Abstract
Previously, several individuals with X-linked SCID (SCID-X1) were treated by gene therapy to restore the missing IL-2 receptor γ (IL2RG) gene to CD34+ BM precursor cells using gammaretroviral vectors. While 9 of 10 patients were successfully treated, 4 of the 9 developed T cell leukemia 31–68 months after gene therapy. In 2 of these cases, blast cells contained activating vector insertions near the LIM domain–only 2 (LMO2) proto-oncogene. Here, we report data on the 2 most recent adverse events, which occurred in patients 7 and 10. In patient 10, blast cells contained an integrated vector near LMO2 and a second integrated vector near the proto-oncogene BMI1. In patient 7, blast cells contained an integrated vector near a third proto-oncogene, CCND2. Additional genetic abnormalities in the patients’ blast cells included chromosomal translocations, gain-of-function mutations activating NOTCH1, and copy number changes, including deletion of tumor suppressor gene CDKN2A, 6q interstitial losses, and SIL-TAL1 rearrangement. These findings functionally specify a genetic network that controls growth in T cell progenitors. Chemotherapy led to sustained remission in 3 of the 4 cases of T cell leukemia, but failed in the fourth. Successful chemotherapy was associated with restoration of polyclonal transduced T cell populations. As a result, the treated patients continued to benefit from therapeutic gene transfer.
Introduction
Full correction of genetic diseases has been achieved in 2 forms of SCID, X-linked SCID (SCID-X1; also referred to as common γ chain [γc] deficiency), and adenosine deaminase (ADA) deficiency. In each case, transduction was carried out by ex vivo gene transfer into explanted CD34+ hematopoietic cells (1–4) followed by transplantation back into patients. In each trial, integrating vectors derived from gammaretroviruses were used to allow covalent integration of the therapeutic gene in patient cell DNA. The clinical benefit of gene therapy has been tempered by the occurrence of leukemia in 4 SCID-X1 patients in our previously reported trial (5). In patient 4 and patient 5 (P4 and P5, respectively), the first 2 cases initially described in 2003 (5), the gammaretroviral vector used for transduction integrated near the LIM domain–only 2 (LMO2) proto-oncogene and activated its transcription, thereby promoting clonal T cell proliferation. In addition, chromosomal translocation events were found in blast cells in each case as well, consistent with the idea that multiple hits contributed to transformation.
Several studies have probed the integration target site selection by gammaretroviruses, providing perspective on the origin of the adverse events in SCID-X1 gene therapy. Gammaretroviral vectors preferentially integrate near transcriptional start sites and associated features such as CpG islands and DNAse I hypersensitive sites (6–8). Integration site placement has been characterized previously for both the SCID-X1 and the ADA trials and showed the same favoring for transcription start sites seen in cell culture studies (6–9). This enables the viral long-terminal repeat (LTR) enhancer to alter expression of nearby cellular genes, including proto-oncogenes (6–9).
Here we report a detailed analysis of the 2 most recent cases of leukemia that occurred in our SCID-X1 gene therapy trial, P7 and P10, and a comparison with the previous 2 events (5). The genetic data, taken together, revealed a regulatory network important for growth control in T cell progenitors. We also report the outcome of chemotherapy regimens treating the adverse events, which were successful in the 2 most recent cases.
Results
Clinical findings.
We previously reported the occurrence of lymphoproliferative diseases in 2 patients, P4 and P5, in our SCID-X1 gene therapy trial (5). Two further children, P7 and P10 — aged 11 and 8 months, respectively, at the time of enrollment in the trial — also developed leukemia 68 and 33 months after gene therapy, respectively (M+68 and M+33). Initial therapy involved ex vivo transduction of autologous BM-derived CD34+ cells using the LTR-driven MFG vector containing the gene encoding γc, IL-2 receptor γ (IL2RG). A total of 4.3 × 106 and 11.3 × 106 CD34+γc+ cells/kg body weight, respectively, were infused (overall range, 1.1 × 106 to 22 × 106 cells; median, 4.3 × 106 cells; Table 1). The outcome after infusion was satisfactory. Preexisting infections cleared after gene therapy, and patients were healthy thereafter. A functional and diversified T cell compartment was generated and persisted (ref. 2 and S. Hacein-Bey-Abina, unpublished observations).
Table 1 .
Patient characteristics

P7 developed massive hepatosplenomegaly abruptly at M+68. The patient’s wbc counts increased to 108,000 wbc/μl, with 85% lymphoblasts in peripheral blood and 95% in the BM. Treatment was initiated under the French Acute Lymphoblastic Leukaemia (FRALLE2000BT) protocol for T cell acute lymphoblastic leukemia (T-ALL) (10). The induction regimen consisted of a prednisone prephase followed by vincristine, l-asparaginase, daunorubicine, cyclophosphamide, and intrathecal methotrexate. P7 was stratified into the consolidation T1 group according to his presenting features and prednisone response. After 2 intensification cycles, P7, now under supportive treatment, was in complete remission (CR) at M+84, 16 months after chemotherapy initiation. P10 presented with diffuse lymphadenopathy and leukocytosis (41,200 wbc/μl) at M+33. Physical examination revealed hepatosplenomegaly, and CT scan showed an enlarged hilar mass in the anterior mediastinum. BM contained 90% lymphoblast cells. P10 was treated according to the International BFM Study Group (I-BFM-SG) protocol for T-ALL (11) and showed a complete response to initial 1-week corticosteroid therapy. CR was achieved with no residual detection of abnormal cells in BM. A complete course of chemotherapy was achieved. At the termination of the present study period, P10 was in CR, 40 months after the diagnosis of T-ALL and 16 months after the completion of chemotherapy.
The clinical courses of leukemia in the first 2 adverse events, P4 and P5, have been reported previously (5). Following an unsatisfactory response to chemotherapy in P4 (12), a BM transplant was carried out using BM from a matched, unrelated donor. A relapse occurred 6 months later, initially characterized by the growth of nonblastic T cells carrying the same Vγ9δ1 TCR and by the same integration site in the LMO2 locus as initially detected (5). Following leukemia recurrence, despite further chemotherapy, treatment with a monoclonal anti TCRδ1 antibody, and a second transplantation from the same donor, the child died at M+60, 26 months after leukemia occurrence, with refractory leukemia. P5 was treated according to the Children’s Cancer Study Group T-ALL protocol (13, 14). CR was achieved by the end of the first month of chemotherapy. Remission has persisted for more than 5 years after detection of leukemia, including 3.5 years off therapy.
The 4 patients were treated for their leukemic complication based on the national protocol used in their respective countries of residence. In all cases, therapy consisted of the standard protocol for T-ALL as a function of cell characteristics. Assays for replication competent retrovirus in samples from the 4 patients were negative (15).
Phenotypic characteristics of patients’ T cell clones.
