections were then counterstained with propidium iodide to demonstrate nuclei and examined with a confocal microscope. For electron microscopic study, the tissues were fixed with 2.5% glutaraldehyde in 0.1 M sodium phosphate buffer (pH 7.2), postfixed with 1% OsO4 in 0.1 M phosphate buffer (pH 7.2), and embedded in Polybed 812 (Polysciences). Ultrathin sections were stained with uranyl acetate and lead citrate and examined with a transmission electron microscope.
RESULTS
Targeted Gene Disruption to Eliminate the Myosin II-B Heavy Chain.
Fig. 1A outlines the gene targeting strategy used in these experiments. DNA corresponding to the mouse NMHC-B exon 2 locus was isolated from a genomic DNA library and a replacement-type targeting vector was used to disrupt it as shown. Southern blot analysis of one of the five independent recombinant clones obtained following transfection of ES cells is shown in Fig. 1B (lane 1). These ES cells were used to produce the chimeric males that gave rise to germ-line transmission of the mutant allele, analyses of which are also shown in Fig. 1B (lanes 2–5). This blot was also probed with a neomycin-resistance fragment and the results were consistent with a single recombination event (data not shown). Mice heterozygous (+/−) for the NMHC-B mutation were phenotypically indistinguishable from +/+ littermates and were born in the expected Mendelian ratio. However, approximately 65% of the −/− mice died in utero. Those that were born were moribund and, therefore, were sacrificed 1–10 h after birth to obtain undegraded tissue.
Northern blot analysis of +/+ mouse brains revealed the expected mRNA band at 7.5 kb (Fig. 1C). This figure also demonstrates the appearance of a 9.0-kb message in +/− and −/− mice due to the insertion of 1.5 kb of neomycin-resistance mRNA in the NMHC-B transcript. When an NMHC-B probe (Fig. 1C Left) or a neomycin-resistance probe (Fig. 1C Right) was used, the only message detected in −/− mice was at 9.0 kb. Because this message contained a stop codon in all three reading frames, ablation of NMHC-B protein was expected.
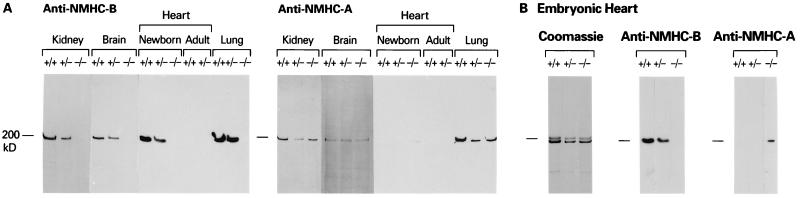
Fig. 2A is an immunoblot of extracts from newborn kidney, brain, and lung, and newborn and adult hearts. The blot demonstrates that −/− newborn mice lack NMHC-B (Fig. 2A Left) and that, with the exception of the heart, all +/+, +/−, and −/− mouse tissues contain NMHC-A (2A Right). In general, +/− mice contained less NMHC-B than did +/+ mice (Fig. 2A Left). As demonstrated in the immunoblot, the +/+ and +/− newborn (Fig. 2A) and embryonic hearts (Fig. 2B) differ from other tissues in that they do not contain NMHC-A. Staining with Coomassie blue confirmed that embryonic mouse hearts, including those from −/− mice, contain α and β cardiac myosin heavy chains (Fig. 2B Left). In contrast to newborn and embryonic mouse hearts, NMHC-B (and NMHC-A) are absent from +/+ and +/− adult mouse hearts (Fig. 2A). However, careful inspection of the anti-NMHC-A blot of the −/− newborn heart occasionally revealed a signal for NMHC-A. We, therefore, undertook a study of NMHC-A and NMHC-B protein in the hearts of embryonic mice as detected by immunoblot analyses. Fig. 2B shows an example of NMHC-A protein expression in the heart of a NMHC-B−/− mouse. Analysis of 35 embryos between E14 and E18 revealed 4 of 8 NMHC-B−/− embryos that showed evidence of up-regulation of expression of NMHC-A protein (see Discussion). None of the other tissues examined demonstrated up-regulation of NMHC-A.
