Abstract
The HSV major tegument structural protein VP22 resides in multiple subcellular regions during productive infection. During an analysis of the molecular determinants of these localizations, we observed that a transfected fusion of the C-terminal portion of VP22, containing its pat4 nuclear localization signal, with GFP lacked nucleolar sparing compared to GFP alone. Thus, the initial goal was to determine whetherr VP22 associates with nucleoli. Using an optimized indirect immunofluorescence system to visualize nucleolin and viral proteins, we observed that VP22 present in VP22-expressing Vero (V49) cells “surrounded” nucleolin. These two initial findings implied that VP22 might associate directly with nucleoli. We next analyzed HSV infected cells and observed that at late times, anti-nucleolin immune reactivity was dispersed throughout the nuclei while it retained uniform, circular staining in mock-infected cells. Time course infection experiments indicated that nucleolin initiated its transition from uniform to dispersed structures between 2 and 4 hpi. Comparison of Hoechst stained nuclei showed bright anti-nucleolin staining localized to regions of marginalized chromatin. These effects required de novo infected cell protein synthesis. A portion of VP22 detected in nuclei at 4 and 6 hpi localized to these areas of altered nucleolin and marginalized chromatin. VP22 was excluded from viral replication compartments containing the viral regulatory protein ICP22. Finally, altered nucleolin and marginalized chromatin were detected with a VP22-null virus, indicating that VP22 was not responsible for these nuclear architecture alterations. Thus, we conclude that nuclear VP22 targets unique subnuclear structures early (< 6 hpi) during HSV-1 infection.
Keywords: Herpes simplex virus 1, VP22, nucleoli, nucleolin, chromatin
1. Introduction
Herpes simplex virus (HSV) infections occur worldwide and the virus is only transmitted between humans {reviewed in (Roizman and Knipe, 2001)}. Once HSV has infected an individual, it will remain in their body for life, hidden in a dormant state inside the trigeminal ganglion. Most of what is known about the replicative cycle of HSV comes from work in tissue culture systems using herpes simplex virus 1 (HSV-1). The expression of HSV-1 genes is coordinately regulated and ordered in a sequential cascade (Honess and Roizman, 1974; Honess and Roizman, 1975). The first class of genes which are expressed after infection are the IE genes. These genes do not require de novo viral protein synthesis for their expression (Costanzo et al., 1977) and are induced by a structural component of the virion, VP16 (Batterson and Roizman, 1983; Campbell et al., 1984; O’Hare and Goding, 1988). Expression of the later classes of genes, E and L, requires functional IE proteins, especially the infected cell protein number 4 (ICP4) (DeLuca et al., 1985; DeLuca and Schaffer, 1985; Dixon and Schaffer, 1980; Gelman and Silverstein, 1987; O’Hare and Goding, 1988; Preston, 1979a; Preston, 1979b). The molecular mechanism of HSV-1 tegument and envelope assembly is poorly understood. Most of the major tegument proteins, especially VP22, exhibit a nuclear distribution late in infection (Albright and Jenkins, 1993; Chang and Roizman, 1993; Conley et al., 1981; Cunningham et al., 2000; Daikoku et al., 1998; Daikoku et al., 1997; MacLean et al., 1987; Morrison et al., 1998; Naldinho-Souto et al., 2006; Pomeranz and Blaho, 1999; Taus et al., 1998) and there is general agreement that primary envelopment occurs as the capsid exits the nucleus (Browne et al., 1996; Gershon et al., 1994; Granzow et al., 1997; Stackpole, 1969; Whiteley et al., 1999) {reviewed in (Enquist et al., 1998; Mettenleiter, 2002)}.
VP22 is one of the most abundant tegument proteins in HSV-1 virus particles (Heine et al., 1974). However, the role of VP22 during HSV-1 infection remains unclear. Full-length VP22 is required for efficient viral cell-to-cell spread during infection in animals and cultured cells (Duffy et al., 2006; Pomeranz and Blaho, 2000). VP22 of HSV-1 possesses two nuclear localization signals (NLS) (Kotsakis et al., 2001) and the protein accumulates to high levels in the nuclei of either infected cells (Pomeranz and Blaho, 1999) or VP22-expressing stable cell lines (Pomeranz and Blaho, 2000). While characterizing a fusion of GFP with an NLS of VP22, we observed that this chimera was not excluded from nucleoli in transfected cells. The nucleolus is a highly structured, specialized organelle that is the site of rRNA synthesis and the assembly/processing of preribosomal ribonucleoprotein particles which disperses and reforms during mitosis {reviewed in (Melese and Xue, 1995; Pederson and Politz, 2000)}. Very little is known about nucleoli during HSV-1 infection (Roizman and Furlong, 1974; Roizman and Knipe, 2001). The initial goal of this study was to determine whether VP22 associates with nucleoli. We developed an indirect immunofluorescence system to visualize nucleolin during the course of infection. We observed that nucleolin was redistributed in HSV-1-infected cells. Nucleolin alteration occurred between 2–4 hpi and correlated with chromatin marginalization. VP22 targeted these areas of dispersed nucleolin and chromatin. Although newly synthesized infected cell protein was required, VP22 was not necessary for nucleolin alteration.
2. Materials and methods
2.1 Cells and viruses
African green monkey kidney (Vero) cells were obtained from the American Type Culture Collection and maintained in Dulbecco’s modified Eagle medium supplemented with 5% fetal bovine serum. VP22 expressing Vero (V49) cells were described previously and were maintained in the presence of 0.5 mg/ml geneticin (Pomeranz and Blaho, 2000). The HSV-1(F) virus is the prototype wild type strain used in our studies. HSV-1(Bac-F) virus represents HSV-1(F) virus (Duffy et al., 2006) generated from a bacterial artificial chromosome (Bac) construct (Tanaka et al., 2003). HSV-1(Bac-ΔVP22) virus is a VP22-null mutant and HSV-1(Bac-ΔVP22Repair) virus has the VP22 gene (UL 49) engineered back in (Duffy et al., 2006). All Bac-ΔVP22 stocks were grown on V49 cells, as described (Duffy et al., 2006). To obtain virus stocks, subconfluent monolayer Vero cells (~ 3 × 10 6 cells) were inoculated at a multiplicity of infection (MOI) of 0.01 for 2 hours at 37°C in 199V (Life Technologies) supplemented with 1% newborn calf serum or 2% newborn calf serum. The inoculum was then removed, fresh Dulbecco’s modified Eagle medium supplemented with 5% new born calf serum was added, and cells were incubated at 37° C in 5% CO2 for 2–3 days. Virus stocks were prepared once the infection reached a cytopathic effect of 100%, the virus titers on Vero and V49 cells were determined, and aliquots were stored at −80° C. All the MOIs were derived from the number of PFU on Vero and V49 cells.
2.2 DNA transfections
Plasmid pJB177 contains the last 15 a.a. of VP22 containing the pat4 NLS fused to GFP (Pomeranz and Blaho, 2000), termed VP22pat4GFP. pJB49 expresses a nonfused GFP only and was used as a control (Blouin and Blaho, 2001). DNAs of both plasmids were used in a modified CaCl2 transfection protocol to transfect Vero cells (Pomeranz and Blaho, 2000). Approximately 5.0 μg of plasmid DNA was combined with sterile distilled H2O to a total volume of 250 μl. Then, 250 μl of 280 mM NaCl, 10 mM KCl, 1.5 mM Na2HPO4, 12 mM glucose, 50 mM HEPES, pH 5.95 was added and mixed prior to addition of 25 μl of 2.5 M CaCl2, followed by incubation at 25°C for 15 min. This mixture was added dropwise to 25 cm2 flasks of Vero cells at approximately 70% confluency which were passaged the previous day and maintained in 5 ml of 5% NBCS. After a 3 h incubation at 37°C in 5% CO2, the cells were shocked with DMEM containing 10% glycerol for 90 s. Glycerol-containing medium was quickly aspirated and cells were washed once in DMEM and incubated for 24 h in 5% NBCS. Cells were examined at 24 h post transfection.
