Abstract
X-linked SCID (SCID-X1) is amenable to correction by gene therapy using conventional gammaretroviral vectors. Here, we describe the occurrence of clonal T cell acute lymphoblastic leukemia (T-ALL) promoted by insertional mutagenesis in a completed gene therapy trial of 10 SCID-X1 patients. Integration of the vector in an antisense orientation 35 kb upstream of the protooncogene LIM domain only 2 (LMO2) caused overexpression of LMO2 in the leukemic clone. However, leukemogenesis was likely precipitated by the acquisition of other genetic abnormalities unrelated to vector insertion, including a gain-of-function mutation in NOTCH1, deletion of the tumor suppressor gene locus cyclin-dependent kinase 2A (CDKN2A), and translocation of the TCR-β region to the STIL-TAL1 locus. These findings highlight a general toxicity of endogenous gammaretroviral enhancer elements and also identify a combinatorial process during leukemic evolution that will be important for risk stratification and for future protocol design.
Introduction
X-linked SCID (SCID-X1) accounts for approximately 40%–50% of all SCIDs and is caused by mutations in the gene encoding the IL-2 receptor γ chain (IL2RG) (1, 2). This molecule is a key subunit of the cytokine receptor complex for IL-2, IL-4, IL-7, IL-9, IL-15, and IL-21 and is designated the common cytokine receptor gamma chain (γc). In the absence of γc signaling, many aspects of immune cell development and function are compromised, leading to absence of T and NK cells and persistence of dysfunctional B cells (T–B+NK– SCID). Two independent studies using similar gammaretroviral vectors have demonstrated major therapeutic benefits following gene therapy in infants with this disease (3–5). However, in 1 of these trials, 4 out of 9 patients developed leukemia between 3 and 6 years after treatment primarily as a result of inadvertent vector-mediated upregulation of host cellular oncogenes (6). Here, we report a similar event in a second trial and show that leukemogenesis likely arises from insertional mutagenesis as the initiating event but requires the acquisition of secondary genetic lesions.
Results
Clinical presentation and findings.
The index patient was one of 10 entered into a trial of gene therapy for SCID-X1 at Great Ormond Street Hospital and is designated P8. He presented at the age of 10 months with a history of failure to thrive and Pneumocystis jiroveci pneumonia. Immunophenotypic analysis revealed classical T–B+NK– SCID. A missense C182Y mutation in IL2RG was confirmed by genomic sequencing, which resulted in markedly abrogated expression of the common cytokine receptor γc. Gene therapy was administered at the age of 13 months following independent counseling with regard to the potential risks of leukemogenesis, ethical and regulatory approval from the Gene Therapy Advisory Committee (GTAC) and the Medicines and Healthcare Regulatory Authority (MHRA), and fully informed parental consent. The gibbon ape leukemia virus–pseudotyped (GALV-pseudotyped) gammaretroviral vector encoding the IL2RG cDNA regulated by intact murine leukemia virus (MLV) long terminal repeats (LTRs) and the transduction protocol have been described previously (4). A total of 7 × 106 CD34+γc+ cells/kg were infused in the absence of conditioning. Immunological recovery was uncomplicated, and the patient attained normal T cell numbers (Figure 1A), diversity, and function within 6 months of therapy. As in other patients, natural killer cell numbers remained low in number although they were detectable after therapy. Subcutaneous immunoglobulin replacement was discontinued after 16 months but recommenced after 22 months due to persisting low levels of IgG and recurrent respiratory tract infections. He was reviewed in clinic at 23 months, at which time clinical health was improved and hematological and immunological parameters were normal. At 24 months (d717), he presented with a 2- to 3-week history of pallor, bruising, malaise, and abdominal distension. Peripheral blood count revealed a high leukocyte count with circulating blasts (>400 × 109/l), thrombocytopenia, and profound anemia consistent with T cell acute lymphoblastic leukemia (T-ALL). He was commenced on standard chemotherapy according to UK T-ALL 2003, version 5 (International Standard Randomized Controlled Trial Number 07355119; http://www.ctsu.ox.ac.uk/projects/leuk/ukall2003).
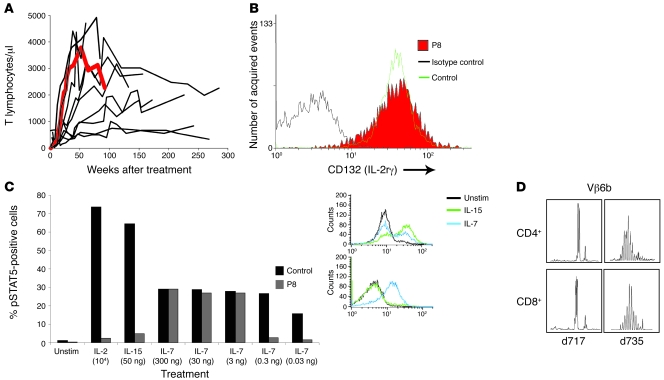
Figure 1. Normal γc surface expression and a lack of constitutive signaling through the IL receptor complex.