Figure 2.
Immunoblot analyses of tissues from +/+, +/−, and −/− mice (A) and evidence for up-regulation of NMHC-A (B). (A) The blot on the left was analyzed with antibodies specific for the carboxyl-terminal sequence of NMHC-B and the one on the right with antibodies specific for the carboxyl-terminal sequence of NMHC-A (13). All samples, except those marked adult, are from newborn mice. (B) These samples are from an E18 mouse heart. The Coomassie stain shows that +/+, +/−, and −/− mice contain α and β cardiac myosin and the immunoblot demonstrates up-regulation of NMHC-A in the −/− heart.
Evidence for Cardiac Defects in NMHC-B−/− Mice.
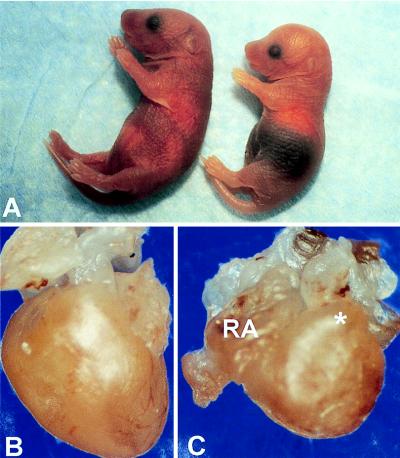
Fig. 3A shows photographs of a +/+ (Left) and a −/− (Right) newborn mouse. The −/− mouse is distinguished by a dark purple, congested liver, easily seen through the skin, and an abnormal, dome-shaped head. Fig. 3 B and C show intact hearts from a +/+ (B) and a −/− (C) newborn mouse. The −/− heart has an abnormal, rounded shape (see Fig. 3 legend).
Figure 3.
Gross features of newborn mice. (A) Newborn +/+ (left) and −/− (right) mice from the same litter. Note the darkened abdominal region, indicative of a congested liver, and the dome-shaped head, due to hydrocephalus involving the lateral ventricles, in the −/− mouse. The size of the newborn −/− mice varied, and all of them failed to nurse. (B and C) Unopened +/+ (B) and −/− (C) hearts. The −/− heart is more rounded in shape and shows right atrial (RA) dilatation and bulging of the right ventricular outflow tract (∗).
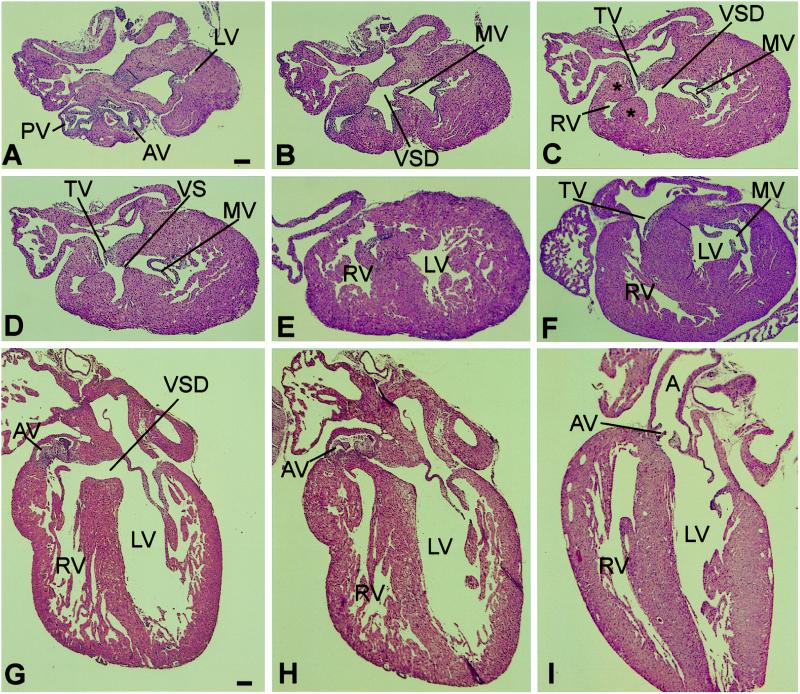
Fig. 4 shows a series of transverse (A–E) and sagittal (G and H) sections through the hearts of two newborn −/− mice. Included for comparison are single transverse (F) and sagittal (I) sections of +/+ hearts of the same age. The sections demonstrate the following features (see also figure legend): (i) the aorta and the pulmonary artery arise as separate, independent vessels and have normal septation (A); (ii) the aortic root is malpositioned