2.3 Synchronized infections
Synchronized infections are defined as uniform staining in all cells in a microscopic field at a given time post infection, as determined by indirect immunofluorescence with specific antibodies for unique HSV-1 polypeptides (Pomeranz and Blaho, 1999). Vero cells were grown the day before infection in six well dishes containing 22-mm2 coverslips for indirect immunofluorescence. Cells were incubated on ice at 4° C for 20 min prior to the addition of virus. After the virus was allowed to adsorb for 1 h, the cells were removed from the ice, 37°C medium was added immediately, and cells were returned to a 37°C incubator. Induction of synchronous infection by adsorption of the inoculum at 4°C is routine and has little or no effect on cells in culture (Pomeranz and Blaho, 1999).
2.4 Chemical treatment during infection
To inhibit de novo protein synthesis, cycloheximide (CHX, Sigma) was added to the medium 30 min prior to synchronous infection at a final concentration of 10 μg/ml of medium. This concentration was previously shown to be sufficient to completely block viral protein synthesis in HSV-1-infected Vero cells (Nguyen et al., 2005). CHX was maintained in the medium at this concentration until the infection was terminated.
2.5 Immunological reagents
RGST49 is a rabbit polyclonal antibody specific for VP22 (Pomeranz and Blaho, 1999) and was used at a dilution of 1:500 for indirect immunofluorescence. Monoclonal antibody (C-23 D-6), specific for nucleolin, was used at a dilution of 1:500 and obtained from Santa Cruz Biotechnology, Inc. RGST22 is a polyclonal antibody specific for ICP22 (Blaho et al., 1997) and was used at a dilution of 1:500. All antibodies dilutions were made in 1% bovine serum albumin (BSA, Sigma). Fluorescein isothiocyanate (FITC)-conjugated anti-rabbit immunoglobulin G (IgG heavy plus light chains) and Alexa Fluor® 568 goat anti-mouse IgG was purchased from Molecular Probes, Inc. and used at a dilution of 1:500 in 1% BSA.
2.6 Indirect immunofluorescence and microscopy
Vero and V49 cells were prepared for indirect immunofluorescence as previously described (Pomeranz and Blaho, 1999). Cells were rinsed twice with phosphate buffered saline (PBS), fixed in 2.5% methanol-free formaldehyde (Polysciences, Inc.) for 20 min at room temperature, rinsed twice again in PBS, and permeablized with either 100% acetone at −20°C or 2% Triton X-100 at 4°C for 4 min. Infected cells were incubated for at least 10 h in 1% BSA supplemented with 10 μg of pooled human Ig, which was shown by us to be sufficient to neutralize Fc binding by the viral glycoprotein E (gE) and gI (Pomeranz and Blaho, 1999). The primary antibodies used for indirect immunofluorescence studies were diluted as described above and added for 1h. After extensive rinsing with PBS, the appropriate secondary antibody was added and incubated for 45 min. Finally, the cells were preserved with Prolong Antifade (Molecular Probes) as an anti-bleaching agent, mounted on a fresh glass slide, and sealed with nail polish. Hoechst 33258 DNA dye (Sigma) was used for staining nuclei at a final concentration of 0.05 μg/ml. For live nucleoli staining, SYTO RNASelect (Molecular Probes), a cell-permanent stain selective for RNA, was added to the medium for 30 min at 37°C, removed, and replaced by Hoechst 33258 containing medium for another 30 min prior to microscopic examination. All cells were visualized (magnification, 60× or 100×) with an Olympus 1X70/IXFLA inverted fluorescence microscope and images were acquired with a Retiga2000R camera from Q Imaging Co. connected to a Dell PC workstation.
3. Results
3.1 Transfected GFP excludes intracellular nuclear structures while VP22pat4GFP does not
Several previous publications have suggested that the carboxy-terminus of VP22 may facilitate the nuclear translocation of fused GFP in transient expression assays (Brignati et al., 2003; Elliott and O’Hare, 1997; Fang et al., 1998). We previously reported that this VP22 region possesses a pat4 NLS (Kotsakis et al., 2001), Thus, we set out to characterize the behavior of a GFP fusion possessing the VP22 pat4 motif (VP22pat4GFP) inside live cells in absence of other viral proteins. Vero cells were transfected with either plasmid pJB177, expressing VP22pat4GFP, or control plasmid pJB49 which expresses GFP alone. Cells were stained with Hoechst dye at 24 h post transfection and visualized live by fluorescence microscopy as described in Material and Methods. The results (Fig. 1A) were as follows.
Fig. 1.
Schematic representation of the recombinant VP22pat4GFP construct used (A). Line 1 - the HSV-1 (F) genome with the unique long (UL), unique short (US), and terminal repeat segments (a, b, c, a′, b′, and c′) indicated. Coordinates of the BamF fragment are indicated. Line 2 - gene UL49 encoding full length VP22 showing approximate locations of pat4 and pat7 nuclear localization signal motifs. Solid arrowhead shows the direction of transcription. Line 3 - orientation of GFP indicating that a portion of UL49 remains at its carboxy terminus and the potential for a GFP carboxy-terminal VP22 (GFP-CTermVP22) chimera expressed by pJB177. Line 4 -sequence of the in frame amino acids of VP22pat4GFP between GFP and 15 aa of the VP22 carboxy terminus, including linker residues. The pat4 nuclear localization signal motifs is shown. Transfected VP22pat4GFP does not spare subnuclear circular structures (B). Vero cells were transfected with either plasmid pJB177 expressing VP22pat4-GFP or pJB49 expressing GFP, nuclei were stained with Hoechst dye, and cells were visualized live by fluorescence microscopy. Merged images (overlay) were prepared to emphasize nuclear staining. Arrows mark staining patterns of GFP sparing of intranuclear structures in representative cells. Panels were enlarged for better visualization. Subnuclear structures bind RNA dye (C). Live Vero cells were stained with Hoechst and SYTO RNASelect dyes and cells were visualized by fluorescence microscopy. Arrows mark consistent SYTO staining with a Hoechst void pattern. Subnuclear structures contain nucleolin (D). Vero cells were fixed/permeablized, stained with antibody specific for nucleolin, and cells were visualized by fluorescence microscopy. Arrows mark nucleolin staining patterns. Phase contrast (Phase) and merged (overlay) images are also shown. Magnification, 60×.
VP22pat4GFP and GFP transiently expressed in Vero cells were detected in both the cytoplasm and nuclei. It appeared that there was a slight increase in the amount of nuclear VP22pat4GFP compared to GFP, suggesting that pat4 alone might serve as an NLS. This general trend was highly reproducible over numerous independent transfections using multiple conditions (data not shown). The VP22pat4GFP linker contains Ser-Gly-Leu-Arg-Ser (Fig. 1) and, while this does not match any known nuclear or nucleolar localization motif, we cannot exclude its potential effect on the phenotype of this chimera. Close inspection of the cells expressing GFP from plasmid pJB49 revealed prominent exclusions of fluorescence in circular intranuclear structures in some cells. In contrast, VP22pat4GFP nuclear fluorescence appeared more evenly distributed and these exclusions were not observed. These differences were clearly visualized in the overlay images (compare the expanded Fig. 1A Panels). These results indicate that VP22pat4GFP and GFP differ in their subnuclear localizations.
In a second series of studies, we characterized what these circular, intranuclear structures might be. Based on the size and shape of the structures, we predicted that they might be nucleoli. The nucleolus is a highly structured and specialized organelle that functions as the site of both the synthesis of rRNA and the assembly and processing of preribosomal ribonucleoprotein particles (Melese and Xue, 1995). The nucleolin protein represents approximately 10% of the total nucleolar protein content (Hiscox, 2002). Uninfected Vero cells were stained with Hoechst and SYTO RNA, a dye specific for nucleolar RNA, for 30 min and visualized live by fluorescence and phase contrast microscopy, as described in Materials and Methods. Finally, the cells were fixed for indirect immunofluorescence using anti-nucleolin antibody. The results (Fig. 1B and 1C) were as follows.