(A) Representation of T cell recovery following gene therapy in 10 patients with SCID-X1. The immunological reconstitution of patient 8 (P8) to normal levels is indicated in red up to 3 weeks prior to diagnosis of leukemia. (B) Expression of γc on the surface of leukemic blasts measured by flow cytometry was within the normal range of control peripheral blood T cells. (C) Phosphorylation of the tyrosine residue of STAT5 is indicative of signaling through the JAK-STAT pathway when relevant ILs bind to cell-surface receptor complexes containing γc. Insert shows the presence of phosphorylated STAT5 by flow cytometry in response to IL-7 and IL-15 from the patient and PBMCs from a control. The response to different cytokines and different concentrations of IL-7 is shown in the main panel. Constitutive phosphorylation of STAT5 is not detectable and is also not induced by IL-2 or IL-15. Unstim, unstimulated. (D) Spectratype analysis revealed a dominant TCR Vβ6b clone in both CD4+ and CD8+ cells (d717, at time of leukemia diagnosis), which disappeared following chemotherapy (d735), allowing the emergence of a normal distribution of clones (see also Supplemental Figure 1).
This treatment comprises a standard 4-drug induction with vincristine, dexamethasone, daunorubicin, and asparaginase as well as consolidation with cyclophosphamide, cytarabine, and mercaptopurine. The patient was in clinical and molecular remission 4 months after commencement of therapy. At this point, although total T cell numbers were low, he had a fully diverse T cell repertoire with normal TCR spectratype (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI35798DS1) and distribution of naive/memory T cells (data not shown).
Characterization of T-ALL and γc expression.
Immunophenotyping and molecular spectratyping of peripheral blood revealed a single TCRVβ6b diploid leukemic clone CD3–(cytoplasmic CD3+), CD4+CD8+, CD7+CD10+, TdT+. Surface expression of γc was comparable to that of normal peripheral blood T cells (Figure 1B). STAT5 phosphorylation was inducible by stimulation with IL-7 but not with IL-2 or IL-15, and there was no detectable constitutive activation of γc signaling (Figure 1C). The dominant TCRVβ6b clone in leukemic cells resolved after chemotherapy, and the spectratype of all Vβ families regained a normal Gaussian distribution (Figure 1D and Supplemental Figure 1). No replication-competent retrovirus was observed in transduced CD34+ progenitor samples or in follow-up samples from this patient (data not shown).
Molecular pathogenesis.
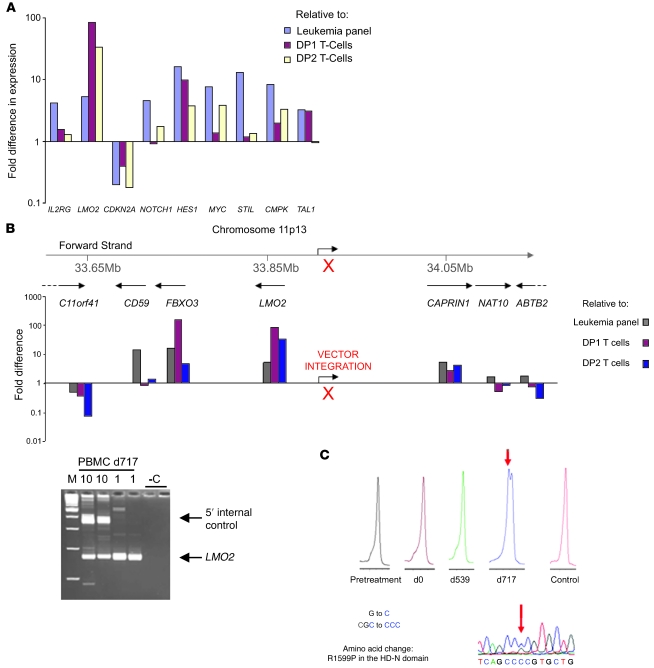
Global gene expression levels in peripheral blood (>99% leukemic cells) at the time of presentation were compared with a series of 104 different childhood leukemia samples or with CD4+CD8+ double-positive (DP) stage 1 (DP1) (CD3–) or stage 2 (DP2) (CD3+) human thymocytes, which approximate to the developmental stage of the clone. Notably, the patient’s cells overexpressed LIM domain only 2 (LMO2), NOTCH1, and downstream regulatory targets of NOTCH1 signaling, including HES1 and c-MYC and genes within the STIL-TAL (STIL, stem cell leukemia gene/TAL-1–interrupting locus gene; TAL, T cell acute lymphocytic leukemia) locus (STIL, TAL1, CMPK) (Figure 2A). Expression of LMO2 was the highest of all leukemia samples. Markedly reduced expression of the tumor suppressor gene p16(INK4A)/p14(ARF1), encoded by the cyclin-dependent kinase 2A (CDKN2A) gene locus, was also noted (Figure 2A). Linear amplification–mediated PCR (LAM-PCR) and direct sequencing of the regions flanking the vector genome indicated a single copy insertion in the leukemic clone 35 kb upstream of the transcription start site of the LMO2 protooncogene in an antisense orientation (Figure 2B). A specific tracking PCR for the LMO2 insertion was positive at d539 in a preleukemic sample but not beforehand (sensitivity <0.1%) (Supplemental Figure 3A). Following chemotherapy treatment (commenced on d735 after gene therapy), the clone containing the insertion close to LMO2 was no longer detectable by LAM-PCR or specific tracking PCR (Supplemental Figure 3, A and B). Genes surrounding LMO2 were also upregulated, reflecting the broad influence that LTR enhancer activity had on neighboring chromosomal loci (Figure 2B). Any influence that the insertion may have on the locus appears to be limited because upregulation of expression was not observed in genes located further from LMO2 (C11orf41, NAT10, ABTB2, and beyond). Screening for acquired mutations in NOTCH1 and FBXW7 revealed a single R1559P NOTCH1 mutation in the heterodimerization–N domain (HD-N domain) at high levels, probably affecting the whole clone (Figure 2C). Within the limits of resolution of the assay (approximately 0.1%–1%), the R1559P NOTCH1 mutation was not detected at d539, indicating that this was not a constitutive or preexisting anomaly (Supplemental Figure 2). Genome-wide SNP array revealed a loss of heterozygosity (LOH) affecting most of the short arm of chromosome 9, which includes the CDKN2A locus (Figure 3). An additional region of LOH close to the TCR-β locus was suggestive of further chromosomal rearrangement.