to the right, so that it straddles the ventricular septum and the septal defect (A and B) or emanates almost entirely from the right ventricle (G); (iii) the defect is present in the membranous portion of the ventricular septum (C and G); (iv) the right ventricular outflow tract is narrowed by encroachment of hypertrophic muscle upon its lumen (C and D); and (v) the muscular portion of the ventricular septum is intact (E, G, and H). Six of seven −/− newborn mouse hearts analyzed by serial sectioning showed these features.
Figure 4.
Histologic sections of newborn −/− and +/+ hearts. (A–E) Selected serial transverse sections, from cephalad to caudad, of a −/− heart. (F) Transverse section of a +/+ heart at approximately the same level as C. (G and H) Sagittal sections of another −/− heart. (I) Sagittal section of a +/+ heart. (Hematoxylin and eosin stain; scale bar = 100 μm.) (A) Cross-sectional view of the aorta and the pulmonary artery at the levels of the semilunar valves, showing the positions of the two vessels, and the uppermost portion of the lateral wall of the left ventricle (LV). AV, aortic valve; PV, pulmonary valve. (B) This section shows the uppermost portion of the ventricular septal defect (VSD). The mitral valve (MV) is also shown. Comparison with A shows that the VSD is located beneath the aortic valve. (C) Shown are the VSD, the MV, the tricuspid valve (TV), and the right ventricular outflow tract (RV), which is narrowed by hypertrophic muscle (∗). (D) The edge of the membranous portion of the ventricular septum (VS), the MV, and the TV are demonstrated. (E) The left and right ventricular walls and cavities are evident, as is the muscular portion of the VS. (F) Transverse section of +/+ heart. (G) Sagittal sections of a −/− heart showing the VSD and the rightward malposition of the AV. (H) View showing the relationships of the AV to the left ventricular outflow tract in the area adjacent to the VS. (I) Sagittal section of +/+ heart showing the normal relationships of the VS to the AV and aorta (A).
