Abstract
Dopamine is a neuromodulator involved in the control of key physiological functions. Dopamine-dependent signal transduction is activated through the interaction with membrane receptors of the seven-transmembrane domain G protein-coupled family. Among them, dopamine D2 receptor is highly expressed in the striatum and the pituitary gland as well as by mesencephalic dopaminergic neurons. Lack of D2 receptors in mice leads to a locomotor parkinsonian-like phenotype and to pituitary tumors. The D2 receptor promoter has characteristics of a housekeeping gene. However, the restricted expression of this gene to particular neurons and cells points to a strict regulation of its expression by cell-specific transcription factors. We demonstrate here that the D2 receptor promoter contains a functional retinoic acid response element. Furthermore, analysis of retinoic acid receptor-null mice supports our finding and shows that in these animals D2 receptor expression is reduced. This finding assigns to retinoids an important role in the control of gene expression in the central nervous system.
Dopamine (DA) is one of the major neuromodulators in the central nervous system (CNS), controlling key physiological functions from coordination of movements to hormone synthesis and secretion. DA functions are exerted through the interaction with five distinct membrane receptors, which belong to the family of seven-transmembrane domain G protein-coupled receptors (1). We are particularly interested in the DA D2 receptor (D2R), which is highly expressed in the CNS and in the pituitary gland. There are two isoforms of this receptor, D2L and D2S, which are generated by alternative splicing from the same gene (2). Both isoforms are expressed in the same tissues and present a similar pharmacological profile (3). However, they couple to different G proteins (4–6). In contrast to the wide expression of dopaminergic receptors throughout the CNS, DA is synthesized in a small group of mesencephalic neurons located in the substantia nigra and ventral tegmental area. Interestingly, D2Rs are located both pre- and postsynaptically (7), indicating a key role not only in mediating events in the target cells of dopaminergic neurons but also in controlling the release of DA.
We have shown previously that knockout of the D2R gene results in a locomotor parkinsonian-like phenotype and in pituitary tumors in the mouse (8, 9). These results underline the importance of the expression of this gene in the control of different physiological functions, whereas alterations of its expression might be the basis of some human pathologies. This has led us to investigate the mechanisms controlling its expression at the transcriptional level. Sequence analysis of the D2R promoter has revealed features of a housekeeping promoter (10, 11). The D2R promoter lacks TATA and CAAT boxes, whereas multiple Sp1 binding sites are present. Thus, the control of the expression of the D2R gene must involve cell-specific transcription factors.
We show here that the transcription of the D2R gene is induced upon treatment of pituitary cells with retinoids. We have defined the/a promoter fragment that is responsible for retinoic acid (RA) inducibility in transfected cells. A functional RA-response element (RARE) is present in the D2R promoter that readily binds retinoic acid receptor/retinoid X receptor (RAR–RXR) heterodimers. Importantly, D2R transcripts are reduced in striatal tissue from certain RAR- and RXR-null mice, thus revealing a novel role for retinoids in the regulation of CNS functions.
MATERIALS AND METHODS
Expression Vectors.
The C1 reporter construct was obtained by subcloning the HindIII fragment spanning the 5′ promoter region of the D2R gene (positions −495 to +273), in the reporter plasmid pBLCAT3 (12). The C2 reporter plasmid was constructed by subcloning the SmaI–XhoI fragment (positions −76 to +273) in the same position. The C3 and C4 reporters were generated by cloning a 29-bp synthetic oligonucleotide corresponding, respectively, to the natural rat D2R RARE sequence and the mutated one into pBLCAT2 (12). The polylinker in pBLCAT2 is located upstream of the herpes simplex virus 1 (HSV1) thymidine kinase (TK) promoter, from position −105 to +51. To measure transfection efficiency, the plasmid pCH110 carrying the β-galactosidase gene (Pharmacia) was used. Mouse RARα, -β, and -γ, as well as mouse RXRα, -β, and -γ, expression vectors were derived from the parental pSG5 vector (13).