Figure 2. Gene expression levels and retroviral insertion site.
(A) Microarray analysis was used to compare levels of gene expression in the patient’s cells to those found in thymocytes or a panel of leukemias. DP1 and DP2 represent arrays performed on 2 different developmental stages of normal human CD4+CD8+ thymocytes. The y axis shows the fold change in expression, with values above 1 representing an increase and those below 1 a decrease. When compared with thymocytes and other leukemias, LMO2, NOTCH1, HES1, STIL, TAL1, and CMPK gene expression is upregulated. In contrast, expression levels of tumor suppressor genes p14(ARF1) and p16(INK4a) were reduced. γc mRNA was not overexpressed relative to thymocytes (consistent with surface expression data), although it was in comparison with the heterogeneous group of leukemias. (B) The vector insertion site is 35087 bp upstream of the LMO2 transcription start site in the opposite orientation (red X). LAM-PCR analysis (inset) of 10 ng and 1 ng of d717 PBMC DNA identified 1 dominant clone. M, 100-bp ladder; –C, 100 ng nontransduced DNA. When using limiting amounts of DNA, the internal control is out-competed by the LMO2 amplicon. The genomic locus and expression levels of genes surrounding LMO2 are shown in comparison with a dataset of arrays performed on other childhood leukemias or DP1 and DP2 human thymocytes. (C) Sequencing of denaturing HPLC analysis revealed a R1599P substitution in the HD-N domain of NOTCH1 in the leukemic cells (d717) but not before. No other NOTCH1 mutations were found.
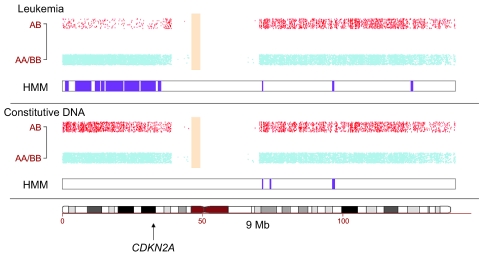
Figure 3. SNP array analysis shows LOH.
A genome-wide SNP array was used to examine gross deletions or rearrangements within the patient’s chromosomes. Signal from the array showing homozygosity (AA/BB) and heterozygosity (AB) of alleles on each chromosome are shown by blue or red dots, respectively. Strikingly, analysis of chromosome 9 revealed a LOH of a large region of the short arm in leukemia cells (top panel) when compared with DNA from pre–gene therapy cells (middle panel). LOH as identified by a hidden Markov model (HMM) is represented graphically in purple to show the area altered between constitutive and leukemic DNA. The tumor suppressor gene CDKN2A, which encodes p14(ARF1) and p16(INK4a), is located at 9p21, shown in the lower panel by an arrow. Alterations within this genomic locus are probably the cause for downregulation of CDKN2A expression (as described in Figure 2A).
Cytogenetic analysis of abnormal cells revealed the following karyotype:
Karyotype 1

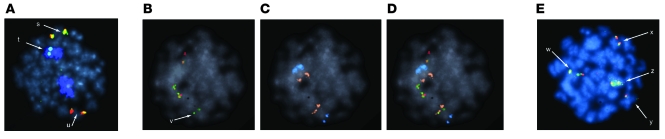
Primary analysis of the peripheral blood cultures demonstrated 31/35 (88.5%) of metaphase cells and 168/200 (84%) of interphase cells as showing a rearrangement of the TCR-β locus, with a breakpoint upstream of the TCR-β breakpoint cluster region on chromosome 7. The translocation was further characterized to reveal an insertion of the TCR-β region into what appeared to be the STIL-TAL1 locus of chromosome 1 at 1p32 and 1p36 (Figure 4), likely accounting for overexpression of TAL1, CMPK, and STIL (Figure 2A).
Figure 4. FISH analysis reveals a chromosomal rearrangement.
Metaphase cells showing a rearrangement of the TCR-β locus by FISH. (A) Normal copy of chromosome 7 with intact TCR-β (colocalization of a red-green probe) (s). Rearrangement partner chromosome 1 (blue) showing the translocated portion of the probe containing the remainder of the 732-kb region upstream of the TCR-β breakpoint cluster (t). Derivative chromosome 7 with intact portion of the probe (red signal) covering 320 kb downstream of the TCR-β breakpoint cluster region with a residual signal from the probe (green signal) covering the proximal region 732 kb upstream from the TCR-β breakpoint cluster region (u). The insertion was located in the STIL-TAL1 region (1p32–1p36). Additional probes show (B) the red/green signal of the normal TCR-β region probe with the inserted fragment (v, green only), (C) the position of chromosome 1 using the 1p36 (false-colored orange) and 1q25 probe sets (false-colored blue), and (D) an overlay of the 2 previous pictures, locating the insertion proximal to 1p36. (E) Red/green signals of the normal (w) and abnormal (x) TCR-β region probes and normal-sized STIL-TAL1 (y) and large STIL-TAL probe signal apparently encompassing the inserted green TCR-β region probe signal (z). Original magnification, ×100.