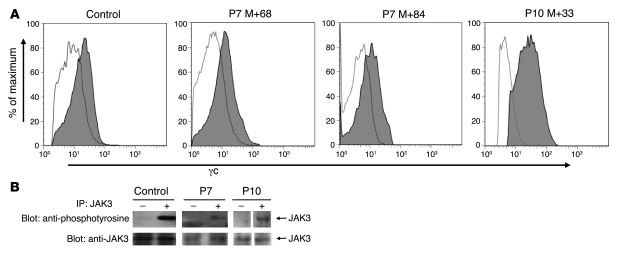
P7 lymphoblasts had an immature T cell phenotype i.e., CD5+, CD1+, CD3+, TCR αβ+, CD4–, and CD8–. The leukemic population was CD10–TdT–CD7low, which is unusual for primary T-ALL. For P10, peripheral blood and BM mononuclear blast cells expressed TdT, CD7, CD2, CD5, CD1, and CD10. TCR αβ and CD3 expression was low, while most of the cells were CD4–CD8–. Therefore, both the P7 and the P10 T-ALL had a late cortical T cell phenotype. The lymphoproliferations in P5, P7, and P10 differed from that of P4, which consisted of more mature T cells (5). Among the potential cofactors suspected to have contributed to leukemia development is the transgene IL2RG itself, which could represent a leukemogenic event. Surface expression of γc on patients’ blast cells was within the normal range and comparable to that of control peripheral blood T cells (Figure 1A). To further rule out an abnormal IL2RG signal through the activation of the specifically associated tyrosine kinase JAK3, phosphorylation status of JAK3 was analyzed. As shown in Figure 1B, no constitutive phosphorylation of JAK3 was observed in nonactivated P10 and P7 blast cells (5).
Figure 1. Expression of γc and phosphorylation status of JAK3 in blast cells.
(A) Flow cytometric analysis of γc expression on P7 and P10 cells revealed a normal range of expression on blast cells of P7 at M+68 and P10 at M+33 as well as on normal P7 T cells recovered at M+84, after 16 months of chemotherapy. (B) Tyrosine phosphorylation status of JAK3 in P7 and P10 leukemic blasts after stimulation with 10 ng/ml IL-7 (+) or without stimulation (–; resting cells). Top: Immunoblot used an antibody to phosphotyrosine. Bottom: The same prestripped blot was reprobed with an antibody to JAK3.
Monitoring of minimal residual disease.
At the time of morphologic remission (i.e., at the end of induction therapy), minimal residual disease (MRD) was detected for P7 and P10; the level of residual blasts in the BM was low, at 5 × 10–4 and 2 × 10–4 for P10 and P7, respectively. MRD testing of all other BM samples remained negative, because no residual cells were detected following consolidation therapy (sensitivity threshold under 1 × 10–5).
Monitoring T cell clonality by analysis of TCR locus structure.
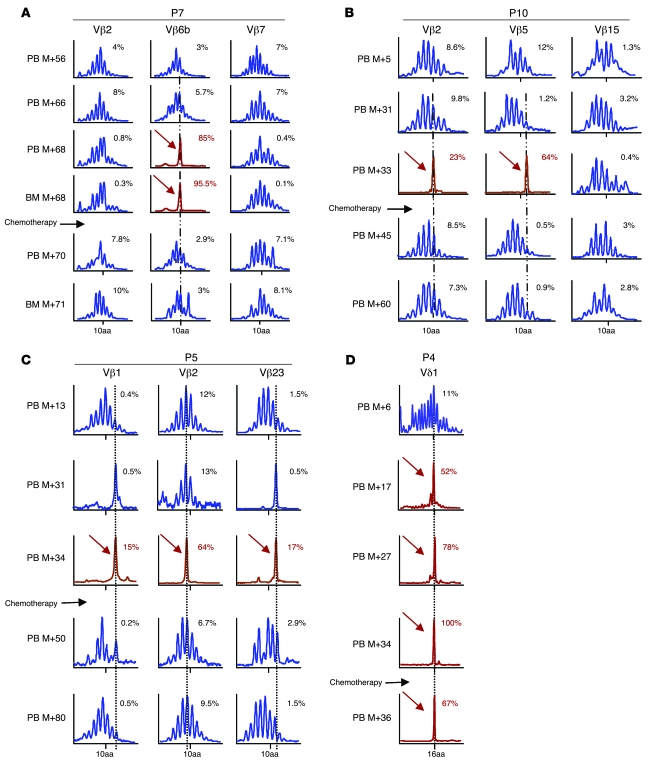
For P7, analysis of the TCR DNA rearrangement (16) present in blast cells at the time of the adverse event revealed a predominant T cell clone expressing a TCR αβ gene that had undergone rearrangement of a TCR Vβ6b gene. The TCR gene encoded a CDR3 length of 10 aa residues. This clone accounted for 85% of the Vβ rearrangements in the mononuclear cells isolated from the peripheral blood and 95% of the cells from the BM (Figure 2A). After therapy, the Vβ6b family expression returned to a normal polyclonal distribution.
Figure 2. Longitudinal immunoscope study of Vβ T lymphocytes.
(A) A loss of polyclonality was found in the Vβ6b family at M+68 in peripheral blood (PB) and BM of P7. After the induction course of chemotherapy, quantitative amplification revealed a disappearance of the pathological clone. (B) In P10 blast cells, 2 clones were detected, Vβ5 and Vβ2, which disappeared 1 month after chemotherapy. (C) Immunoscope analysis of P5 cells (Vβ1, Vβ2, and Vβ23) indicated that 4 years after chemotherapy, the 3 clones displayed a gaussian distribution. (D) Immunoscope analysis of P4 cells indicated that the Vδ1 clone, which had been detected retrospectively 17 months before leukemia diagnosis, remained dominant at M+36, 2 months after chemotherapy. Percentages within graphs denote percent of the Vβ rearrangements.
For P10, 2 rearrangements of the β chain were found in blast cells at M+33. One was a functional Vβ5-J2.5-02 rearrangement encoding a 13-aa CDR3. This clone accounted for 64% of the Vβ population. The second rearranged allele encoded a nonfunctional Vβ2-J2.5 rearrangement accounting for 23% of Vβ expression. We inferred that both rearrangements were present within each blast cell, because it is known that initial failure of TCR rearrangement triggers rearrangement of the second allele to produce a functional allele (17). These rearrangements were not detected following chemotherapy (Figure 2B). For P5, immunoscope analysis of lymphocytes 4 years after the leukemia diagnosis revealed a polyclonal distribution of the Vβ families without resurgence of the 3 Vβ1+, Vβ2+, and Vβ23+ abnormal clones (Figure 2C). For P4, the Vδ1 clone that had been detected retrospectively 17 months before leukemia diagnosis remained dominant 2 months after chemotherapy, at M+36 (Figure 2D).
Integration site analysis.
Vector integration site sequences were characterized for P7 and P10 from cell samples before, during, and after the adverse events (Figures 3 and 4). We monitored longitudinal samples in order to identify integration sites that expanded during the adverse event but declined in abundance with successful therapy. DNA bar coding and pyrosequencing were optimized for each patient and used to obtain a total of 6,891 sequence reads corresponding to 617 unique integration sites. Integration site sequences were also characterized by directed ligation PCR and Sanger sequencing. As has been reported previously for gammaretroviral vectors (6–8), the integration sites were found to be enriched near transcription start sites and associated features such as CpG islands and DNAseI hypersensitive sites (P < 0.0001 for all comparisons; Supplemental Table 1; supplemental material available online with this article; doi: 10.1172/JCI35700DS1). In P4 and P5, the 2 patients with previously characterized adverse events, blast cells harbored a predominant integration site near the transcription start site of the LMO2 gene (Figure 3B), and expression analysis showed that LMO2 transcription increased (5).