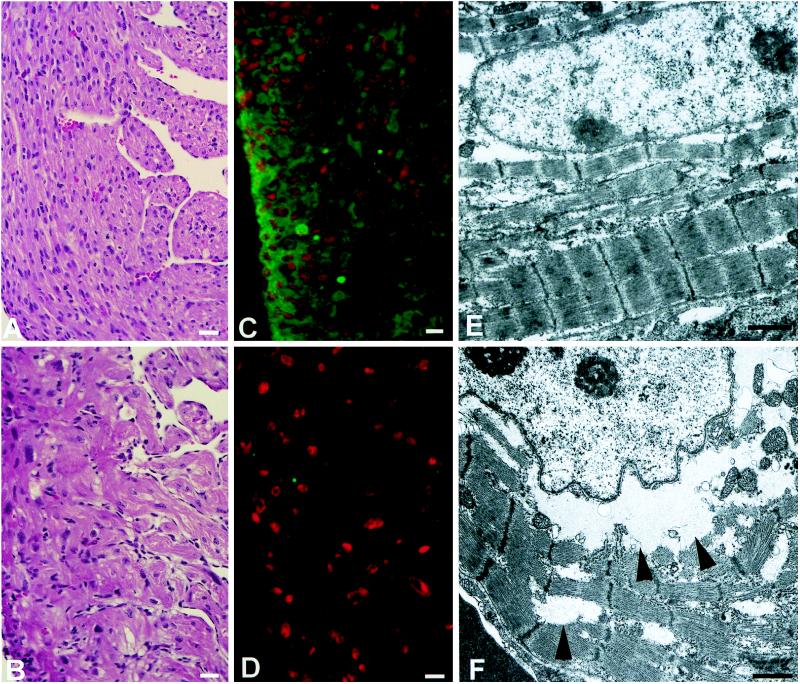
Fig. 5 compares the histologic (A and B), immunofluorescence (C and D), and electron microscopic (E and F) findings in +/+ (A, C, and E) and −/− (B, D, and F) hearts. The histologic sections revealed marked hypertrophy of the −/− myocytes throughout the ventricles and atria, compared with the +/+ and +/− myocytes (+/− not shown). Histological studies also revealed mild to moderate degrees of myofibrillar disarray as well as clear areas of cytoplasm (Fig. 5B). The latter corresponded to accumulations of glycogen particles, as confirmed by specific staining (data not shown). The occurrence of cardiac hypertrophy in the newborn −/− mice was documented by the quantitative measurements of myocyte diameters (see Table 1). The table also shows that cardiac myocyte hypertrophy was present as early as E12.5 and thus preceded the development of the VSD and overriding of the aorta. Immunostaining demonstrated the presence of NMHC-B in the hearts of E18.5 +/− (not shown) and +/+ mice, as well as the absence of this protein in the hearts of −/− embryos (Fig. 5 C and D).
Figure 5.
Histology (A and B), immunocytochemistry (C and D), and ultrastructure (E and F) of +/+ (A, C, and E) and −/− (B, D, and F) hearts. (A and B) Myocytes from the right ventricular wall of newborn hearts. Note the disarray and larger size of the myocytes in the −/− heart (B). (Hematoxylin and eosin stain; bar = 20 μm.) (C and D) Fluorescence photomicrographs of heart sections from E18.5 embryos. The green color indicates the presence of NMHC-B in the +/+ heart (C) and the lack of green, its absence in the −/− heart (D). Nuclei are stained red with propidium iodide. (Bar = 10 μm.) (E and F) Electron micrographs demonstrating myofibrillar disarray in the −/− heart (F) compared with the normal heart (E). Although there is myofibrillar disarray, the myofibrillar components appear normal in the −/− heart. Arrowheads indicate the perinuclear and interfibrillar areas where glycogen particles are present. (Bar = 1 μm.)
Table 1.
Measurement of myocyte sizes in gene-ablated and control mice
| Mice | Myocyte diameter, μm
|
||
|---|---|---|---|
| Newborn
|
E12.5
|
||
| RV | LV | RV + LV | |
| NMHC-B+/+, -B+/− | 7.8 ± 1.8 (4) | 7.8 ± 1.3 (4) | 4.5 ± 0.2 (4) |
| NMHC-B−/− | 14.7 ± 1.1* (8) | 13.8 ± 2.3* (8) | 7.2 ± 0.6† (3) |
Numbers in parentheses are number of mice examined. RV, right ventricle; LV, left ventricle. ∗, P < 0.001; †, P < 0.01.
Electron microscopic examination of the hearts of newborn −/− mice confirmed the findings of hypertrophy of the ventricular myocytes and disclosed focal disarray of the myofilaments (Fig. 5 E and F); however, the structure of the various components of the sarcomeres, the intercellular junctions, the sarcoplasmic reticulum, and the mitochondria appeared normal. The right atrial myocytes of −/− mice were also enlarged and contained more numerous cisterns of endoplasmic reticulum and Golgi complexes, as well as more abundant glycogen particles, than did normal controls (data not shown). The endocardial and capillary endothelial cells appeared normal, as did the smooth muscle cells of the great vessels and the cardiac interstitial fibroblasts.