Discussion
Several clinical trials have demonstrated clinical efficacy of somatic gene therapy for patients with primary immunodeficiency (4, 5, 7–10). However, inadvertent vector-mediated insertional activation of cellular oncogenes has caused significant toxicity (6). Although mutagenesis is presumed to be a primary precipitating event, other contributing factors have not been clearly identified. These may be important for determining disease-specific risks and for design of future strategies.
Here, we have shown in a second clinical study of gene therapy for SCID-X1 that conventional murine gammaretroviral vectors (based on MLVs) present a high risk of leukemogenesis as a result of their powerful enhancer sequences in the intact LTR regions. In the index patient, a single insertion in an antisense orientation 35 kb upstream of the transcription start site caused substantial overexpression of LMO2 and neighboring genes. This activity reflects the mechanism of oncogenesis utilized by many wild-type slow-transforming murine gammaretroviruses that themselves do not encode oncogenes and highlights the potential for mutagenesis of these elements irrespective of favored integration much closer to transcription start sites (11–14). Remarkably, 4 out of the 5 patients in the 2 SCID-X1 gene therapy trials who have now developed lymphoproliferative disease as a complication of gene therapy in both trials have been found to have an activating insertion in the LMO2 locus. This indicates that either this is a preferred site for integration of gammaretroviral vectors or more likely that integration at this site imparts a powerful preleukemic predisposition (15).
Unsurprisingly, differences in vector pseudotype and culture conditions prior to cell engraftment appear not to be of major importance in the context of these 2 SCID-X1 studies, as suggested by the general similarity of insertion patterns noted between the 2 studies (12, 14). Integration site analysis at present may be too immature to allow prediction of adverse events, as there are several confounding factors. First, it is not possible to predict whether a specific insertion will necessarily cause dysregulated gene expression. Second, only a fraction of all insertions can at present be sampled and characterized by routine methodology. Third, these events are arising in the thymus, whereas clonality studies are based on sampling of peripheral blood. Insertions that disturb thymopoiesis may therefore be underrepresented in the periphery. Theoretically, this is true for LMO2 upregulation. Development of high throughput screening of the complete inventory may help, but the issue of thymic sampling remains.
Activation of LMO2 is known to participate in human leukemogenesis by chromosomal translocation (often through deletion of a negative regulatory region) and results in the development of T cell lymphoproliferation and leukemia in mice, albeit with a long latency (16–18). Interestingly, translocations at this locus have also been found in normal human thymus tissue, although the precise site of translocation may be critical for determining the extent of gene activation (17, 19). Overexpression of LMO2 in mice or in vitro results in impaired T cell development (18, 20). It is therefore likely that other contributing factors are required for terminal leukemic events to manifest. In the patient reported on in this study, gene expression analysis revealed overexpression of NOTCH1 and 1 of its transcriptional targets, HES1, suggesting either an additional insertional event or a secondary acquired mutation. Activating NOTCH1 mutations have been found in over 50% of patients with T-ALL (21) (although it has been unclear whether they are early initiating events or secondary events). In rodents, Notch1 mutation can directly induce leukemia but also can participate as a secondary collaborator (22, 23). Although prenatal NOTCH1 mutation can also act as an initiating factor in human T-ALL, there is good evidence to support this as a secondary event, therefore suggesting that the temporal sequence of genetic lesions is less important than the combinatorial effect (24, 25). Here, it appears likely that an activating mutation in the HD-N domain of NOTCH1 is indeed a secondary event following upregulated expression of LMO2, in accordance with mouse models of T-ALL induced by constitutive LMO2 expression (16). The fact that it appears to be present at high levels in the majority of cells would indicate its importance for the pathophysiology. The R1599P mutation has previously been described in human patients with T-ALL and is a potent activator of NOTCH1 signaling leading to ligand-independent cleavage of the receptor (26). Another frequently observed acquired molecular participant in T-ALL is deletion or inactivation of the tumor suppressor locus CDKN2A on chromosome band 9p21, which encodes p16(INK4A), a negative regulator of cyclin-dependent kinases, and p14(ARF1), an activator of p53. SNP array analysis indicated LOH over a large proportion of chromosome 9p. Similarly, a nonreciprocal rearrangement of the TCR-β region of chromosome 7 inserting into or close to the STIL-TAL1 region on chromosome 1 is also likely to participate. These data highlight an accumulation of gain-of-function and loss-of-function genetic lesions during evolution of leukemia.
The index patient presented within 2 years of treatment and did not have any obvious preceding risk factors when compared with other patients in the same trial with longer follow-up. Whether constitutive expression of γc itself in thymocytes or thymocyte precursors is a risk factor remains controversial, although there is good evidence that the SCID-X1 background is important (27, 28). It is possible, for example, that there is an accumulation of cells susceptible to mutagenesis or that additional mutagenic events occur as a result of nonphysiological rapid expansion of thymocytes following release of precursors from their developmental block. Conversely, outside the context of gene therapy, it has been proposed that IL-7 is an important extrinsic factor that supports viability and proliferation of leukemic cells so that aberrant expression could circumvent normal cellular mechanisms to shut down dysregulated cellular growth (29).