Cell Culture and Transient Expression Assay.
MMQ, a rat pituitary cell line (14, 15), was grown in RPMI medium 1640 supplemented with 10% fetal calf serum and 5% horse serum. Time course induction assays with all-trans-RA (t-RA) and 9-cis-RA at 0.1 μM were performed in RPMI medium 1640 supplemented with charcoal-treated serum.
COS-1 cells were used for transient transfection assays. Cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 5% fetal calf serum.
Several hours before transfection, the medium was replaced with fresh medium containing 5% charcoal-treated fetal calf serum. Cells (106 per 10-cm plate) were transfected by the calcium phosphate method using 2 μg of the reporter plasmid, 0.1 μg of RAR expression vectors, and 0.5 μg of pCH110. In all transfection assays, the Bluescript (SK−) plasmid was used as a carrier to reach 20 μg of total transfected DNA per plate. Twenty-four hours after transfection, cells were washed, and the medium was replaced for an additional 24 hr in either the presence or the absence of t-RA or 9-cis-RA ligand (0.1 μM). Before chloramphenicol acetyltransferase (CAT) assays, extracts were normalized for β-galactosidase activity (16).
DNA Binding Assays.
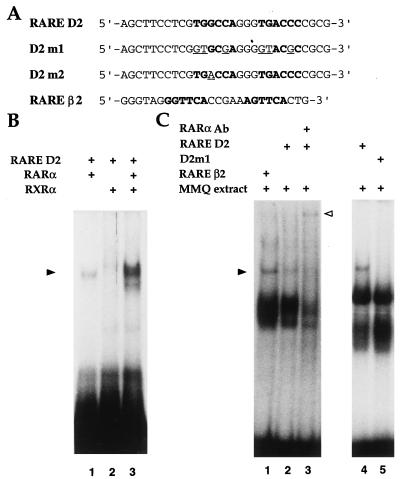
A 29-bp synthetic oligonucleotide corresponding to the RARE region in the D2R promoter (from positions −76 to −49), in addition to two mutated oligonucleotides (see Fig. 3A) were used in gel retardation assays. A synthetic oligonucleotide corresponding to the RARE-β2 (17) was used as positive control. Bacterially expressed purified mouse RARα (ΔAB) and RXRα (ΔAB) proteins were kindly provided by H. Gronemeyer. Nuclear extracts from MMQ cells were prepared by lysing the cells in extraction buffer (20 mM Tris⋅HCl (pH 8.0)/2 mM DTT/400 mM KCl/20% glycerol) by three cycles of freeze–thawing (18). Gel retardation assays were performed as described (19). For competition reactions, unlabeled double-stranded oligonucleotides were added to the binding reaction 20 min before RAR–RXR or the MMQ extracts. Supershift assay was performed by adding the mouse RARα monoclonal antibody (20) to the binding reaction 20 min before labeled D2R RARE oligonucleotides.
Figure 3.
DNA binding analysis of RARα and RXRα to the D2R RARE. (A) Nucleotide sequences of the D2R RARE (RARE D2) and the two mutants D2 m1 and D2 m2 oligonucleotides. RARE D2 contains the native RARE-like element of the D2R promoter, highlighted in bold. The mutated base pairs are underlined. RARβ2 oligonucleotide containing the DR5 RARE present in the promoter of RARβ2 gene is also represented. (B). Experiments were performed using a labeled RARE D2 oligonucleotide as probe, in the presence of recombinant retinoid receptors RARα and/or RXRα as indicated. The shifted complexes are indicated by the arrowhead. (C) Gel shift analysis performed using RAREβ2 or RARE D2 as labeled probe together with 6 μg of MMQ cell nuclear extracts. The nature of the DNA–protein complex obtained with RARE D2 was verified by supershift experiments using a monoclonal antibody against RARα. The open arrowhead indicates the supershifted complex.