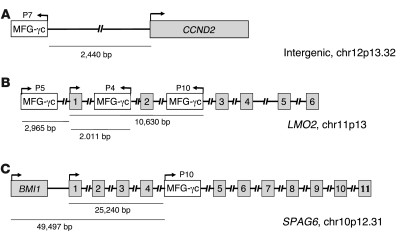
Figure 3. Structure of vector integration sites in blast cells of P7 and P10.
(A) Sequence mapping to the human genomic database indicated a 100% match to the 5′ CCND2 genomic DNA locus on chromosome 12p13.32 at position 4250758. (B) Sequence mapping to the human genomic database indicated a 100% match to the LMO2 genomic DNA locus on chromosome 11p13 at position 33859782. (C) Sequence mapping to the human genomic database indicated a 100% match to the SPAG6 genomic DNA locus on chromosome 10p12.31 at position 22699645. Integrated retrivorus vectors and genes are represented by white and gray boxes, respectively; numbered boxes represent exons; right- and left-facing arrows indicate forward and reverse orientation, respectively, of the inserted vector.
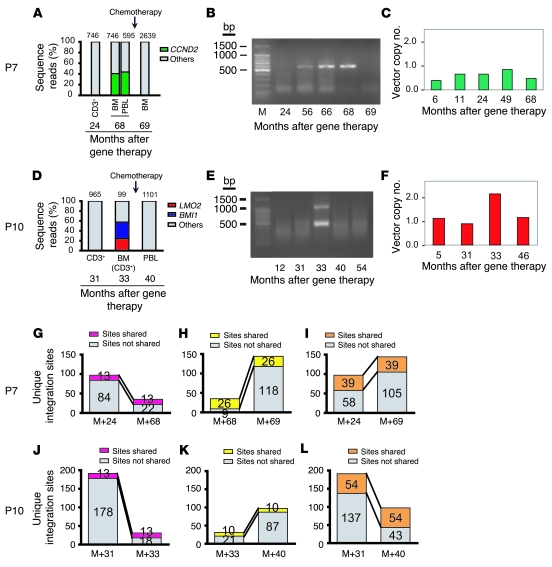
Figure 4. Longitudinal analysis of vector integration sites in P7 and P10.
(A and D) DNA samples from patient cells were digested with restriction enzymes and ligated to linkers, after which integration site sequences were determined by PCR amplification and pyrosequencing. Numbers above bars denote total integration site reads, with composition broken down. PBL, peripheral blood lymphocytes. Longitudinal analysis of samples from P7 (A) and P10 (D). Time of chemotherapy is indicated by the arrow. (B and E) Analysis of genomic DNA samples by ligation-mediated PCR, separation by electrophoresis on an agarose gel, and staining with ethidium bromide. (B) In P7, the intense band in the M+68 sample had the mobility expected for the CCND2 amplification product. (E) In P10, bands of the expected sizes were observed for the LMO2 and BMI1 amplification products in the M+33 sample. (C and F) Longitudinal vector copy number analysis. Quantitative real-time PCR analysis of PBMC DNA from P7 and P10 obtained at the indicated times after gene therapy. (C) P7 showed 1 integrated copy in the leukemic blasts at M+68, concordant with the unique detected integration site. (F) In P10, 2 integrated copies at M+33 were detected, contrasting with the 1 copy detected before and after chemotherapy. These data confirmed that there is 1 leukemic clone bearing the 2 proviral integration sites, LMO2 and BMI1. (G–L) Number of integration sites shared (pink, yellow, and orange) versus not shared (gray) at the indicated time points for P7 (G–I) and P10 (J–L).
For P7, the prevalence of a single integration site increased to approximately 42% of all sites at the time of the adverse event, then dropped sharply in abundance following chemotherapy. The sequencing analysis showed that the integrated vector was located 2,440 bp upstream of the CCND2 gene transcription start site (Figure 3A). CCND2 encodes cyclin D2, a known oncogene in lymphoid cells (18). These data thus identify what we believe to be a new proto-onocogene that potentially can be activated by insertional mutagenesis in these cells and thereby contribute to transformation. Figure 4A plots the CCND2 site as a proportion of all sites in longitudinal sequence reads. Figure 4B presents an analysis of ligation-mediated PCR products by gel electrophoresis, showing the appearance of a predominant amplification product at the time of the adverse event with the mobility expected for the CCND2 product.
For P10, 2 integration sites increased sharply in abundance at the time of the adverse event, then dropped following successful chemotherapy (Figure 4D). One vector integration site was located within the second intron of LMO2, 10.6 kb from the transcription start site (Figure 3B). The second integration site was in the fourth intron of SPAG6, 49,497 bp downstream of the BMI1 gene transcription start site (Figure 3C). Figure 4E shows detection of the LMO2 and BMI1 sites by ligation-mediated PCR and gel electrophoresis at M+33.
The contribution of specific transduced cell clones, as marked by integration sites, could be followed longitudinally in the pyrosequencing data. For both P7 and P10, the overlap of clones shared between the adverse event and time points before (Figure 4, G and J) or after (Figure 4, H and K) was modest, because the blast cell clone crowded out the other clones in the population. However, for the time points before and after the adverse events, the number of clones recovered was larger and overlapped to a greater degree (Figure 4, I and L). Thus, many of the clones present prior to the adverse event survived chemotherapy and remained active after the adverse event.
Longitudinal γc provirus integration analysis.
We studied the vector copy number by quantitative real-time PCR as previously described (2) in P7 and P10 PBMCs. As shown in Figure 4, C and F, we detected no more than 1 vector copy at any time point, except in P10 blast cells, in which 2 copies were repeatedly found. This result contrasts with the 1 copy detected before and after chemotherapy (Figure 4F).
Expression analysis of the targeted genes.
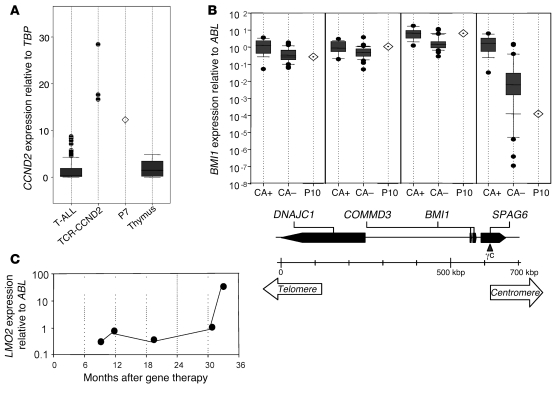
Expression of genes near integration sites in the blast cells was quantified by quantitative real-time RT-PCR (RQ-PCR). For comparison, primary T-ALL samples were also assayed. CCND2 gene expression was analyzed in P7 blast cells and compared with a series of primary T-ALL samples, including cases with CCND2 gene translocations. In the P7 leukemic cells, CCND2 was expressed at an abnormally high level, near the levels in the CCND2-rearranged T-ALL samples. These expression levels were higher than in the other primary T-ALL samples or normal human thymic subpopulations (Figure 5A).