Live Hoechst staining showed uniform nuclear signals but slightly-dark, round structures were visualized within them (Fig. 1B). While visualization of “negative staining” of RNA-rich, DNA-poor nucleoli in live images of Hoechst stained nuclei is technically challenging, it is observed. These slight, dark regions corresponded with the raised spheres observed in the phase contrast image. While SYTO exhibited a faint signal throughout the nucleus, intense staining was restricted to these spherical structures. The merged image showed that SYTO staining occurred in the structures in which the Hoechst staining was reduced. These live images indicate that the subnuclear structures are rich in RNA, likely devoid of DNA, and can be visualized in phase contrast. In fixed cells stained with the anti-nucleolin antibody, spherical structures with slightly darker centers were observed which corresponded directly with the subnuclear structures observed in phase (Fig. 1C). This was readily observed in the merged image. Based on these findings, we conclude that the spherical subnuclear structures that we observed above (Fig. 1A) were nucleoli. In subsequent studies, we use nucleolin staining as our marker for nucleoli.
3.2 VP22 derived from VP22-expressing V49 cells surrounds nucleolin
The results presented above suggest that the VP22 pat4 NLS motif prevented GFP from being excluded from nucleoli in transfected Vero cells. The goal of this experiment was to determine whether the VP22 protein itself associates with nucleoli. First, we validated our anti-nucleolin antibody staining methodology. Vero were stained with Hoechst, then fixed with formaldehyde and permeablized with either acetone or Triton X-100 as described in Materials and Methods. Indirect immunofluorescence was performed using anti-nucleolin antibody and cells were visualized by fluorescence microscopy. Representative images are shown in Fig. 2A. Two staining patterns were observed. Bright ring-like structures were observed with both permeablization conditions, consistent with results in Fig. 1C. In addition, bright non-ring patterns were observed which were sometimes smaller in size. Since most non-ring structures existed in pairs, they may represent disassembling/reassembling nucleoli due to cell division (Pederson and Politz, 2000). We attempted to quantitate the ratio of the rings to non-rings by counting approximately 100 cells for each condition. With acetone, 43% of nuclei had circular nucleolin staining (rings) while with Triton X-100, 40% were rings. These results indicate that the circular, ring anti-nucleolin staining was not an artifact of acetone permeablization. All subsequent studies used paraformaldehyde/acetone treatment conditions.
Fig. 2.
The circular nucleolin staining pattern is independent of fixing conditions (A). Vero cells were fixed with 2.5% paraformaldehyde and then permeablized using two different conditions; either acetone (−20°C) or Triton X-100 (4°C). Phase contrast (Phase) and merged (overlay) images are shown. VP22 surrounds nucleolin in VP22-expressing V49 cells (B). Vero and V49 cells were immunostained with antibodies specific for VP22 and nucleolin. Arrows mark a portion of VP22 staining in V49 cells which is circular and resembles that of nucleolin. Nuclei were visualized following Hoechst staining (A and B). Magnification, 100×.
Next (Fig. 2B), VP22-expressing Vero (V49) cells were double stained with anti-VP22 and anti-nucleolin antibodies as described in Materials and Methods. As expected, no VP22 staining was detected in Vero cells and circular nucleolin staining was observed. In V49 cells, we observed that while VP22 was present in nuclei (Pomeranz and Blaho, 2000), a portion of it appeared as a ring with in the nuclei. This VP22 ring-like staining pattern might even colocalize with nucleolin staining. When we assessed the patterns of nucleolin staining in V49 cells, we found that 93% were rings. The remaining 7% of nuclei consistently had either (i) a diffuse, nonstructured nucleolin staining, or (ii) a very faint VP22 staining pattern. An example of the diffuse nucleolin pattern can be observed in the representative image in Fig. 2B. Taken together, these results suggest that VP22 may associate with nucleolin in the absence of other viral proteins. That the vast majority of VP22-expressing V49 cells have circular, ring nucleolin patterns suggests that VP22 may influence the physiology of these structures.
3.3 HSV-1 infection alters nucleolin
Having now defined the behavior of nucleolin in Vero and V49 cells, we next characterized its structures during the course of productive viral infection. Vero cells were synchronously mock-infected or infected with HSV-1(F) (MOI = 15 PFU/cell) and at 13 and 24 hpi, the cells were stained with Hoechst and visualized by indirect immunofluorescence using anti-nucleolin and -VP22 antibodies. The results (Fig. 3) were as follows.
Fig. 3.

Altered nucleolin staining observed at late infection time points. Vero cells were synchronously mock- or HSV-1(F)-infected. At 13 and 24 hpi, cells were fixed/permeablized and immunostained with antibodies specific for VP22 and nucleolin. Nuclei were visualized following Hoechst staining. Magnification, 100×.
As in Fig. 1, mock-infected cells at both times showed ring-like nucleolin staining. VP22 predominantly localized in nuclei at both 13 and 24 hpi. However, at both times, the anti-nucleolin immune reactivity in the infected cells was totally dispersed throughout the nuclei. Some infected cells had subnuclear regions of bright nucleolin staining which seems to coincide with foci of bright Hoechst staining. Importantly, this altered nucleolin pattern was different from what we defined as “non-ring” in the uninfected Vero cells in Fig. 1. Thus, at late times post infection, productive HSV-1 replication results in a change in nucleolin. This, our specific indirect immunofluorescence study corroborates earlier electron microscopic analyses (Roizman and Furlong, 1974; Roizman and Knipe, 2001). From these experiments, we can conclude that this change in nuclear structures begins earlier than 13 hpi.
3.4 Nucleolin alteration occurs between two and four hours post infection
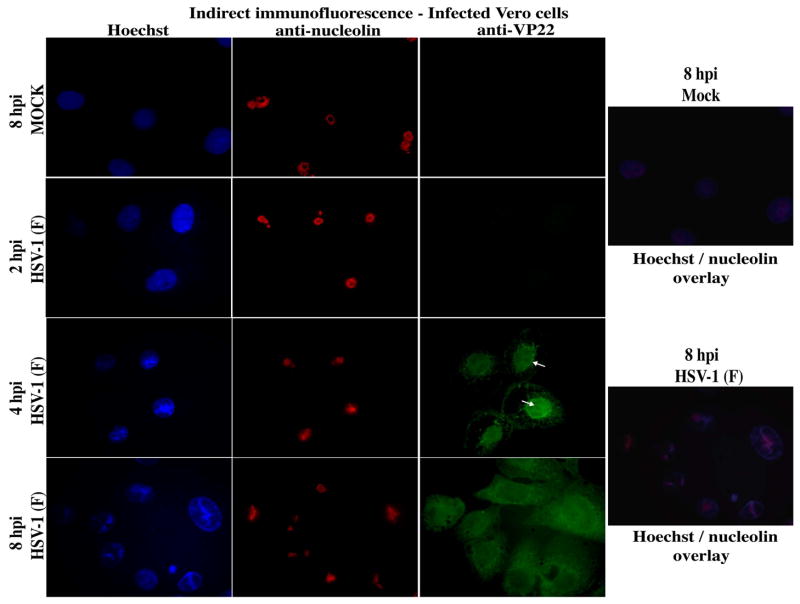
To further define the kinetics of the HSV-1-dependent nucleolin alteration, we performed similar indirect immunofluorescence analyses in HSV-1(F)-infected Vero cells at 2, 4, and 8 hpi (Fig. 4). At 8 hpi, VP22 was diffuse as expected (Pomeranz and Blaho, 1999), but the anti-nucleolin immune reactivity was not completely dispersed throughout the cell. It was, however, significantly altered compared to the ring-like structures observed in mock-infected cells. Again, the altered, bright nucleolin at 8 hpi seemed to align with areas of bright Hoechst staining. This was readily seen in the infected overlay images where red nucleolin is confined to areas of bright (condensed) white-blue Hoechst staining; mock shows only red nucleolin rings in evenly-distributed blue nuclei. At 4 hpi, VP22 was detected in nuclei and the anti-nucleolin staining was altered as observed at 8 hpi. Close inspection of the infected cells at 4 hpi seemed to indicate that a subset of nuclear VP22 “surrounded” the regions of bright nucleolin staining. Although VP22 appears to be mostly present throughout the nucleus, as we have reported previously (Kotsakis et al., 2001; Pomeranz and Blaho, 1999; Yedowitz et al., 2005), regions exist in which there is slightly more intense VP22 staining over this background. In contrast, VP22 was undetectable at 2 hpi and the anti-nucleolin staining was uniform and circular, as in mock-infected cells. From these experiments, we conclude that nucleolin initiates its transition from uniform to dispersed structures between 2 and 4 hpi.