Substantial evidence supports an important role for NK cells in immunosurveillance of cancer, as most recently demonstrated in mice deficient in an NK cell–activating receptor, NKG2D (30). In humans, NK cells have been shown to participate in leukemia clearance in KIR-mismatched allogeneic stem cell transplantation, at least for acute myeloid leukemias. In SCID-X1 patients treated by gene therapy, NK cell numbers usually remain low due to the small number of transduced hematopoietic stem cells that engraft. It is therefore conceivable that there is a deficiency in tumor surveillance in this specific setting. Similar patterns of NK engraftment and persistence are observed after unconditioned allogeneic transplantation for SCID-X1, and leukemogenesis has not to our knowledge been reported here. However, these patients do not have a primary mutagenic insult as a result of vector insertion.
Elucidation of vector-mediated and combinatorial mechanisms that lead to leukemia in human gene-therapy patients provides a unique insight into the genetic events causing cancer. Crucially, it also provides the rationale for development of vectors with reduced mutagenicity and enhanced safety (31, 32). Recent studies have clearly demonstrated their potential in diseases such as SCID-X1 and lead the way for further clinical trials in patients for whom toxicities associated with conventional allogeneic therapy remain unacceptable (33, 34).
Methods
Microarray analysis.
Total RNA was prepared from cells using TRI Reagent (Sigma-Aldrich) and quality assessed using a NanoDrop spectrophotometer (Thermo Scientific) and Agilent Bioanalyzer 2100 (Agilent Technologies). Labeled cRNA was prepared according to standard Affymetrix procedures for hybridization to U133plus2.0 arrays. After scanning, expression summaries were generated by MAS5 and data transferred to GeneSpring 7.3 for comparison with either a panel of 104 leukemias including T-ALL, B cell acute lymphoblastic leukemia (B-ALL), and others (35) using the GeneSpring default normalization or to the average gene expression levels of 2 independent microarray experiments performed using 5 thymi of sorted human CD4+CD8+CD3– (DP1) and CD4+CD8+CD3+ (DP2) thymocyte stages of T cell development (17). Where comparisons were made between data produced on U133A and U133plus2.0 arrays, probe sets missing from the U133A array were not included in the analysis.
So we could examine chromosomal changes, genomic DNA was prepared from PBMCs with a DNeasy Tissue Kit (QIAGEN) following manufacturer’s instructions. DNA was labeled according to standard Affymetrix protocols and hybridized to 250K Nsp and 250K Sty SNP arrays.
Statistics: expression microarrays.
The mean expression values taken from 2 arrays performed for the gene therapy patient were compared with the mean values of a control panel of 104 leukemia patients. A stringent filter of a 20-fold increase or decrease in normalized values was applied, and genes of interest within this group were examined in more detail.
Statistics: SNP arrays.
The SNP array data were processed using BioConductor 2.7.0 (36) with the package oligo 1.4.0. The Affymetrix 250K SNP and 250K Sty chips were processed separately. To identify copy number variation, the arrays were normalized to a reference distribution based on 270 HapMap samples and summarized using SNPRMA (SNP robust multiarray average), after which a CRLMM (corrected robust linear model with maximum-likelihood based distances) was fitted according to default parameters as described before (37). The CRLMM algorithm performs regressions to adjust for average intensity and fragment length in the log-scale ratio. log likelihood ratio thresholds of 5,5,1 were used for AA, BB, and AB genotype calls, respectively. A hidden Markov model (Bioconductor package VanillaICE 1.20) was then applied to the collection of genotype calls to identify longer stretches of homozygosity than would be expected by chance, which shows LOH. The algorithm used was instructed to look for 2 states, N (normal) and L (loss), with estimated emission probabilities of 0.99 and 0.7. The predicted areas of LOH were then shown for each chromosome in the normal and leukemic sample.
Notch mutation detection.
The HD-N domain, the TAD and PEST domains of Notch1, and the WD40 domain (exons 8 to 12) of FBXW7 were amplified by PCR and analyzed by denaturing HPLC (WAVE; Transgenomic) as previously (24). Positive chromatograms were sequenced (CEQ8000; Beckman Coulter).
For sensitive identification of the Notch1 G4796C mutation (R1599P), a 2-round amplification refractory mutation system (ARMS) was developed. The first round consisted of a 35-cycle PCR of exon 26 with 20 ng genomic DNA, BioTAQ DNA polymerase 0.5 units (Bioline), MgCl2 (1 mM), deoxynucleotide triphosphates (dNTPs) (200 mM), primers 26-F (5′-GGAAGGCGGCCTGAGCGTGTC-3′) and 26-R (5′-ATTGACCGTGGGCGCCGGGTC-3′), with an annealing temperature of 67°C. PCR products were visualized after agarose gel electrophoresis. One microliter of the first-round PCR product was used as template in an ARMS PCR using a fluorescently labeled common forward primer (dye D4 5′-GGAAGGCGGCCTGAGCGTGTC-3′), and a common reverse (26R) and mutation-specific reverse primer (5′-CACGTTGGTGTGCAGCACGG-3′). The reaction was carried out in a final volume of 20 μl using BioTAQ DNA polymerase (0.5 units), MgCl2 (1 mM), and dNTPs (200 mM) with 30 cycles of amplification and an annealing temperature of 67°C. Second-round PCR products were sized by fragment analysis (CEQ 8000 Genetic Analysis System; Beckman Coulter).
Sensitivity was determined using dilution mixes of mutant and wild-type TOPO-cloned PCR fragments (TOPO TA; Invitrogen) and confirmed by mixes of mutant and wild-type genomic DNA.
Spectratyping.