Figure 5. Expression of proto-oncogenes.
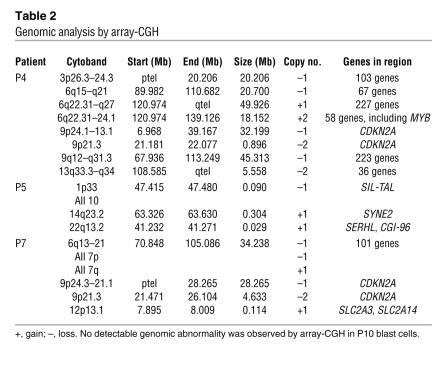
(A) CCND2 expression analysis in P7 blast cells compared with a series of primary T-ALL cases from different oncogenic subgroups — CCND2-rearranged cases (TCR-CCND2; n = 3) and other T-ALL primary cases (T-ALL; n = 86) — and with normal thymus (n = 3). CCND2 expression levels are shown as copy number value in a given sample related to TBP copy number in the series, as determined using the ΔCt method. CCND2 levels in 11 normal human thymic subpopulations were previously analyzed for comparison and showed varied gene expression depending on the cells’ stage of thymic differentiation (18). Results are presented as a box plot: the box boundaries indicates the twenty-fifth and seventy-fifth percentiles; the line within the box denotes the median; whiskers denote tenth and ninetieth percentiles; outliers are indicated as dots. (B) RQ-PCR of SPAG6, BMI1, COMMD3, and DNAJC1 genes in P10 leukemic sample at M+33 compared with CALM-AF10–positive (CA+; n = 14) and –negative (CA–; n = 31) samples. The ratio of gene expression relative to ABL was calculated for each sample. Below, the genomic organization around the AF10 breakpoint is presented along with the location of the γc retrovirus insertion. (C) Longitudinal expression of LMO2 in P10 as determined by RQ-PCR.
Expression of BMI1 was compared among the P10 leukemic cells and a series of primary T-ALL samples, including cells from myeloid and/or T cell leukemias harboring the CALM-AF10 translocation, where the BMI1 gene is highly expressed (19, 20). As shown in Figure 5B, high-level expression of BMI1 was detected in P10 blast cells, comparable to that of CALM-AF10, and approximately 3-fold higher than that of other T-ALL samples. We conclude that the insertion of the vector led to activation of BMI1 transcription (Figure 5B). Expression of the nearby genes DNAJC1, COMMD3, and SPAG6 was also analyzed, showing high-level expression of COMMD3 (of unknown significance) as in CALM-AF10 samples (Figure 5B). LMO2 expression was found to be high in P10 blast cells at M+33 (Figure 5C). The magnitude of expression was 30-fold higher than in polyclonal CD3+ cells. LMO2 overexpression was detected only at M+33, the time of cell overgrowth.
Expression and mutations of the common T-ALL oncogenes.
We also carried out targeted tests in patient blast cells of possible overexpression or mutation of proto-oncogenes known to be associated with T-ALL. Expression of TLX1 (also known as HOX11), TLX3 (also known as HOX11L2), CALM-AF10, BCR-ABL, and NUP214-ABL were tested by RQ-PCR. None of these genes were overexpressed in the blast cells of the 4 cases (Table 1). However, strong TAL1 expression was detected in P5, consistent with detection of a SIL-TAL rearrangement (ref. 5 and see below). Sequencing of the NOTCH1 genes in the blast cells from the 4 patients revealed heterozygous mutations in regions encoding the heterodimerization domain in P5 and P10 (Table 1).
Genome-wide chromosomal copy number analysis.
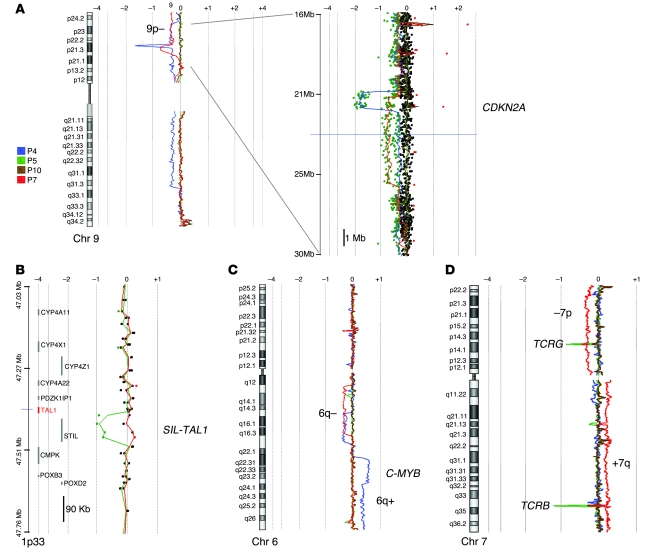
Blast cell DNA samples were analyzed for copy number alterations using high-density 105K oligonucleotide array–comparative genomic hybridization (array-CGH). The abnormal genomic profiles identified are described in Table 2. Deletions at 9p21, including biallelic deletions targeting the CDKN2A (also referred to as p16 and ARF) locus were found in P4 and P7 (Figure 6A). A SIL-TAL microdeletion at 1p32 was found in P5 (Figure 6B), explaining the SIL-TAL fusion transcript that was previously detected by RT-PCR (5). Large interstitial deletions at 6q14-21 were detected in P4 and P7. A duplication at 6q22-23 was also detected in P4 (Figure 6C), consistent with the unbalanced translocation t(6;13) that was previously reported (5). The 6q22-23 region includes the MYB locus, which has been recently shown to be duplicated in some T-ALL cases (21). Several other abnormalities were found (Figure 6D and Table 2), which suggests that acquired oncogenic rearrangements occurred in a multistep oncogenesis process. In addition, deletion within the TCRG, TCRA/D, and TCRB loci were found, as expected from cells of the T cell lineage (Figure 6D).
Table 2 .
Genomic analysis by array-CGH
Figure 6. Genome-wide copy number analysis by array-CGH.
(A) Array-CGH analysis of chromosome 9. The moving average ratio between leukemic and germline DNA is shown by colored lines. Biallelic deletions, including the CDKN2A locus, were detected in P4 and P7. A 14-Mb region at 9p21 is shown magnified at right. (B) A 90-kb deletion was observed between the SIL and TAL1 genes at 1p33 in P5 (green). This microdeletion corresponded to a SIL-TAL1 oncogenic rearrangement. (C) Copy number analysis of chromosome 6. Interstitial 6q deletion in P4 and P7, and gain (4 copies, and then 3 copies) in P4 are shown. (D) Copy number analysis of chromosome 7. A loss of the short arm (–7p) and gain of the long arm (+7q) in P7 (red) is shown. As expected in clonal T cell proliferation, the TCRG and TCRB loci were deleted at 7p13 and 7q34, respectively.