Examination of all other organs of newborn −/− mice revealed primary abnormalities only in the brain and retina (see Discussion). Evidence for tissue changes secondary to congestive heart failure were seen in the liver, kidney, and spleen, but, unlike the findings in the heart, no primary cellular abnormalities were seen.
DISCUSSION
Cardiac failure was thought to be the cause of death in the NMHC-B−/− mice. This failure appeared to be a consequence of the cardiac hypertrophy and the cardiac malformations. The moderate myofibrillar disarray could also contribute to the severity of the failure, as has been noted previously (23). Two other abnormalities were observed in these mice, retinal dysplasia with rosette formation, indicating a failure in the development of the bipolar layer of the retinal ganglion cells (not shown) and hydrocephalus, which involved the lateral ventricles of the brain and resulted in a dome-shaped skull (see Fig. 3). All of the other organs showed no evidence of any primary abnormalities. Because these organs are known to express both myosin isoforms (see Fig. 2), the most logical explanation is that NMHC-A can compensate for NMHC-B during embryonic development of these tissues. This does not appear to be true for the cardiac myocytes. In contrast to these cells, cardiac fibroblasts showed no obvious abnormalities in number or ultrastructural morphology.
The most striking structural defects in the hearts of −/− mice are the VSD and cardiac outflow tract abnormalities. Despite their relative frequency among congenital heart defects, the pathogenesis of these cardiac defects remains unclear. In the chick, a number of manipulations have been shown to interfere with the processes of convergence and wedging of the cardiac inflow and outflow tracts, which determine the final relative positions of the ventricular outflow tracts and the great vessels. These manipulations include venous or arterial occlusion and partial ablation of the cardiac neural crest (for a review, see ref. 24). The mechanism for the misalignment is unknown. A possible cause for the structural defects observed in these mice is that the myocyte hypertrophy in the −/− embryos causes an abnormal feedback to the developing myocardium, which in turn is unable to complete the cardiac looping process normally. The normal septation between the aorta and the pulmonary artery argues against the presence of a primary defect in the neural crest cells (24).
In mice, the occurrence of a VSD and abnormalities of the great vessels has been reported after targeted disruption of a number of genes, including NF1 (VSD; disoriented, poorly developed myocytes; ref. 25), RXRα receptors (hypoplastic chambers; VSD that may consist of multiple fenestrations, double outlet right ventricle; ref. 26) and the growth factor neurotrophin 3 (atrial and VSDs; ref. 27). Three previous studies, two on cultured chicken cardiac myocytes (20, 21) and one on embryonic, newborn, and adult mouse hearts (8), reported that cardiac myocytes contained NMHC-B, but not NMHC-A. Although our results do not confirm the previously postulated role for NMHC-B in myofibrillogenesis (20), because the hypertrophied cardiac myocytes do contain well developed myofibrils (Fig. 5), they do suggest that cardiac myocytes require NMHC-B for normal development. Because mild to moderate myofibrillar disarray is seen in the −/− mouse heart (see Fig. 5F), we cannot rule out a defect in myofibrillar structure subsequent to the assembly of filaments. In addition, we cannot rule out a contribution by the nonmuscle cells, such as fibroblasts, that are present in the developing heart.
At present, the cause of the hypertrophy of the cardiac myocytes in NMHC-B−/− mice is unknown. The finding that the abnormal transverse diameters were similar in the left and right ventricular myocytes of newborn −/− mice (see Table 1), as well as their presence in both atria (data not shown), suggests that a generalized cellular defect contributed to the pathogenesis of this hypertrophy. In addition, the timing of the hypertrophy is unusual in that it was seen as early as E12.5—i.e., at a time when cardiac myocytes are still capable of undergoing cytokinesis and would be expected to undergo hyperplasia rather than hypertrophy. Thus, normal embryonic and newborn cardiac myocytes, which contain NMHC-B, can undergo cytokinesis, but this ceases soon after birth.