Complementarity determining region-3 (CDR3) TCR spectratyping was performed as previously described (38). In brief, RNA was extracted and cDNA prepared from CD4+ and/or CD8+ T cells. Twenty-four Vβ-specific primers were used with a fluorescent-labeled constant region–specific (Cβ-specific) primer to RT-PCR amplify the CDR3 region of the TCR-β chain. Products were run on an AB 3130 Genetic Analyzer (Applied Biosystems) and analyzed using GeneMapper v4.0 software (Applied Biosystems).
Detection of 3′ vector genome junctions by LAM-PCR.
LAM-PCR was performed as described previously (12, 14, 39). In brief, the 3′ vector-genome junctions were preamplified using 2 biotinylated vector-specific primers in a linear PCR. (LTRI, 5′-TCCGATTGACTGAGTCGC-3′; LTRII, 5′-GGTACCCGTGTATCCAATA-3′). After a magnetic capture (MC) step of the biotinylated PCR fragments using streptavidin-coupled paramagnetic beads (Dynal), double strands were synthesized, and after restriction digestion, a linker cassette ligation was performed. The double strands were denatured and 2%–40% of the LAM-PCR product was introduced in a first exponential PCR using a biotinylated vector (LTRIII, 5′-TCTTGCAGTTGCATCCGACT-3′) and a linker cassette–specific primer (LKI, 5′-GACCCGGGAGATCTGAATTC-3′). After MC of the first exponential PCR product, a second exponential PCR with nested primers (LTRIV, 5′-GTGGTCTCGCTGTTCCTT-3′; LKII, 5′-GATCTGAATTCAGTGGCACAG-3′) was performed on 5% of the MC product. 20% of the LAM-PCR product was separated on a Spreadex high-resolution gel (Elchrom Scientific). The remaining LAM-PCR product was purified (PCR Purification Kit; QIAGEN), cloned (TOPO-TA Cloning Kit; Invitrogen), and sequenced.
Integration site tracking.
To track the integration site close to LMO2 in different samples, 10 ng of patient DNA was preamplified using the GenomiPhi DNA Amplification Kit (GE Healthcare). 0.5%–5% of the product was introduced into a PCR with a vector-specific primer (LTRV, 5′-TCTTGCAGTTGCATCCGACT-3′) and a primer specific for flanking genomic sequence (FPI, 5′-CCATTGAGAGGTGAAGACTA-3′). 2% of the first PCR was introduced into a second nested PCR using primers LTRVI (5′-GTGGTCTCGCTGTTCCTT-3′) and FPII (5′-CTTGAACCTCATGATTTGTC-3′). Both PCRs were performed using an initial denaturation at 95°C for 2 minutes and 35 cycles of denaturation at 95°C for 45 seconds, annealing at 56–58°C for 45 seconds and extension at 72°C for 60 seconds, which were followed by a final extension at 72°C for 5 minutes. The expected fragment length resulting from the LMO2 integration clone was 195 bp.
Detection of STAT5 tyrosine phosphorylation and γc expression by flow cytometry.
1 × 104 IU/ml IL-2 (Chiron), 50 ng/ml IL-15 (R&D Systems), or 0.03–300 ng/ml IL-7 (R&D systems) was added to 100 μl of whole blood and incubated at 37°C for 10 minutes. 2 ml of prewarmed FACS lyse/fix (BD Biosciences) was then added to the blood, mixed, and incubated at 37°C for 10 minutes. The cells were pelleted and washed once with PBS (Invitrogen) containing 1% fetal calf serum (STAT wash). The cells were resuspended in cold Perm Buffer III (BD Biosciences) and incubated at 4°C for 30 minutes. The cells were then washed once with STAT wash before 5 μl of antibodies (STAT5 ptyr and CD4 PerCP; BD Biosciences) was added, and the cells were incubated at room temperature for 30 minutes in the dark, washed with STAT wash, and fixed (FACSFix; BD Biosciences). 10,000 lymphocyte events were acquired (FACSCalibur; BD Biosciences) and analyzed using CellQuest software (BD Biosciences). To determine the amount of γc expressed on the cell surface of PBMCs, cells were stained with a PE-labeled anti-CD132 antibody (clone TUGh4; BD Biosciences) and analyzed as above.
Cytogenetics and fluorescence in situ hybridization.
Cultures of peripheral blood cells were either incubated in RPMI-1640 with 20% fetal calf serum at 37°C for times varying between 1 hour and 24 hours or grown in coculture with an OP9 stromal layer to improve viability (34). Snap-frozen mononuclear cells were incubated for 24 hours at 37°C. All cells were exposed to 0.05 mg/ml colcemid (Gibco; Invitrogen) at 37°C, with the RPMI cultures being exposed for 45 minutes and the stromal layer culture being exposed for 18 hours. All cell cultures were harvested using routine cytogenetic methods. Metaphase chromosomes were GTG banded by a conventional trypsin-Giemsa technique, and 20 metaphase cells were karyotyped and reported using the International System for Human Cytogenetic Nomenclature (ISCN 2005). For FISH, chromosome spreads were prepared directly from the same samples. Slides were pretreated in 2× SSC at 37°C for 1 hour and dehydrated through a series of ethanol washes. Whole-chromosome paints for chromosomes 1, 7, and 9 (Cytocell), a split-signal DNA FISH probe for detection of the TCR-β region at chromosome 7q34 (Dako), a dual-color DNA FISH probe for detection of the STIL and TAL1 regions at chromosome 1p32 (Dako), and a dual-color DNA FISH probe for the detection of the TP73 and ANGPTL regions at chromosomes 1p36 and 1q25, respectively (Abbott) were prepared and hybridized according to the manufacturers’ instructions. Further sequential FISH was carried out using the TCR-β probe in conjunction with whole-chromosome paints for several targets, including chromosome 1 and the STIL-TAL1 probe and the 1p/1q probe, which were all hybridized according to published Dako protocols. Cell images were captured using an Olympus BX61 microscope, and image analysis was performed using CytoVision software (Applied Imaging). For analysis by whole-chromosome paint, 50 metaphase cells were examined for rearrangements between chromosomes 1, 7, and 9. Primary analysis of the TCR-β region was performed on 200 interphase cells and on 35 metaphase cells. Analysis of the multiplexed TCR-β probe FISH was carried out on 5 metaphase cells per hybridization.