Fig. 4.
Altered nucleolin staining detected at 4 h post infection. Vero cells were synchronously mock- or HSV-1(F)-infected. At 2, 4, and 8 hpi, cells were fixed/permeablized and immunostained with antibodies specific for VP22 and nucleolin at 4 hpi. Nuclei were visualized following Hoechst staining. Overlay images of Hoechst and nucleolin at 8 hpi show colocalization of altered nucleolin with marginalized chromatin in infected cells. Arrows mark a portion of VP22 staining which is circular and surrounds altered nucleolin. Magnification, 100×.
3.5 Nuclear VP22 targets areas of altered nucleolin and condensed chromatin
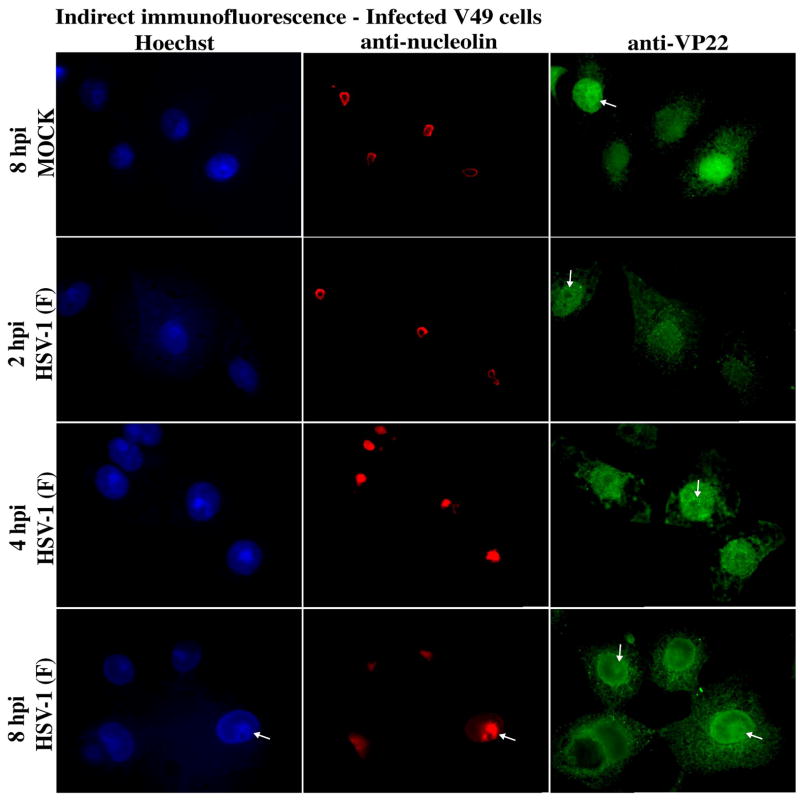
The results above (Fig. 1 and 4) show that a portion of nuclear VP22 in V49 cells or in infected Vero cells at 4 hpi was observed at/near nucleolin. Here, we asked whether infecting VP22-expressing V49 cells might alter the kinetics of HSV-1-dependent nucleolin alteration. V49 cells were synchronously infected and at 2, 4, and 8 hpi, stained with Hoechst and visualized by indirect immunofluorescence using anti-nucleolin and -VP22 antibodies (Fig. 5). As above, we observed uniform ring-like anti-nucleolin staining at 2 hpi which resembled the mock-infected cells at 8 hpi. Consistent with Fig. 2, VP22 from the V49 cells in mock surrounds nucleolin. Again, nucleolin was altered at 4 and 8 hpi. At 4 hpi, a portion of VP22 surrounded the altered nucleolin. Thus, the presence of preexisting VP22 in nuclei in the V49 cells did not appear to affect the kinetics of nucleolin alteration. At 8 hpi, nuclear VP22 appeared to colocalize with the areas of bright nucleolin staining. It is of interest to note that almost all VP22 (both endogenous V49- and de novo virus-derived) in the infected V49 cells targeted nucleolin at 8 hpi (compare nuclear VP22 in mock and infected cells), suggesting that this association maybe an intrinsic property of VP22. However, we acknowledge that some VP22 does exist in other areas, including the perinuclear area. Confirmation of these observations by confocal microscopy should clarify the significance of this finding using these experimental conditions. We consistently observed in this (Fig. 2) and previous studies (Blouin and Blaho, 2001; Pomeranz and Blaho, 2000; Yedowitz et al., 2005) that the staining pattern of VP22 in V49 cells is homogeneous and remains stable at different times post seeding. Increased levels of VP22 staining at 4 and 8 hpi results from de novo VP22 expression from virus. Thus, the difference between the VP22 staining at 8 hpi between infected and mock illustrates that the nuclear structural changes induced by the virus are able to sequester all VP22 and influence its subnuclear localization.
Fig. 5.
Altered nucleolin in infected VP22 expressing V49 cells. V49 cells were synchronously mock- or HSV-1(F)-infected. At 2, 4, and 8 hpi, cells were fixed/permeablized and immunostained with antibodies specific for VP22 and nucleolin. Nuclear chromatin was visualized following Hoechst staining. Arrows mark a portion of VP22 staining which surrounds nucleolin; at 4 and 8 hpi VP22 targets altered nucleolin and marginalized chromatin. Magnification, 100×.
3.6 VP22 is not required for the alteration of nucleolin during HSV-1 infection
The studies above define the behavior of nucleolin during the course of productive HSV-1 infection. Our observations may be summarized as follows. Prior to 2 hpi, nucleolin staining is observed as bright rings, indicative of intact nucleoli. In uninfected VP22-expressing cells, a portion of VP22 surrounds nucleolin. At 4 hpi, nucleolin becomes altered and some VP22 can be observed surrounding these structures. The altered nucleolin observed between 4 and 8 hpi is characterized by bright, non-ring structures that colocalize with condensed chromatin. A portion of nuclear VP22 targets these regions. At late infection times (> 12 hpi), nucleolin is completely dispersed throughout nuclei and this represents phenotypes reported earlier using electron microscopy. In the next series of experiments, our goal was to determine whether VP22 plays a role in the nucleolin alteration process in HSV-1 infected cells.