The hypertrophic response of the NMHC-B−/− embryonic and newborn cardiac myocytes is reminiscent of the abnormal enlargement of cells found in Dictyostelium discoideum that were rendered null for the single isoform of nonmuscle myosin II (14). These cells were defective in cytokinesis but underwent karyokinesis and became enlarged and multinucleated. The hypertrophied myocytes in −/− mice, however, clearly were capable of dividing and were not multinucleated. Thus, any defect in cytokinesis must have been compensated for, albeit incompletely, by other proteins. Four of eight NMHC-B−/− mice showed evidence for up-regulation of NMHC-A protein in the heart during embryogenesis. The variation in this expression may be due, in part, to the mixed genetic background of the mice under study. We also cannot exclude the possibility that small amounts of NMHC-A, below the limits of detection, are present in the cardiac myocytes of NMHC-B−/− mice, which could suffice for cytokinesis, but not for rescue of the phenotype.
Congenital heart defects in humans are present in nearly 1% of all newborns and continue to be a significant cause of death in infancy (28). Of particular interest is the possible relevance of the cardiac abnormalities in the NMHC-B−/− mice to human diseases such as tetralogy of Fallot and double outlet right ventricle. Both of these disorders involve the cardiac outflow tract. In the former, the aorta straddles the left and right ventricle, whereas in the latter it originates entirely from the right ventricle. However, intermediate forms are known to exist (29). Our present studies, which are directed toward understanding the function of NMHC-B in the pathogenesis of these defects, should aid in elucidating this connection.
Acknowledgments
We acknowledge many useful discussions with James Sellers and members of the Laboratory of Molecular Cardiology, Margaret Kirby (Medical College of Georgia), Kenneth Chien (University of California at San Diego), Beth Goens (Johns Hopkins School of Medicine), and Jerrold Ward (National Cancer Institute). Catherine S. Magruder provided expert editorial assistance.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: En, embryonic day n; ES, embryonic stem; NMHC, nonmuscle myosin heavy chain; VSD, ventricular septal defect; Neor, resistant to neomycin.
References
- 1.Sellers J R, Goodson H V. Protein Profile. 1995;2:1323–1423. [PubMed] [Google Scholar]
- 2.Goodson H V, Spudich J A. Proc Natl Acad Sci USA. 1993;90:659–663. doi: 10.1073/pnas.90.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saez C G, Myers J C, Shows T B, Leinwand L A. Proc Natl Acad Sci USA. 1990;87:1164–1168. doi: 10.1073/pnas.87.3.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simons M, Wang M, McBride O W, Kawamoto S, Yamakawa K, Gdula D, Adelstein R S, Weir L. Circ Res. 1991;69:530–539. doi: 10.1161/01.res.69.2.530. [DOI] [PubMed] [Google Scholar]
- 5.Katsuragawa Y, Yanagisawa M, Inoue A, Masaki T. Eur J Biochem. 1989;184:611–616. doi: 10.1111/j.1432-1033.1989.tb15057.x. [DOI] [PubMed] [Google Scholar]
- 6.Murakami N, Mehta P, Elzinga M. FEBS Lett. 1991;278:23–25. doi: 10.1016/0014-5793(91)80074-d. [DOI] [PubMed] [Google Scholar]