Supplementary Material
Acknowledgments
We would like to acknowledge assistance from Nipurna Jina and Kerra Pearce of the Institute of Child Health Gene Microarray Centre in performing the microarrays, Dale Moulding in preparing the figures, and the following funding sources: the Wellcome Trust (to A.J. Thrasher), United Kingdom Department of Health, FP6 grant Concerted Safety and Efficacy Evaluation of Retroviral Transgenesis in Gene Therapy of Inherited Diseases (CONSERT 005242), the Medical Research Council, and the Great Ormond Street Hospital National Institutes of Health Biomedical Research Centre.
Footnotes
Nonstandard abbreviations used: γc, gamma chain; CDKN2A, cyclin-dependent kinase 2A; DP, double positive; HD, heterodimerization; IL2RG, IL-2 receptor γ chain; LAM-PCR, linear amplification–mediated PCR; LMO2, LIM domain only 2; LOH, loss of heterozygosity; LTR, long terminal repeat; MC, magnetic capture; MLV, murine leukemia virus; SCID-X1, X-linked SCID; TAL, T cell acute lymphocytic leukemia; T-ALL, T cell acute lymphoblastic leukemia; STIL, stem cell leukemia gene/TAL-1 interrupting locus gene.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 118:3143–3150 (2008). doi:10.1172/JCI35798
References
- 1.Noguchi M., et al. Interleukin-2 receptor gamma chain mutation results in X-linked severe combined immunodeficiency in humans. Cell. 1993;73:147–157. doi: 10.1016/0092-8674(93)90167-O. [DOI] [PubMed] [Google Scholar]
- 2.Takeshita T., et al. Cloning of the gamma chain of the human IL-2 receptor. Science. 1992;257:379–382. doi: 10.1126/science.1631559. [DOI] [PubMed] [Google Scholar]
- 3.Cavazzana-Calvo M., et al. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science. 2000;288:669–672. doi: 10.1126/science.288.5466.669. [DOI] [PubMed] [Google Scholar]
- 4.Gaspar H.B., et al. Gene therapy of X-linked severe combined immunodeficiency by use of a pseudotyped gammaretroviral vector. Lancet. 2004;364:2181–2187. doi: 10.1016/S0140-6736(04)17590-9. [DOI] [PubMed] [Google Scholar]
- 5.Hacein-Bey-Abina S., et al. Sustained correction of X-linked severe combined immunodeficiency by ex vivo gene therapy. N. Engl. J. Med. 2002;346:1185–1193. doi: 10.1056/NEJMoa012616. [DOI] [PubMed] [Google Scholar]
- 6.Hacein-Bey-Abina S., et al. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N. Engl. J. Med. 2003;348:255–256. doi: 10.1056/NEJM200301163480314. [DOI] [PubMed] [Google Scholar]
- 7.Aiuti A., et al. Correction of ADA-SCID by stem cell gene therapy combined with nonmyeloablative conditioning. Science. 2002;296:2410–2413. doi: 10.1126/science.1070104. [DOI] [PubMed] [Google Scholar]
- 8.Aiuti A., et al. Immune reconstitution in ADA-SCID after PBL gene therapy and discontinuation of enzyme replacement. Nat. Med. 2002;8:423–425. doi: 10.1038/nm0502-423. [DOI] [PubMed] [Google Scholar]
- 9.Gaspar H.B., et al. Successful reconstitution of immunity in ADA-SCID by stem cell gene therapy following cessation of PEG-ADA and use of mild preconditioning. Mol. Ther. 2006;14:505–513. doi: 10.1016/j.ymthe.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 10.Ott M.G., et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat. Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- 11.Wu X., Li Y., Crise B., Burgess S.M. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 12.Schwarzwaelder K., et al. Gammaretrovirus-mediated correction of SCID-X1 is associated with skewed vector integration site distribution in vivo. J. Clin. Invest. 2007;117:2241–2249. doi: 10.1172/JCI31661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitchell R.S., et al. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2:E234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deichmann A., et al. Vector integration is nonrandom and clustered and influences the fate of lymphopoiesis in SCID-X1 gene therapy. J. Clin. Invest. 2007;117:2225–2232. doi: 10.1172/JCI31659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cattoglio C., et al. Hot spots of retroviral integration in human CD34+ hematopoietic cells. Blood. 2007;110:1770–1778. doi: 10.1182/blood-2007-01-068759. [DOI] [PubMed] [Google Scholar]
- 16.Nam C.H., Rabbitts T.H. The role of LMO2 in development and in T cell leukemia after chromosomal translocation or retroviral insertion. Mol. Ther. 2006;13:15–25. doi: 10.1016/j.ymthe.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 17.Dik W.A., et al. New insights on human T cell development by quantitative T cell receptor gene rearrangement studies and gene expression profiling. J. Exp. Med. 2005;201:1715–1723. doi: 10.1084/jem.20042524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larson R.C., Osada H., Larson T.A., Lavenir I., Rabbitts T.H. The oncogenic LIM protein Rbtn2 causes thymic developmental aberrations that precede malignancy in transgenic mice. Oncogene. 1995;11:853–862. [PubMed] [Google Scholar]