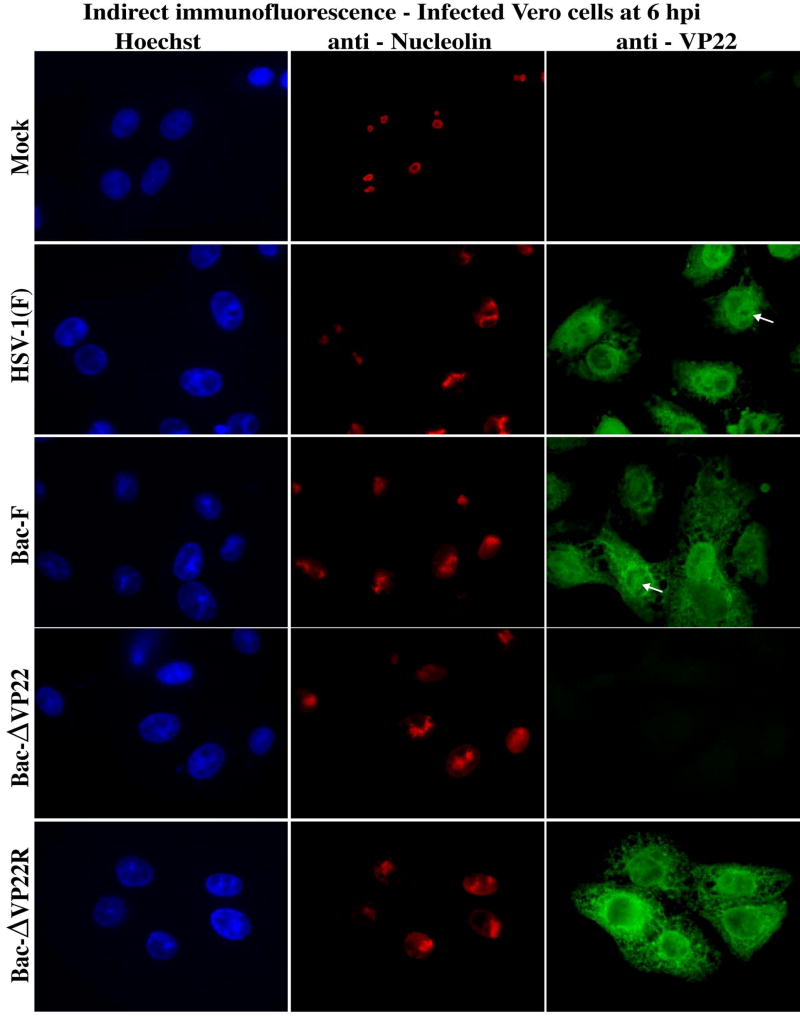
Vero cells were mock-, Bac-F-, and Bac-ΔVP22-infected and analyzed at 6 hpi by indirect immunofluorescence with anti-VP22 and anti-nucleolin antibodies (Fig. 6) as described in Material and Methods. As expected (Pomeranz and Blaho, 1999), VP22 was diffuse in Bac-F-infected cells at this time point. With both Bac-F and Bac-ΔVP22, the anti-nucleolin immune reactivity was altered compared with the mock-infected cells. We also observed areas of marginalized chromatin in the infected cells at this time. Based on this experiment, it appears that VP22 is not required for the alteration of nucleolin.
Fig. 6.
Alteration of subnuclear structures is not dependent on VP22. Vero cells were mock- or HSV-1(F), Bac-F-, and Bac-ΔVP22-infected. At 6 hpi, cells were fixed/permeablized and immunostained with antibodies specific for VP22 and nucleolin. Nuclei were visualized following Hoechst staining. Magnification, 100×.
We extended these studies to include the viral repair of Bac-ΔVP22. In addition, we used an ICP22 antibody as a marker for infection (Fig. 7 and 8). Again, the alteration of nucleolin occurred both in the presence and absence of VP22 in infected Vero cells at 6 hpi (Fig. 7 and 8), while mock-infected Vero cells retained circular nucleolin staining. A portion of nuclear VP22 seemed to target areas of marginalized chromatin/altered nucleolin in the infected cells (Fig. 7). In contrast, bright nuclear anti-ICP22 staining excluded regions of altered nucleolin and marginalized chromatin (Fig. 8). At 2 hpi, VP22 was undetectable and cells retained uniform circular anti-nucleolin staining, as with mock-infected cells (data not shown). Faint ICP22 staining was observed at 2 hpi (data not shown). It is interesting to note that the VP22 in the wild type HSV-1 strains does not surround nucleolin at 6 hpi, implying that this feature of VP22 either occurs in the absence of other viral proteins (Fig. 2) or at infection times prior to 6 hpi (Fig. 4 and 5). From these experiments, we can conclude that VP22 is not required for this alteration in the nuclear architecture. VP22 appears to be excluded from nuclear subdomains containing the viral regulatory protein ICP22, which likely represent viral replication compartments (Leopardi et al., 1997; Pomeranz and Blaho, 1999). This result is consistent with our earlier study that directly compared VP22 and ICP4 staining in the same infected cells by double-label indirect immunofluorescence and showed that these proteins do not colocalize (Pomeranz and Blaho, 1999). It is unlikely that VP22 targeting to marginalized chromatin and altered nucleolin is pure coincidence. This finding may imply an unanticipated role for VP22 in the primary nuclear envelopment of tegument and capsids, thus providing a possible link between replication compartments and assemblons (Simpson-Holley et al., 2005).
Fig. 7.
Alteration of subnuclear structures is not dependent on VP22. Vero cells were synchronously mock- or HSV-1(F), Bac-F-, Bac-ΔVP22-, and Bac-ΔVP22Repair-infected. At 6 hpi, cells were fixed/permeablized and immunostained with antibodies specific for VP22 and nucleolin. Nuclei were visualized following Hoechst staining. Arrows mark a portion of VP22 staining which targets altered nucleolin and marginalized chromatin. Magnification, 100×.
Fig. 8.
Alteration of subnuclear structures is not dependent on VP22. Vero cells were synchronously mock- or HSV-1(F), Bac-F, Bac-ΔVP22-, and Bac-ΔVP22Repair-infected. At 6 hpi, cells were fixed/permeablized and immunostained with antibodies specific for ICP22 and nucleolin. Nuclei were visualized following Hoechst staining. Arrows mark ICP22 exclusion of nucleolin. Magnification, 100×.
3.7 Nucleolin alteration requires de novo infected cell protein synthesis
In the final portion of this study, we set out to determine whether incoming virion protein was responsible for the alteration of nucleolin observed above. Vero cells were mock-, HSV-1(F)- and Bac-ΔVP22-infected in the presence and absence of CHX. At 6 hpi, cells were stained with Hoechst and analyzed by indirect immunofluorescence using anti-VP22 (data not shown), anti-ICP22 and anti-nucleolin antibodies as described in Materials and Methods. ICP22 was used as a marker for viral infection. The results (Fig. 9) were as follows.
Fig. 9.
Alteration of subnuclear structures requires de novo infected cell protein synthesis. Vero cells were synchronously mock- or HSV-1(F), Bac-F-, Bac-ΔVP22-, and Bac-ΔVP22Repair-infected in the presence (plus) and absence (minus) of CHX. At 6 hpi, cells were fixed/permeablized and immunostained with antibodies specific for ICP22 and nucleolin. Nuclei were visualized following Hoechst staining. Merged images (overlay) were prepared to emphasize staining patterns. Arrows mark ICP22 exclusion of nucleolin. Magnification, 100×.
In mock-infected cells, with and without CHX, no ICP22 staining was observed, as expected, and circular nucleolin staining predominated. As above (Fig. 2), nucleoli had reduced Hoechst staining and this was readily observed in the merged images. In both HSV-1(F)- and Bac-ΔVP22-infected cells without CHX, nucleolin was altered, as above (Fig. 7 and 8). In each case, ICP22 staining was nuclear and globular, with clearly defined areas devoid of immune reactivity. These ICP22-containing regions represent viral replication compartments. Inspection of the merged images indicates that nucleolin staining was excluded from the replication compartments. In the overlays, red nucleolin does not colocalize with green ICP22. When the infected cells were treated with CHX, no ICP22 staining was observed. Under these infection conditions, nucleolin had a ring-like staining pattern similar to that observed with mock-infected cells.
In our earlier characterizations of nucleolin (Fig. 2), we observed that uninfected cell structures were approximately equally divided between ring and non-ring structures. The presence of VP22 in cells shifted this balance to almost all rings. We performed a quantitation study similar to that of Fig. 2 with these CHX treated cells. As expected, 50% of untreated mock-infected cells were rings. Addition of CHX to mock resulted in 75% rings. This finding supports a model in which non-ring nucleolin in uninfected cells arise due to disassembling/reassembling nucleoli and this process likely requires new protein synthesis for optimal effect. Also as expected, the infected nuclei without CHX had completely altered nucleolin (0% rings). Addition of CHX to HSV-1(F) infection increased the nucleolin patterns to 92% rings. The presence to CHX in the Bac-ΔVP22-infected cells yielded 87% rings. Based on these results, we conclude the following. Newly synthesized infected cell protein is required for the cellular nucleolin to undergo its alteration. The altered nucleolin is excluded from regions containing the HSV-1 replication compartments.