- 7.Kawamoto S, Adelstein R S. J Cell Biol. 1991;112:915–924. doi: 10.1083/jcb.112.5.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murakami N, Trenkner E, Elzinga M. Dev Biol. 1993;157:19–27. doi: 10.1006/dbio.1993.1108. [DOI] [PubMed] [Google Scholar]
- 9.Kawamoto S. J Biol Chem. 1996;271:17613–17616. [PubMed] [Google Scholar]
- 10.Itoh K, Adelstein R S. J Biol Chem. 1995;270:14533–14540. doi: 10.1074/jbc.270.24.14533. [DOI] [PubMed] [Google Scholar]
- 11.Takahashi M, Kawamoto S, Adelstein R S. J Biol Chem. 1992;267:17864–17871. [PubMed] [Google Scholar]
- 12.Rochlin M W, Itoh K, Adelstein R S, Bridgman P C. J Cell Sci. 1995;108:3661–3670. doi: 10.1242/jcs.108.12.3661. [DOI] [PubMed] [Google Scholar]
- 13.Phillips C L, Yamakawa K, Adelstein R S. J Muscle Res Cell Motil. 1995;16:379–389. doi: 10.1007/BF00114503. [DOI] [PubMed] [Google Scholar]
- 14.De Lozanne A, Spudich J A. Science. 1987;236:1086–1091. doi: 10.1126/science.3576222. [DOI] [PubMed] [Google Scholar]
- 15.Edwards K A, Kiehart D P. Development. 1996;122:1499–1511. doi: 10.1242/dev.122.5.1499. [DOI] [PubMed] [Google Scholar]
- 16.Guo S, Kemphues K J. Nature (London) 1996;382:455–458. doi: 10.1038/382455a0. [DOI] [PubMed] [Google Scholar]
- 17.Kelley C A, Sellers J R, Gard D L, Bui D, Adelstein R S, Baines I C. J Cell Biol. 1996;134:675–687. doi: 10.1083/jcb.134.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maupin P, Phillips C L, Adelstein R S, Pollard T D. J Cell Sci. 1994;107:3077–3090. doi: 10.1242/jcs.107.11.3077. [DOI] [PubMed] [Google Scholar]
- 19.Choi O H, Park C-S, Itoh K, Adelstein R S, Beaven M A. J Muscle Res Cell Motil. 1996;17:69–77. doi: 10.1007/BF00140325. [DOI] [PubMed] [Google Scholar]
- 20.Rhee D, Sanger J M, Sanger J W. Cell Motil Cytoskeleton. 1994;28:1–24. doi: 10.1002/cm.970280102. [DOI] [PubMed] [Google Scholar]
- 21.Conrad A H, Jaffredo T, Conrad G W. Cell Motil Cytoskeleton. 1995;31:93–112. doi: 10.1002/cm.970310203. [DOI] [PubMed] [Google Scholar]
- 22.Li E, Bestor T H, Jaenisch R. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 23.Dyson E, Sucov H M, Kubalak S W, Schmid-Schönbein G, Delano F A, Evans R M, Ross J, Jr, Chien K R. Proc Natl Acad Sci USA. 1995;92:7386–7390. doi: 10.1073/pnas.92.16.7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirby M L, Waldo K L. Circ Res. 1995;77:211–215. doi: 10.1161/01.res.77.2.211. [DOI] [PubMed] [Google Scholar]
- 25.Brannan C I, Perkins A S, Vogel K S, Ratner N, Nordlund M L, Reid S W, Buchberg A M, Jenkins N A, Parada L F, Copeland N G. Genes Dev. 1994;8:1019–1029. doi: 10.1101/gad.8.9.1019. [DOI] [PubMed] [Google Scholar]
- 26.Gruber P J, Kubalak S W, Pexieder T, Sucov H M, Evans R M, Chien K R. J Clin Invest. 1996;98:1332–1343. doi: 10.1172/JCI118920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Donovan M J, Hahn R, Tessarollo L, Hempstead B L. Nat Genet. 1996;14:210–213. doi: 10.1038/ng1096-210. [DOI] [PubMed] [Google Scholar]
- 28.Gelb B D. In: Molecular Biology of Cardiovascular Disease. Marks A R, Taubman M B, editors. New York: Dekker; 1997. pp. 111–140. [Google Scholar]
- 29.Anderson R H. Prog Cardiol. 1978;7:1–54. [Google Scholar]