Network analysis of affected genes.
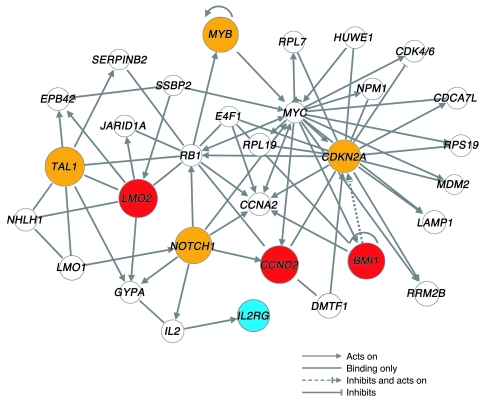
The genes marked by insertional activation, copy number variations, chromosomal translocations, and point mutations in the 4 patients — LMO2, BMI, CCND2, STIL (also known as SIL), TAL, NOTCH1, MYB, and CDKN2A — specify regulatory molecules involved in the adverse events. We therefore sought to analyze relationships among these genes. When analyzed by pathway analysis software (see Methods), these genes clustered in a network containing multiple direct and indirect interactions (Figure 7). An initial analysis of the ontology of genes in this network showed enrichment for the term acute lymphocytic leukemia (P < 0.001), which emphasizes that many of the identified genes are known proto-oncogenes in childhood leukemia and developmental processes in blood cells (P < 0.001), consistent with the cell type of origin. Particularly densely connected nodes involved CDKN2A, one of the genes identified by genome-wide CGH, and also RB1, CCNA2 (which encodes cyclin A), and MYC. As we discuss below, these findings suggest pathways involved in regulating proliferation and persistence in the patients’ cells, and also mark genes for surveillance during potential future adverse events.
Figure 7. Pathways involved in the 4 adverse events.
Interactions among genes affected in the 4 adverse events were determined as described in Methods. Red symbols denote genes identified by insertional activation in the 4 adverse events. Orange symbols denote genes identified by copy number alterations or point mutations. The therapeutic transgene IL2RG is shown in blue. For ease of visualization, the network was limited to genes directly connected to 1 of the 7 functionally identified genes or the transgene IL2RG; all except STIL were connected in a tightly linked network.
Discussion
Ex vivo retroviral vector–mediated γc gene transfer into CD34+ cells resulted in successful gene therapy for SCID-X1 (1, 2). However, 4 of the 10 treated patients developed a lymphoproliferative disease at M+31 to M+68. Chemotherapy eliminated 3 of the 4 leukemic clones, leading to sustained CR. In each of the 3 cases, a well-differentiated polyclonal γc+ T cell population reappeared in the blood following chemotherapy.
In all 4 adverse events, proto-oncogenes were activated as a consequence of vector integration. Activation of LMO2 was involved in 3 of 4 cases. The role of LMO2 in oncogenesis has been well established from analysis of primary T cell leukemia (22) and murine models. In P10, 2 insertions were found, one in LMO2 locus and the other in BMI1. The Bmi1 gene was first identified in mice as a target of retroviral insertional activation in B cell lymphoma in association with c-Myc mutation (23, 24). The BMI1 product is known to regulate normal and leukemic stem cell proliferation, including through repression of the Cdkn2a locus (25, 26). It is likely that LMO2 and BMI1 overexpression both contributed to the P10 leukemic cell proliferation. Cyclin D2 overexpression, as observed in P7, has recently been reported in cases of primary T-ALL as a result of TCRB or TCRA/D-related translocations (18). CCND2 is normally active in the earliest thymic subpopulations but downregulated during development, suggesting an oncogenic mechanism for CCND2 overexpression in T-ALL (18, 27).
A variety of genetic alterations were detected in the 4 cases in addition to vector integration, such as the SIL-TAL1 fusion in P5. This rearrangement, common in T-ALL, consists of the juxtaposition of the enhancer region of the SIL gene to the proto-oncogene TAL1, which boosts TAL1 expression (28). Mutations in the NOTCH1 heterodimerization domain were identified in P5 and P10 blast cells, consistent with the frequent gain-of-function NOTCH1 mutations in T-ALL (29). Biallelic deletion of the CDKN2 locus was found in blast cells of P4 and P7. The inactivation of this tumor suppressor gene is the most prevalent genomic abnormality in common T-ALL (30). Large chromosomal losses at 6q14-16 were observed in P4 and P7. Deletions of this region have previously been reported in T-ALL, although no tumor suppressor gene has been identified to date (31). A gain at 6q22-23 was also detected in P4. This region contains the MYB gene, overexpression of which has been implicated in T-ALL cases (21, 32). Other abnormalities were found in P4, P5, and P7, which suggests that further oncogenic rearrangements could have occurred in the selected clones. Collectively, these data fit with multistep oncogenesis of T-ALL (33, 34), in which oncogenes were first activated by vector insertional mutagenesis, followed by accumulation of secondary genome rearrangements, including point mutations as well as gene deletions and amplifications.
The network analysis in Figure 7 highlights further interactions among the functionally identified genes and suggests that these interactions may be mediated, at least in part, through a small number of growth control genes at densely connected nodes, including RB1, CCNA2, and MYC. LMO2 and IL2RG have previously been reported to cooperate in inducing hematopoietic tumors by studies of insertional mutagenesis in mice (35). The data thus begin to outline a signaling cascade, involving the cytoplasmic membrane proteins (IL2RG and NOTCH1), transcriptional control proteins (LMO2, TAL1, MYB, MYC, RB1, and BMI1) and cell cycle control proteins (CCND2, CCNA2, and CDKN2A), that regulates persistence and proliferation of the T cell progenitors. It is striking that, in contrast to the gene therapy developed for SCID, no clonal lymphoproliferation has been reported to date in patients treated for ADA deficiency (36), despite the observation of a similar frequency of integration near LMO2 and other proto-oncogenes (36, 37). Woods et al. have argued, based on an experimental model of gene transfer in γc– mice, that γc overexpression itself could be oncogenic (38). According to one hypothesis, γc expression could enable progenitor cells to respond to cytokines such as IL-7 and thereby promote survival and proliferation. However, several other studies have indicated that γc is not an oncogene per se (39–41). Furthermore, there was no evidence of γc overexpression or constitutive JAK3/STAT5 activation in the leukemic cells from the 4 patients. Alternatively, it is possible that the SCID-X1 background constitutes a risk factor for tumorigenesis. Data from Shou et al. suggest the presence of an expanded population of primitive progenitors highly prone to growth-promoting integration in the BM of SCID-X1 mutant mice (42). In the ARF–/– mouse model, there is a higher incidence of tumors when ARF–/–IL2RG–/– cells are transduced with an IL2RG-containing vector than in treated ARF–/–IL2RG+/+ mice (42). This hypothesis of cooperation between proto-oncogene activation and γc function is attractive, but has not received a molecular explanation.