RNA Analysis.
Total RNA from MMQ cells and striatal tissues was prepared by using the urea/LiCl procedure (21). Northern blot analysis was performed using 7 μg of total RNA. Restriction fragments derived from the mouse D2R cDNA (360-bp AccI fragment) and β-actin cDNA (700-bp HindIII–EcoRI fragment) were used to generate 32P-labeled probes by random priming. Hybridizations were performed under standard conditions in 50% formamide at 42°C, followed by washes in 0.3 M NaCl/30 mM sodium citrate/0.1% SDS at 50°C.
The intensity of bands on x-ray autoradiographs was quantified by using an imaging densitometer (Bio-Rad).
RESULTS
The D2R Gene Promoter Is Induced by Retinoids.
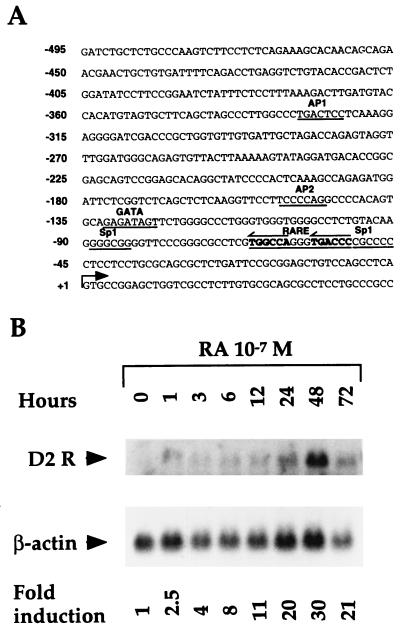
Despite the specific expression of the dopamine D2R, which is restricted to certain neurons of the central nervous system and cells of the pituitary gland, the D2R gene promoter has housekeeping characteristics and does not contain functional TATA and CAAT boxes. However, the presence of a positive cis-acting element located between nucleotides −75 and −30, and of two negative cis-acting elements, between nucleotides −116 and −76 and −160 and −135, has been reported (10, 22). In addition, putative consensus target sequences for Sp1, AP1, AP2, and GATA transcription factors can be identified (Fig. 1A). The analysis of the D2R promoter sequence revealed the presence of a putative RARE centered at position −68. This led us to investigate whether RA might play a role in the control of the D2R gene expression.
Figure 1.
(A) Nucleotide sequence of the rat D2 receptor gene promoter. Nucleotides are numbered according to the transcription initiation site (+1), indicated by the arrow. Putative response elements for AP1, AP2, Sp1, Sp2, and GATA factors are underlined. A putative RA/vitamin D receptor response element (RARE) is in bold and underlined. (B) Time course analysis of D2R transcripts from MMQ cells after induction with 10−7 M t-RA and 9-cis-RA. Induction periods are represented in hours. D2R and β-actin transcripts are indicated. The increase in D2R messenger is represented as fold induction with respect to the transcript level in the absence of RA treatment, after normalization for β-actin mRNA levels.
We took advantage of the availability of a pituitary cell line (MMQ) expressing D2Rs (14) to test whether RA treatment of these cells would result in an increased expression of the D2R gene. Northern blot analysis of RNA extracted from these cells after treatment with t-RA and 9-cis-RA (10−7 M) showed a 2-fold increase in D2R mRNA after 1 hr of treatment with a peak of 30-fold induction after 48 hr (Fig. 1B). The increase in D2R mRNA while reduced was still significant after 72 hr of induction (Fig. 1B). These data suggested that retinoids might be involved in the regulation of D2R gene expression at the transcriptional level.
Retinoids Induce the Activity of the D2R Promoter in Transfected Cells.