- 19.Marculescu R., Le T., Simon P., Jaeger U., Nadel B. V(D)J-mediated translocations in lymphoid neoplasms: a functional assessment of genomic instability by cryptic sites. J. Exp. Med. 2002;195:85–98. doi: 10.1084/jem.20011578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pike-Overzet K., et al. Ectopic retroviral expression of LMO2, but not IL2Rgamma, blocks human T-cell development from CD34+ cells: implications for leukemogenesis in gene therapy. Leukemia. 2007;21:754–763. doi: 10.1038/sj.leu.2404563. [DOI] [PubMed] [Google Scholar]
- 21.Weng A.P., et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 22.Grabher C., von Boehmer H., Look A.T. Notch 1 activation in the molecular pathogenesis of T-cell acute lymphoblastic leukaemia. Nat. Rev. Cancer. 2006;6:347–359. doi: 10.1038/nrc1880. [DOI] [PubMed] [Google Scholar]
- 23.O’Neil J., et al. Activating Notch1 mutations in mouse models of T-ALL. Blood. 2006;107:781–785. doi: 10.1182/blood-2005-06-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mansour M.R., et al. Notch-1 mutations are secondary events in some patients with T-cell acute lymphoblastic leukemia. Clin. Cancer Res. 2007;13:6964–6969. doi: 10.1158/1078-0432.CCR-07-1474. [DOI] [PubMed] [Google Scholar]
- 25.Eguchi-Ishimae M., Eguchi M., Kempski H., Greaves M. NOTCH1 mutation can be an early, prenatal genetic event in T-ALL. Blood. 2008;111:376–378. doi: 10.1182/blood-2007-02-074690. [DOI] [PubMed] [Google Scholar]
- 26.Malecki M.J., et al. Leukemia-associated mutations within the NOTCH1 heterodimerization domain fall into at least two distinct mechanistic classes. Mol. Cell. Biol. 2006;26:4642–4651. doi: 10.1128/MCB.01655-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shou Y., Ma Z., Lu T., Sorrentino B.P. Unique risk factors for insertional mutagenesis in a mouse model of XSCID gene therapy. Proc. Natl. Acad. Sci. U. S. A. 2006;103:11730–11735. doi: 10.1073/pnas.0603635103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dave U.P., Jenkins N.A., Copeland N.G. Gene therapy insertional mutagenesis insights. . Science. 2004;303:333. doi: 10.1126/science.1091667. [DOI] [PubMed] [Google Scholar]
- 29.Barata J.T., Cardoso A.A., Boussiotis V.A. Interleukin-7 in T-cell acute lymphoblastic leukemia: an extrinsic factor supporting leukemogenesis? Leuk. Lymphoma. 2005;46:483–495. doi: 10.1080/10428190400027852. [DOI] [PubMed] [Google Scholar]
- 30.Guerra N., et al. NKG2D-deficient mice are defective in tumor surveillance in models of spontaneous malignancy. Immunity. 2008;28:571–580. doi: 10.1016/j.immuni.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montini E., et al. Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration. Nat. Biotechnol. 2006;24:687–696. doi: 10.1038/nbt1216. [DOI] [PubMed] [Google Scholar]
- 32.Modlich U., et al. Cell-culture assays reveal the importance of retroviral vector design for insertional genotoxicity. Blood. 2006;108:2545–2553. doi: 10.1182/blood-2005-08-024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang F., et al. Lentiviral vectors containing an enhancer-less ubiquitously acting chromatin opening element (UCOE) provide highly reproducible and stable transgene expression in hematopoietic cells. Blood. 2007;110:1448–1457. doi: 10.1182/blood-2006-12-060814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thornhill S.I., et al. Self-inactivating gammaretroviral vectors for gene therapy of X-linked severe combined immunodeficiency. Mol. Ther. 2008;16:590–598. doi: 10.1038/sj.mt.6300393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Delft F.W., et al. Prospective gene expression analysis accurately subtypes acute leukaemia in children and establishes a commonality between hyperdiploidy and t(12;21) in acute lymphoblastic leukaemia. Br. J. Haematol. 2005;130:26–35. doi: 10.1111/j.1365-2141.2005.05545.x. [DOI] [PubMed] [Google Scholar]
- 36.Gentleman R.C., et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carvalho B., Bengtsson H., Speed T.P., Irizarry R.A. Exploration, normalization, and genotype calls of high-density oligonucleotide SNP array data. Biostatistics. 2007;8:485–499. doi: 10.1093/biostatistics/kxl042. [DOI] [PubMed] [Google Scholar]
- 38.King D.J., Gotch F.M., Larsson-Sciard E.L. T-cell re-population in HIV-infected children on highly active anti-retroviral therapy (HAART). Clin. Exp. Immunol. 2001;125:447–454. doi: 10.1046/j.1365-2249.2001.01616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmidt M., et al. High-resolution insertion-site analysis by linear amplification-mediated PCR (LAM-PCR). Nat. Methods. 2007;4:1051–1057. doi: 10.1038/nmeth1103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.