The demonstrated sustained efficacy of retroviral-mediated gene therapy of SCID-X1 stresses the need to match efficacy with safety. A variety of improvements in vector design have been suggested and may reduce genotoxicity (43–45). However, despite the setbacks, at the termination of the present study, 3 of the 4 patients who experienced severe adverse events were in CR and continued to benefit from therapeutic gene transfer.
Methods
Patients.
Ten patients without an HLA genoidentical donor were enrolled in the γc gene therapy trial at Hôpital Necker–Enfants Malades — Paris between March 1999 and April 2002. SCID-X1 diagnosis was based on peripheral blood lymphocyte counts and confirmed by γc gene mutation analysis. The protocol was approved by the French Drug Agency and the Comité de protection des personnes of Hôpital Cochin (Paris, France), and written informed consent was obtained from the parents including information that an alternative treatment (BM transplantation) was available. All of the patients were in sterile isolation and received nonabsorbable antibiotics and anti-infectious agents, including intravenous immunoglobulins.
Retrovirus-mediated transduction.
The IL2RG cDNA has previously been introduced in the amphotropic-pseudotyped gammaretroviral vector derived from the Moloney murine leukemia virus (1). BM (30–150 ml) was harvested under general anesthesia, and the positively selected CD34+ cells were prestimulated in X-vivo 10 medium (BioWhittaker) containing 4% fetal calf serum (Stem Cell Technologies) in the presence of Stem Cell Factor (300 ng/ml; provided by Amgen) Flt-3 ligand (300 ng/ml; provided by Immunex), IL-3 (60 ng/ml; Novartis) and polyethylene glycol–Megakaryocyte Growth and Differentiation Factor (100 ng/ml; provided by Amgen). The cells were then transduced with viral supernatant on sterile bags (PL-2417; provided by Nexell-Therapeutics) coated with human recombinant fibronectin (50 ng/ml, CH-296; provided by Takara Shuzo), in the presence of the same cytokines and protamine sulfate (4 ng/ml; Choay Sanofi). Viral supernatant was replaced every 24 hours during the 3-day transduction period.
Monitoring of MRD.
MRD PCR targets were identified, sequenced, and analyzed according to standardized protocols. Screening DNA PCR included TCRD (complete and incomplete), TCRG, and IgH rearrangements. MRD level in follow-up samples was quantified by RQ-PCR according to guidelines from the European Study Group on MRD detection in ALL (ESGMRD-ALL; ref. 46). The MRD-based risk group stratification required the availability of target with sensitivity of at least 1 × 10–4. Follow-up time points were at the end of induction treatment (4–5 weeks), before consolidation, and before the first intensification cycle.
Quantitative analysis of TCR repertoire and longitudinal immunoscope.
PBMC fractions were obtained from patients at different follow-up time points, and BM mononuclear cells were isolated from patients at the time of leukemia occurrence. Total RNA was then prepared using the RNeasy kit (Qiagen). RNA was converted into cDNA following standard protocols by using Superscript II from Invitrogen, oligo dT primers (Eurogentech). PCR reactions were carried out by combining a primer and a specific fluorophore-labeled probe for the constant region with 1 of 24 primers covering the different Vβ chains. Reactions were carried out using the TaqMan 7730 system (Applied Biosystems) and standard reagents from Applied Biosystems. For immunoscope profiles, products were then subjected to runoff reactions with a nested fluorescent primer specific for the constant region for 4 cycles. The fluorescent products were separated and analyzed using an ABI-PRISM 3730 DNA analyzer. The size and intensity of each band were analyzed with Immunoscope software that was adapted to the capillary sequencer. Fluorescence intensities were plotted in arbitrary units on the y axis, and CDR3 lengths were plotted in aa on the x axis.
Isolation and analysis of integration site sequences.
Ligation-mediated (47, 48) PCR was used to sequence vector integration sites. Genomic DNA was extracted from peripheral blood lymphocytes, CD3+ cells, lymph nodes, or BM cells. The restriction enzymes used to cleave genomic DNA samples were chosen after performing pilot studies of the genes involved in order to ensure good recovery, despite the strong recovery biases caused by uneven distribution of restriction enzyme cleavage site in the human genome (49). MseI was found to be optimal for P7 and ApoI for P10. The digested DNA samples were ligated to linkers, cleaved with SpeI to minimize amplification of a vector internal fragment, and then amplified by nested PCR (7, 8, 49–54). In order to sequence patient samples in pools, primers that contained unique 4- or 8-bp barcodes were used in the second PCR step, as described previously (49). The PCR products were gel purified, pooled, and then subjected to pyrosequencing as implemented by 454 Life Sciences (55). To minimize bar code crossover between patients, samples from the same patient were pooled and run in the same quadrant on the sequencing plate. Integration sites were determined to be authentic if the sequences contained the correct barcodes, began within 3 bp of vector LTR ends, had either a greater than 98% sequence match or no more than 1 bp mismatch if the read length was less than 50 bp, and had a unique best hit when aligned to the draft human genome (hg18) using BLAT. All integration site sequences are available on NCBI GenBank (accession no. FI129363–FI129979). The network analyses were generated through the use of Ingenuity Pathways Analysis (Ingenuity Systems), which investigates the relationships among a set of genes and their relationships to further genes based on peer-reviewed biomedical literature.
RQ-PCR for proto-oncogene expression analysis.
For CCND2 expression analysis, the P7 sample, along with 89 primary T-ALL samples (3 CCND2-rearranged and 86 other T-ALL), was quantified for CCND2 expression using Taqman RQ-PCR and TBP as control housekeeping gene, as previously described (18). For BMI1 locus analysis, 45 T-ALL samples (14 CALM-AF10+ T-ALL and 31 CALM-AF10– T-ALL) along with the P10 sample at M+33 were quantified using Taqman RQ-PCR for BMI1, DNAJC1, COMMD3, and SPAG6 (19). The P10 retrovirus insertion in SPAG6 was located in its fourth intron. We quantified expression of the 4 genes. For LMO2 expression analysis, RNA extracted from CD3+ T cells was reverse transcribed using a polydT anchor primer. cDNA quality was checked using ABL as control gene (46). cDNA with ABL Ct values higher than 34 were discarded. Expression of LMO2 gene was defined by calculating the ratio of expression of the gene of interest to the ABL value. RQ-PCR was performed as described previously (46) using an Applied Biosystems 7000 PCR machine.
Array-CGH analysis.
Tumoral genomic DNA was extracted from leukemic cells of the 4 patients, and 500 ng was labeled and cohybridized with normal unrelated control DNA on the 105K Human Genome CGH Microarray (Agilent Technologies). After washing, the arrays were scanned, and data were analyzed using Feature Extraction and CGH Analytics 3.4 software (Agilent Technologies). In P7, tumoral DNA was also cohybridized with paired preleukemic DNA on an additional array to confirm that the genomic abnormalities were indeed acquired but not constitutional polymorphisms known as copy number variations.
Statistics.