4. Discussion
During the course of our transfection studies designed to define the function of the major tegument protein VP22 pat4 NLS, we observed that fusion of this portion of VP22 precluded GFP sparing of nucleoli in live cells. The subsequent purpose of this study was to determine whether VP22 associates with nucleoli. We defined the kinetics of nucleolar alteration during HSV-1 infection and showed that VP22 is not required for these changes in subnuclear architecture. Our key findings may be summarized as follows.
4.1 Nucleolar dynamics are influenced by HSV-1 infection during the course of productive infection
At least four phenotypes exist which we define based on the patterns of nucleolin immune reactivity. In uninfected cells, nucleolin is evenly distributed between rings and bright dense non-ring structures. At early (< 4 hpi) infection times, rings predominate. Between 4 and 8 hpi, nucleolin becomes altered. By 13 hpi, nucleolin becomes diffuse throughout nuclei and remains as such at 24 hpi. At least two novel findings were observed regarding altered nucleolin between 4 and 8 hpi. First, nucleolin is excluded from viral replication compartments which contain ICP22. Second, altered nucleolin appears to localize to specific subdomains of marginalized chromatin. The kinetics of nuclear expansion during HSV-1 infection was recently documented (Simpson-Holley et al., 2005). These findings suggest that viral replication compartment formation may be responsible for the chromatin marginalization observed. A recent study has shown the involvement of the HSV-1 UL24 protein in the dispersal of nucleolin (Lymberopoulos and Pearson, 2007). It remains unclear as to whether the alteration of nucleolin is an active process or a secondary consequence of replication compartment formation and/or chromatin marginalization.
4.2 Newly synthesized infected cell proteins are the responsible for the alteration in nucleolin
This conclusion is based on the finding that during infection in the presence of CHX, no alteration of the nucleolin was observed. This implies that neither incoming virion factors nor the induction of cellular antiviral responders (Mossman et al., 2001; Nicholl et al., 2000) are responsible for the effect. Although HSV-1 infection in the presence of CHX results in apoptosis (Aubert and Blaho, 1999; Koyama and Adachi, 1997) in tumor cells (Nguyen et al., 2007), Vero cells require protein facilitator(s) to stimulate this cell death response (Nguyen et al., 2005). Therefore, the current data cannot eliminate a role for apoptotic cell death induction in the alteration of nucleolin during HSV-1 infection. It is important to note that the facilitators of apoptosis induction needed in Vero cells are produced at approximately 3 hpi in infected Vero cells (Nguyen et al., 2005), which exactly corresponds to the time frame in which the nucleolin alteration occurs. Current investigations are focusing on the association of apoptosis induction in this process. The key question remaining is what viral factor(s) cause these changes in the cells during a productive HSV-1 infection.
4.3 VP22 targets nucleolin
The major virion structural protein VP22 is not required for the alterations in nucleolin, inasmuch as changes in nucleolin occur during Bac-ΔVP22 infection. However, VP22 is observed to “surround” areas of nucleolin staining in either mock-infected VP22-expressing V49 cells or infected V49 cells at 2 hpi. These finding imply that a portion of nuclear VP22 targets to nucleolin. That VP22pat4GFP is not spared from nucleoli suggests that the VP22 pat4 motif may function as a nucleolar targeting signal {reviewed in (Hiscox, 2002)}. At later infection times (4–8 hpi), the majority of nuclear VP22 partitions to regions of marginalized chromatin where the altered nucleolin also resides. Our results indicating that VP22 is excluded from viral replication compartments during infection corroborates the original indirect immunofluorescence characterization of VP22 which showed its lack of colocalization with ICP4 (Pomeranz and Blaho, 1999). The most important question remaining is what might be the physiological relevance to VP22’s association with nucleolin? Our finding that the presence of VP22 in V49 cells reduces the number of non-ring nucleolin patterns seems to imply that VP22 may function to inhibit nucleolar disassembly, thus stabilizing these structures. That the number of ring-like nucleolin patterns increased to almost 100% during HSV-1(F) infection in the presence of CHX suggested that incoming virion components may play a role in the ring to non-ring dynamic. The comparison of HSV-1(F) with Bac-ΔVP22 in the presence of CHX was not dramatically different (92% versus 87%, respectively). Unfortunately, this analysis has a major technical caveat. The Bac-ΔVP22 virus is exquisitely sensitive to the acquisition of second site mutations and absolutely must be grown on complementing V49 cells {(Duffy et al., 2006) and data not shown}. Therefore, the Bac-ΔVP22 used in our studies possesses virion-derived VP22. It is conceivable that the dramatic reduction in cell to cell spreading observed with Bac-ΔVP22 and other VP22-deficit viruses (Pomeranz and Blaho, 2000) may be due to the loss of VP22 association with nucleolin.
These findings raise the significant issue of what role does nucleoli play in HSV-1 infected cells. The nucleolus is necessary for protein translation {reviewed in (Melese and Xue, 1995; Pederson and Politz, 2000)}. It has been proposed that the herpesvirus saimiri ORF57 protein uses nucleoli to export intronless viral messages (Boyne and Whitehouse, 2006; Williams et al., 2005). The HSV-1 ICP27, ICP0, γ134.5, and US11 proteins were reported to possess nucleolar targeting sequences (Cheng et al., 2002; MacLean et al., 1987; Mears et al., 1995; Morency et al., 2005) but the implications of these are unknown. Emerging evidence now argues that nucleoli act as a cellular stress sensor which functions to regulate p53 and cell cycle arrest (Mayer et al., 2005; Rubbi and Milner, 2003). The fate of p53 during HSV infection is unclear as it may be targeted for degradation by ICP0 ubiquitination but it might also be stabilized in certain cell lines (Boutell and Everett, 2003; Boutell and Everett, 2004). The interaction of VP22 with nucleoli may represent a novel way the virus regulates the cellular stress response. The development of appropriate biochemical and molecular genetic systems will be necessary to address these and other important questions regarding the physiology of nucleoli during productive HSV-1 infection.