For comparison of integration site distribution with that of random controls, Fisher’s exact test was used. A P value less than 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank the CIC Biothérapies in which part of these studies was performed (codirected by INSERM and Direction Régionale de la Recherche Clinique, AP-HP, Paris, France) and Elisabeth Gouguenheim and Malika Tiouri for their secretarial assistance. This work was funded by grants from INSERM, by Association Française contre les Myopathies (AFM) contract AT0203, by the Consortium National de Recherche en Génomique (CNRG), by European Community contract QLK3-CT-1999-00859, by Consert contract no. 005242, by Inherinet contract no. QLK3-CT-2001-00427, by Agence Nationale de la Recherche (ANR) grant 05-MRAR-004, by a grant from the Cancéropole Ile-de-France and the CIT “ligue contre le cancer program” to J. Soulier, and by a grant from Amgen-France to M. de Garidel and C. Cailliot. J. Hauer is a fellow of the Deutsche Akademie der Naturforscher Leopoldina (BMBF-LPD 9901/8-149). F.D. Bushman was supported by NIH grants AI52845 and AI66290. G.P. Wang was supported by National Institute for Allergy and Infectious Diseases, NIH, grant T32 AI07634 (Training Grant in Infectious Diseases) and a University of Pennsylvania Department of Medicine Measey Basic Science Fellowship.
Footnotes
Nonstandard abbreviations used: ADA, adenosine deaminase; γc, common γ chain; CGH, comparative genomic hybridization; CR, complete remission; IL2RG, IL-2 receptor γ; LMO2, LIM domain–only 2; LTR, long-terminal repeat; M+68, 68 months after gene therapy; MRD, minimal residual disease; P-, patient; RQ-PCR, quantitative real-time RT-PCR; SCID-X1, X-linked SCID; T-ALL, T cell acute lymphoblastic leukemia.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 118:3132–3142 (2008). doi:10.1172/JCI35700
Alain Fischer and Marina Cavazzana-Calvo are co–senior authors.
References
- 1.Cavazzana-Calvo M., et al. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science. 2000;288:669–672. doi: 10.1126/science.288.5466.669. [DOI] [PubMed] [Google Scholar]
- 2.Hacein-Bey-Abina S., et al. Sustained correction of X-linked severe combined immunodeficiency by ex vivo gene therapy. N. Engl. J. Med. 2002;346:1185–1193. doi: 10.1056/NEJMoa012616. [DOI] [PubMed] [Google Scholar]
- 3.Aiuti A., et al. Correction of ADA-SCID by stem cell gene therapy combined with nonmyeloablative conditioning. Science. 2002;296:2410–2413. doi: 10.1126/science.1070104. [DOI] [PubMed] [Google Scholar]
- 4.Gaspar H.B., et al. Gene therapy of X-linked severe combined immunodeficiency by use of a pseudotyped gammaretroviral vector. Lancet. 2004;364:2181–2187. doi: 10.1016/S0140-6736(04)17590-9. [DOI] [PubMed] [Google Scholar]
- 5.Hacein-Bey-Abina S., et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 6.Wu X., Li Y., Crise B., Burgess S.M. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 7.Mitchell R.S., et al. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2:E234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewinski M.K., et al. Retroviral DNA integration: viral and cellular determinants of target-site selection. PLoS Pathog. 2006;2:e60. doi: 10.1371/journal.ppat.0020060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bushman F., et al. Genome-wide analysis of retroviral DNA integration. Nat. Rev. Microbiol. 2005;3:848–858. doi: 10.1038/nrmicro1263. [DOI] [PubMed] [Google Scholar]
- 10.Oudot C., et al. Prognostic factors for leukemic induction failure in children with acute lymphoblastic leukemia and outcome after salvage therapy: the FRALLE 93 study. J. Clin. Oncol. 2008;26:1496–1503. doi: 10.1200/JCO.2007.12.2820. [DOI] [PubMed] [Google Scholar]
- 11.Schrappe M., Beier R., Burger B. New treatment strategies in childhood acute lymphoblastic leukaemia. Best Pract. Res. Clin. Haematol. 2002;15:729–740. doi: 10.1053/beha.2002.0226. [DOI] [PubMed] [Google Scholar]
- 12.Kamps W.A., et al. Intensive treatment of children with acute lymphoblastic leukemia according to ALL-BFM-86 without cranial radiotherapy: results of Dutch Childhood Leukemia Study Group Protocol ALL-7 (1988-1991). Blood. 1999;94:1226–1236. [PubMed] [Google Scholar]
- 13.Asselin B., et al. Improved event-free survival (efs) with high dose methotrexate (HDM) in T-cell lymphoblastic leukemia (T-ALL) and advanced lymphoblastic lymphoma (T-NHL): a Pediatric Oncology Group (POG) study [abstract]. Proc. Am. Soc. Clin. Oncol. 2001;20:367a. [Google Scholar]
- 14.Goldberg J.M., et al. Childhood T-cell acute lymphoblastic leukemia: the Dana-Farber Cancer Institute acute lymphoblastic leukemia consortium experience. J. Clin. Oncol. 2003;21:3616–3622. doi: 10.1200/JCO.2003.10.116. [DOI] [PubMed] [Google Scholar]
- 15.Printz M., et al. Recombinant retroviral vector interferes with the detection of amphotropic replication competent retrovirus in standard culture assays. Gene Ther. 1995;2:143–150. [PubMed] [Google Scholar]
- 16.Lim A., et al. Combination of MHC-peptide multimer-based T cell sorting with the Immunoscope permits sensitive ex vivo quantitation and follow-up of human CD8+ T cell immune responses. . J. Immunol. Methods. 2002;261:177–194. doi: 10.1016/S0022-1759(02)00004-2. [DOI] [PubMed] [Google Scholar]
- 17.Aifantis I., Buer J., von Boehmer H., Azogui O. Essential role of the pre-T cell receptor in allelic exclusion of the T cell receptor beta locus. Immunity. 1997;7:601–607. doi: 10.1016/S1074-7613(00)80381-7. [DOI] [PubMed] [Google Scholar]
- 18.Clappier E., et al. Cyclin D2 dysregulation by chromosomal translocations to TCR loci in T-cell acute lymphoblastic leukemias. Leukemia. 2006;20:82–86. doi: 10.1038/sj.leu.2404008. [DOI] [PubMed] [Google Scholar]
- 19.Dik W.A., et al. CALM-AF10+ T-ALL expression profiles are characterized by overexpression of HOXA and BMI1 oncogenes. Leukemia. 2005;19:1948–1957. doi: 10.1038/sj.leu.2403891. [DOI] [PubMed] [Google Scholar]
- 20.Soulier J., et al. HOXA genes are included in genetic and biologic networks defining human acute T-cell leukemia (T-ALL). Blood. 2005;106:274–286. doi: 10.1182/blood-2004-10-3900. [DOI] [PubMed] [Google Scholar]
- 21.Clappier E., et al. The C-MYB locus is involved in chromosomal translocation and genomic duplications in human T-cell acute leukemia (T-ALL), the translocation defining a new T-ALL subtype in very young children. Blood. 2007;110:1251–1261. doi: 10.1182/blood-2006-12-064683. [DOI] [PubMed] [Google Scholar]
- 22.Royer-Pokora B., Loos U., Ludwig W.D. TTG-2, a new gene encoding a cysteine-rich protein with the LIM motif, is overexpressed in acute T-cell leukaemia with the t(11;14)(p13;q11). Oncogene. 1991;6:1887–1893. [PubMed] [Google Scholar]
- 23.Haupt Y., Alexander W.S., Barri G., Klinken S.P., Adams J.M. Novel zinc finger gene implicated as myc collaborator by retrovirally accelerated lymphomagenesis in E mu-myc transgenic mice. Cell. 1991;65:753–763. doi: 10.1016/0092-8674(91)90383-A. [DOI] [PubMed] [Google Scholar]
- 24.van Lohuizen M., et al. Identification of cooperating oncogenes in E mu-myc transgenic mice by provirus tagging. Cell. 1991;65:737–752. doi: 10.1016/0092-8674(91)90382-9. [DOI] [PubMed] [Google Scholar]
- 25.Jacobs J.J., et al. Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev. 1999;13:2678–2690. doi: 10.1101/gad.13.20.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lessard J., Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255–260. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- 27.Sicinska E., et al. Requirement for cyclin D3 in lymphocyte development and T cell leukemias. Cancer Cell. 2003;4:451–461. doi: 10.1016/S1535-6108(03)00301-5. [DOI] [PubMed] [Google Scholar]
- 28.Aplan P.D., et al. Disruption of the human SCL locus by “illegitimate” V-(D)-J recombinase activity. Science. 1990;250:1426–1429. doi: 10.1126/science.2255914. [DOI] [PubMed] [Google Scholar]
- 29.Weng A.P., et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 30.Cayuela J.M., Madani A., Sanhes L., Stern M.H., Sigaux F. Multiple tumor-suppressor gene 1 inactivation is the most frequent genetic alteration in T-cell acute lymphoblastic leukemia. Blood. 1996;87:2180–2186. [PubMed] [Google Scholar]
- 31.Jackson A., et al. Deletion of 6q16-q21 in human lymphoid malignancies: a mapping and deletion analysis. Cancer Res. 2000;60:2775–2779. [PubMed] [Google Scholar]
- 32.Lahortiga I., et al. Duplication of the MYB oncogene in T cell acute lymphoblastic leukemia. Nat. Genet. 2007;39:593–595. doi: 10.1038/ng2025. [DOI] [PubMed] [Google Scholar]
- 33.Armstrong S.A., Look A.T. Molecular genetics of acute lymphoblastic leukemia. J. Clin. Oncol. 2005;23:6306–6315. doi: 10.1200/JCO.2005.05.047. [DOI] [PubMed] [Google Scholar]
- 34.Mullighan C.G., et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 35.Dave U.P., Jenkins N.A., Copeland N.G. Gene therapy insertional mutagenesis insights. Science. 2004;303:333. doi: 10.1126/science.1091667. [DOI] [PubMed] [Google Scholar]
- 36.Aiuti A., et al. Multilineage hematopoietic reconstitution without clonal selection in ADA-SCID patients treated with stem cell gene therapy. J. Clin. Invest. 2007;117:2233–2240. doi: 10.1172/JCI31666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deichmann A., et al. Vector integration is nonrandom and clustered and influences the fate of lymphopoiesis in SCID-X1 gene therapy. J. Clin. Invest. 2007;117:2225–2232. doi: 10.1172/JCI31659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woods N.B., Bottero V., Schmidt M., von Kalle C., Verma I.M. Gene therapy: therapeutic gene causing lymphoma. Nature. 2006;440:1123. doi: 10.1038/4401123a. [DOI] [PubMed] [Google Scholar]
- 39.Pike-Overzet K., et al. Ectopic retroviral expression of LMO2, but not IL2Rgamma, blocks human T-cell development from CD34+ cells: implications for leukemogenesis in gene therapy. Leukemia. 2007;21:754–763. doi: 10.1038/sj.leu.2404563. [DOI] [PubMed] [Google Scholar]
- 40.Pike-Overzet K., et al. 2006Gene therapy: is IL2RG oncogenic in T-cell development? Nature. 443 : E5–; discussion E6–E7. [DOI] [PubMed] [Google Scholar]
- 41.Thrasher A.J., et al. 2006Gene therapy: X-SCID transgene leukaemogenicity. Nature. 443E5–E6; discussion E6–E7. . [DOI] [PubMed] [Google Scholar]
- 42.Shou Y., Ma Z., Lu T., Sorrentino B.P. Unique risk factors for insertional mutagenesis in a mouse model of XSCID gene therapy. Proc. Natl. Acad. Sci. U. S. A. 2006;103:11730–11735. doi: 10.1073/pnas.0603635103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zychlinski D., et al. Physiological promoters reduce the genotoxic risk of integrating gene vectors. Mol. Ther. 2008;16:718–725. doi: 10.1038/mt.2008.5. [DOI] [PubMed] [Google Scholar]
- 44.Montini E., et al. Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration. Nat. Biotechnol. 2006;24:687–696. doi: 10.1038/nbt1216. [DOI] [PubMed] [Google Scholar]
- 45.Aker M., et al. Extended core sequences from the cHS4 insulator are necessary for protecting retroviral vectors from silencing position effects. Hum. Gene Ther. 2007;18:333–343. doi: 10.1089/hum.2007.021. [DOI] [PubMed] [Google Scholar]
- 46.Beillard E., et al. Evaluation of candidate control genes for diagnosis and residual disease detection in leukemic patients using ‘real-time’ quantitative reverse-transcriptase polymerase chain reaction (RQ-PCR) - a Europe against cancer program. Leukemia. 2003;17:2474–2486. doi: 10.1038/sj.leu.2403136. [DOI] [PubMed] [Google Scholar]
- 47.Mueller P.R., Wold B. In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science. 1989;246:780–786. doi: 10.1126/science.2814500. [DOI] [PubMed] [Google Scholar]
- 48.Fors L., Saavedra R.A., Hood L. Cloning of the shark Po promoter using a genomic walking technique based on the polymerase chain reaction. Nucleic Acids Res. 1990;18:2793–2799. doi: 10.1093/nar/18.9.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang G.P., et al. DNA bar coding and pyrosequencing to analyze adverse events in therapeutic gene transfer. Nucleic Acids Res. 2008;36:e49. doi: 10.1093/nar/gkn125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang G.P., Ciuffi A., Leipzig J., Berry C.C., Bushman F.D. HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 2007;17:1186–1194. doi: 10.1101/gr.6286907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barr S.D., et al. HIV integration site selection: targeting in macrophages and the effects of different routes of viral entry. Mol. Ther. 2006;14:218–225. doi: 10.1016/j.ymthe.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 52.Ciuffi A., et al. Integration site selection by HIV-based vectors in dividing and growth-arrested IMR-90 lung fibroblasts. Mol. Ther. 2006;13:366–373. doi: 10.1016/j.ymthe.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 53.Lewinski M.K., et al. Genome-wide analysis of chromosomal features repressing human immunodeficiency virus transcription. J. Virol. 2005;79:6610–6619. doi: 10.1128/JVI.79.11.6610-6619.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schroder A.R., et al. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/S0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 55.Margulies M., et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.