Acknowledgments
We thank Leah Kang for expert technical cell culture assistance. These studies were supported in part by grants from the United States Public Health Service (AI 38873 to J.A.B). M.R.L. is a postbaccalaureate trainee and was supported in part by an Institutional Research Training Award (GM 064118). S.W. performed these studies as part of her Master’s diploma thesis at the University of Salzburg, Austria and was supported in part by an International Scholar Grant from the Naturwisseneschaftliche Fakultat.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albright AG, Jenkins FJ. The herpes simplex virus UL37 protein is phosphorylated in infected cells. J Virol. 1993;67(8):4842–7. doi: 10.1128/jvi.67.8.4842-4847.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert M, Blaho JA. The herpes simplex virus type 1 regulatory protein ICP27 is required for the prevention of apoptosis in infected human cells. J Virol. 1999;73(4):2803–2813. doi: 10.1128/jvi.73.4.2803-2813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batterson W, Roizman B. Characterization of the herpes simplex virion-associated factor responsible for the induction of alpha genes. J Virol. 1983;46(2):371–7. doi: 10.1128/jvi.46.2.371-377.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaho JA, Zong CS, Mortimer KA. Tyrosine phosphorylation of the herpes simplex virus type 1 regulatory protein ICP22 and a cellular protein which shares antigenic determinants with ICP22. J Virol. 1997;71(12):9828–32. doi: 10.1128/jvi.71.12.9828-9832.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blouin A, Blaho JA. Assessment of the subcellular localization of the herpes simplex type 1 virion structural protein VP22 in the absence of other viral gene products. 2001 doi: 10.1016/s0168-1702(01)00355-0. submitted. [DOI] [PubMed] [Google Scholar]
- Boutell C, Everett RD. The herpes simplex virus type 1 (HSV-1) regulatory protein ICP0 interacts with and Ubiquitinates p53. J Biol Chem. 2003;278(38):36596–602. doi: 10.1074/jbc.M300776200. [DOI] [PubMed] [Google Scholar]
- Boutell C, Everett RD. Herpes simplex virus type 1 infection induces the stabilization of p53 in a USP7- and ATM-independent manner. J Virol. 2004;78(15):8068–77. doi: 10.1128/JVI.78.15.8068-8077.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyne JR, Whitehouse A. Nucleolar trafficking is essential for nuclear export of intronless herpesvirus mRNA. Proc Natl Acad Sci U S A. 2006;103(41):15190–5. doi: 10.1073/pnas.0604890103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brignati MJ, Loomis JS, Wills JW, Courtney RJ. Membrane association of VP22, a herpes simplex virus type 1 tegument protein. J Virol. 2003;77(8):4888–98. doi: 10.1128/JVI.77.8.4888-4898.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne H, Bell S, Minson T, Wilson DW. An endoplasmic reticulum-retained herpes simplex virus glycoprotein H is absent from secreted virions: evidence for reenvelopment during egress. J Virol. 1996;70(7):4311–6. doi: 10.1128/jvi.70.7.4311-4316.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell ME, Palfreyman JW, Preston CM. Identification of herpes simplex virus DNA sequences which encode a trans-acting polypeptide responsible for stimulation of immediate early transcription. J Mol Biol. 1984;180(1):1–19. doi: 10.1016/0022-2836(84)90427-3. [DOI] [PubMed] [Google Scholar]
- Chang YE, Roizman B. The product of the UL31 gene of herpes simplex virus 1 is a nuclear phosphoprotein which partitions with the nuclear matrix. J Virol. 1993;67(11):6348–56. doi: 10.1128/jvi.67.11.6348-6356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng G, Brett ME, He B. Signals that dictate nuclear, nucleolar, and cytoplasmic shuttling of the gamma(1)34.5 protein of herpes simplex virus type 1. J Virol. 2002;76(18):9434–45. doi: 10.1128/JVI.76.18.9434-9445.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conley AJ, Knipe DM, Jones PC, Roizman B. Molecular genetics of herpes simplex virus. VII Characterization of a temperature-sensitive mutant produced by in vitro mutagenesis and defective in DNA synthesis and accumulation of gamma polypeptides. J Virol. 1981;37(1):191–206. doi: 10.1128/jvi.37.1.191-206.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo F, Campadelli-Fiume G, Foa-Tomasi L, Cassai E. Evidence that herpes simplex virus DNA is transcribed by cellular RNA polymerase B. J Virol. 1977;21(3):996–1001. doi: 10.1128/jvi.21.3.996-1001.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham C, Davison AJ, MacLean AR, Taus NS, Baines JD. Herpes simplex virus type 1 gene UL14: phenotype of a null mutant and identification of the encoded protein. J Virol. 2000;74(1):33–41. doi: 10.1128/jvi.74.1.33-41.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daikoku T, Ikenoya K, Yamada H, Goshima F, Nishiyama Y. Identification and characterization of the herpes simplex virus type 1 UL51 gene product. J Gen Virol. 1998;79(Pt 12):3027–31. doi: 10.1099/0022-1317-79-12-3027. [DOI] [PubMed] [Google Scholar]
- Daikoku T, Shibata S, Goshima F, Oshima S, Tsurumi T, Yamada H, Yamashita Y, Nishiyama Y. Purification and characterization of the protein kinase encoded by the UL13 gene of herpes simplex virus type 2. Virology. 1997;235(1):82–93. doi: 10.1006/viro.1997.8653. [DOI] [PubMed] [Google Scholar]
- DeLuca NA, McCarthy AM, Schaffer PA. Isolation and characterization of deletion mutants of herpes simplex virus type 1 in the gene encoding immediate-early regulatory protein ICP4. J Virol. 1985;56(2):558–70. doi: 10.1128/jvi.56.2.558-570.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca NA, Schaffer PA. Activation of immediate-early, early, and late promoters by temperature- sensitive and wild-type forms of herpes simplex virus type 1 protein ICP4. Mol Cell Biol. 1985;5(8):1997–208. doi: 10.1128/mcb.5.8.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon RA, Schaffer PA. Fine-structure mapping and functional analysis of temperature-sensitive mutants in the gene encoding the herpes simplex virus type 1 immediate early protein VP175. J Virol. 1980;36(1):189–203. doi: 10.1128/jvi.36.1.189-203.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy C, Lavail JH, Tauscher AN, Wills EG, Blaho JA, Baines JD. Characterization of a UL49-null mutant: VP22 of herpes simplex virus type 1 facilitates viral spread in cultured cells and the mouse cornea. J Virol. 2006;80(17):8664–75. doi: 10.1128/JVI.00498-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott G, O’Hare P. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell. 1997;88(2):223–33. doi: 10.1016/s0092-8674(00)81843-7. [DOI] [PubMed] [Google Scholar]
- Enquist LW, Husak PJ, Banfield BW, Smith GA. Infection and spread of alphaherpesviruses in the nervous system. Adv Virus Res. 1998;51:237–347. doi: 10.1016/s0065-3527(08)60787-3. [DOI] [PubMed] [Google Scholar]
- Fang B, Xu B, Kock P, Roth JA. Intercellular trafficking of VP22-GFP fusion proteins is not observed in cultured mammalian cells. Gene Therapy. 1998;5:1420–1424. doi: 10.1038/sj.gt.3300741. [DOI] [PubMed] [Google Scholar]
- Gelman IH, Silverstein S. Dissection of immediate-early gene promoters from herpes simplex virus: sequences that respond to the virus transcriptional activators. J Virol. 1987;61(10):3167–72. doi: 10.1128/jvi.61.10.3167-3172.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershon AA, Sherman DL, Zhu Z, Gabel CA, Ambron RT, Gershon MD. Intracellular transport of newly synthesized varicella-zoster virus: final envelopment in the trans-Golgi network. J Virol. 1994;68(10):6372–90. doi: 10.1128/jvi.68.10.6372-6390.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granzow H, Weiland F, Jons A, Klupp BG, Karger A, Mettenleiter TC. Ultrastructural analysis of the replication cycle of pseudorabies virus in cell culture: a reassessment. J Virol. 1997;71(3):2072–82. doi: 10.1128/jvi.71.3.2072-2082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heine JW, Honess RW, Cassai E, Roizman B. Proteins specified by herpes simplex virus. XII The virion polypeptides of type 1 strains. J Virol. 1974;14(3):640–51. doi: 10.1128/jvi.14.3.640-651.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscox JA. The nucleolus--a gateway to viral infection? Arch Virol. 2002;147(6):1077–89. doi: 10.1007/s00705-001-0792-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honess RW, Roizman B. Regulation of herpesvirus macromolecular synthesis. I Cascade regulation of the synthesis of three groups of viral proteins. J Virol. 1974;14(1):8–19. doi: 10.1128/jvi.14.1.8-19.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honess RW, Roizman B. Regulation of herpesvirus macromolecular synthesis: sequential transition of polypeptide synthesis requires functional viral polypeptides. Proc Natl Acad Sci U S A. 1975;72(4):1276–80. doi: 10.1073/pnas.72.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotsakis A, Pomeranz LE, Blouin A, Blaho JA. Microtubule reorganization during herpes simplex virus type 1 infection facilitates the nuclear localization of VP22, a virion tegument protein. J Virol. 2001;75 doi: 10.1128/JVI.75.18.8697-8711.2001. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama AH, Adachi A. Induction of apoptosis by herpes simplex virus type 1. J Gen Virol. 1997;78(Pt 11):2909–12. doi: 10.1099/0022-1317-78-11-2909. [DOI] [PubMed] [Google Scholar]
- Leopardi R, Ward PL, Ogle WO, Roizman B. Association of herpes simplex virus regulatory protein ICP22 with transcriptional complexes containing EAP, ICP4, RNA polymerase II, and viral DNA requires posttranslational modification by the U(L)13 proteinkinase. J Virol. 1997;71(2):1133–9. doi: 10.1128/jvi.71.2.1133-1139.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lymberopoulos MH, Pearson A. Involvement of UL24 in herpes-simplex-virus-1-induced dispersal of nucleolin. Virology. 2007;363(2):397–409. doi: 10.1016/j.virol.2007.01.028. [DOI] [PubMed] [Google Scholar]
- MacLean CA, Rixon FJ, Marsden HS. The products of gene US11 of herpes simplex virus type 1 are DNA-binding and localize to the nucleoli of infected cells. J Gen Virol. 1987;68(Pt 7):1921–37. doi: 10.1099/0022-1317-68-7-1921. [DOI] [PubMed] [Google Scholar]
- Mayer C, Bierhoff H, Grummt I. The nucleolus as a stress sensor: JNK2 inactivates the transcription factor TIF-IA and down-regulates rRNA synthesis. Genes Dev. 2005;19(8):933–41. doi: 10.1101/gad.333205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mears WE, Lam V, Rice SA. Identification of nuclear and nucleolar localization signals in the herpes simplex virus regulatory protein ICP27. J Virol. 1995;69(2):935–47. doi: 10.1128/jvi.69.2.935-947.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melese T, Xue Z. The nucleolus: an organelle formed by the act of building a ribosome. Curr Opin Cell Biol. 1995;7(3):319–24. doi: 10.1016/0955-0674(95)80085-9. [DOI] [PubMed] [Google Scholar]
- Mettenleiter TC. Herpesvirus assembly and egress. J Virol. 2002;76(4):1537–47. doi: 10.1128/JVI.76.4.1537-1547.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morency E, Coute Y, Thomas J, Texier P, Lomonte P. The protein ICP0 of herpes simplex virus type 1 is targeted to nucleoli of infected cells. Brief report Arch Virol. 2005;150(11):2387–95. doi: 10.1007/s00705-005-0546-5. [DOI] [PubMed] [Google Scholar]
- Morrison EE, Stevenson AJ, Wang YF, Meredith DM. Differences in the intracellular localization and fate of herpes simplex virus tegument proteins early in the infection of Vero cells [In Process Citation] J Gen Virol. 1998;79(Pt 10):2517–28. doi: 10.1099/0022-1317-79-10-2517. [DOI] [PubMed] [Google Scholar]
- Mossman KL, Macgregor PF, Rozmus JJ, Goryachev AB, Edwards AM, Smiley JR. Herpes simplex virus triggers and then disarms a host antiviral response. J Virol. 2001;75(2):750–8. doi: 10.1128/JVI.75.2.750-758.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naldinho-Souto R, Browne H, Minson T. Herpes simplex virus tegument protein VP16 is a component of primary enveloped virions. J Virol. 2006;80(5):2582–4. doi: 10.1128/JVI.80.5.2582-2584.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen ML, Kraft RM, Blaho JA. African green monkey kidney Vero cells require de novo protein synthesis for efficient herpes simplex virus 1-dependent apoptosis. Virology. 2005;336(2):274–90. doi: 10.1016/j.virol.2005.03.026. [DOI] [PubMed] [Google Scholar]
- Nguyen ML, Kraft RM, Blaho JA. Susceptibilty of cancer cells to herpes simplex virus dependent apoptosis. J Gen Virol. 2007;88 doi: 10.1099/vir.0.82868-0. in press. [DOI] [PubMed] [Google Scholar]
- Nicholl MJ, Robinson LH, Preston CM. Activation of cellular interferon-responsive genes after infection of human cells with herpes simplex virus type 1. J Gen Virol. 2000;81(Pt 9):2215–8. doi: 10.1099/0022-1317-81-9-2215. [DOI] [PubMed] [Google Scholar]
- O’Hare P, Goding CR. Herpes simplex virus regulatory elements and the immunoglobulin octamer domain bind a common factor and are both targets for virion transactivation. Cell. 1988;52(3):435–45. doi: 10.1016/s0092-8674(88)80036-9. [DOI] [PubMed] [Google Scholar]
- Pederson T, Politz JC. The nucleolus and the four ribonucleoproteins of translation. J Cell Biol. 2000;148(6):1091–5. doi: 10.1083/jcb.148.6.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomeranz LE, Blaho JA. Modified VP22 localized to the cell nucleus during synchronized herpes simplex virus type 1 infection. J Virol. 1999;73:6769–6781. doi: 10.1128/jvi.73.8.6769-6781.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomeranz LE, Blaho JA. Assembly of infectious herpes simplex type 1 virions in the absence of full length VP22. J Virol. 2000;74(21):10041–10054. doi: 10.1128/jvi.74.21.10041-10054.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston CM. Abnormal properties of an immediate early polypeptide in cells infected with the herpes simplex virus type 1 mutant tsK. J Virol. 1979a;32(2):357–69. doi: 10.1128/jvi.32.2.357-369.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston CM. Control of herpes simplex virus type 1 mRNA synthesis in cells infected with wild-type virus or the temperature-sensitive mutant tsK. J Virol. 1979b;29(1):275–84. doi: 10.1128/jvi.29.1.275-284.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roizman B, Furlong D. The replication of herpesviruses. In: Fraenkel-Conrat H, Wagner RR, editors. Comprehensive Virology. 3. Plenum; New York, NY: 1974. pp. 229–403. [Google Scholar]
- Roizman B, Knipe DM. Herpes simplex viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. Vol. 2. Lippincot, Williams, and Wlkins; Philadelphia: 2001. pp. 2399–2459. [Google Scholar]
- Rubbi CP, Milner J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. Embo J. 2003;22(22):6068–77. doi: 10.1093/emboj/cdg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson-Holley M, Colgrove RC, Nalepa G, Harper JW, Knipe DM. Identification and functional evaluation of cellular and viral factors involved in the alteration of nuclear architecture during herpes simplex virus 1 infection. J Virol. 2005;79(20):12840–51. doi: 10.1128/JVI.79.20.12840-12851.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stackpole CW. Herpes-type virus of the frog renal adenocarcinoma. I Virus development in tumor transplants maintained at low temperature. J Virol. 1969;4(1):75–93. doi: 10.1128/jvi.4.1.75-93.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Kagawa H, Yamanashi Y, Sata T, Kawaguchi Y. Construction of an excisable bacterial artificial chromosome containing a full-length infectious clone of herpes simplex virus type 1: viruses reconstituted from the clone exhibit wild-type properties in vitro and in vivo. J Virol. 2003;77(2):1382–91. doi: 10.1128/JVI.77.2.1382-1391.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taus NS, Salmon B, Baines JD. The herpes simplex virus 1 UL 17 gene is required for localization of capsids and major and minor capsid proteins to intranuclear sites where viral DNA is cleaved and packaged. Virology. 1998;252(1):115–25. doi: 10.1006/viro.1998.9439. [DOI] [PubMed] [Google Scholar]
- Whiteley A, Bruun B, Minson T, Browne H. Effects of targeting herpes simplex virus type 1 gD to the endoplasmic reticulum and trans-Golgi network. J Virol. 1999;73(11):9515–20. doi: 10.1128/jvi.73.11.9515-9520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BJ, Boyne JR, Goodwin DJ, Roaden L, Hautbergue GM, Wilson SA, Whitehouse A. The prototype gamma-2 herpesvirus nucleocytoplasmic shuttling protein, ORF 57, transports viral RNA through the cellular mRNA export pathway. Biochem J. 2005;387(Pt 2):295–308. doi: 10.1042/BJ20041223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yedowitz JC, Kotsakis A, Schlegel EF, Blaho JA. Nuclear localizations of the herpes simplex virus type 1 tegument proteins VP13/14, vhs, and VP16 precede VP22-dependent microtubule reorganization and VP22 nuclear import. J Virol. 2005;79(8):4730–43. doi: 10.1128/JVI.79.8.4730-4743.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]