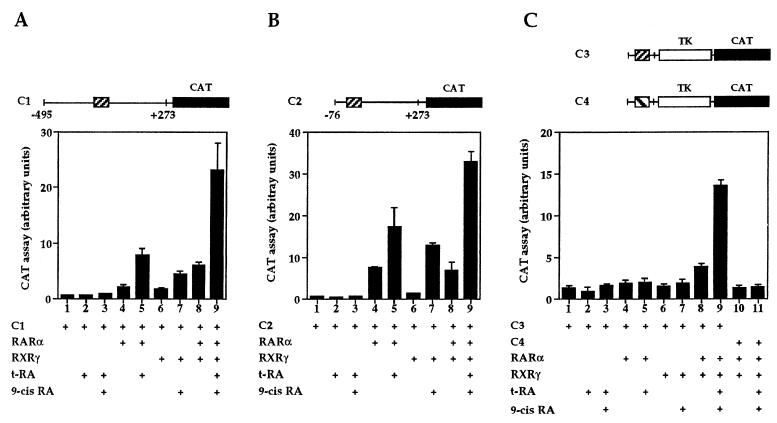
A plasmid was constructed in which a 750-bp fragment (C1), spanning the 5′ flanking region of the gene (11), drives the expression of the CAT reporter gene (Fig. 2A). COS-1 cells, which do not express endogenously the D2R gene, were transfected with this reporter plasmid together with vectors expressing retinoid receptors (Fig. 2A). Two families of nuclear retinoid receptors, binding as heterodimers to RAREs, have been described (RAR and RXR), which bind t-RA and 9-cis-RA with different affinities (23–25). Each of these families comprises three members: α, β, and γ. The ability of RARs and RXRs to induce transcription from the D2R promoter was assessed either when transfected alone or in combination, in the presence or absence of RA.
Figure 2.
(A) Analyses of transcriptional activation by RARα, RXRγ, and RARα–RXRγ on the D2R promoter reporter constructs. Reporter constructs are schematically represented at the top of each figure. The striped box (▨) corresponds to the RARE-like sequence (RARE D2) of the D2R promoter, and the black box (▪) corresponds to the CAT gene. The striped box (▧) in the C4 reporter construct represents the mutated sequence of the RARE-like site D2 m1 (see Fig. 3). COS-1 cells were cotransfected with the indicated expression vectors and the C1 reporter (A), the C2 reporter (B), or the C3 or C4 reporter (C) constructs. t-RA and 9-cis-RA were used at 10−7 M and added as indicated. Bars represent the mean of several independent experiments, and standard deviations are indicated.
The basal activity of the D2R promoter was undetectable (Fig. 2A, lane 1). Similarly, no significant changes in CAT activity were observed after treatment of C1 reporter-transfected cells with 10−7 M either t-RA or 9-cis-RA (Fig. 2A, lanes 2 and 3). Cotransfection of the C1 reporter with an RARα expression vector in cells treated with t-RA resulted into a 16-fold increase of CAT activity (Fig. 2A, lane 5). Similar results were obtained with RARβ or RARγ (data not shown). Cotransfection of RXRγ (similar results were obtained with RXRα and RXRβ) with the C1 reporter led to a 9-fold increase in CAT activity in the presence of 9-cis-RA (Fig. 2A, lane 7), whereas cotransfection of RARα and RXRγ resulted in a 45-fold synergistic activation in the presence of t-RA and 9-cis-RA (Fig. 2A, lane 9). Other combinations of RAR and RXR isotypes yielded similar synergistic activities (not shown). The basal activation of transfected RAR and RXR in the absence of ligand, observed in Fig. 2A, lanes 4, 6, and 8, is most likely due to endogenous RA in the culture media.
To establish whether the RARE-like element found in the D2R promoter might be the RA response element, the C2 and C3 reporters were constructed (Fig. 2 B and C). In the C2 reporter, the region 5′ to the putative RARE site was deleted. This region has also been reported to contain a negative regulator of the D2R promoter in the neuroblastoma cell line NB41A3 (10). In C3, a single copy of a synthetic oligonucleotide (RARE D2) containing the putative D2R RARE was cloned upstream of a heterologous promoter, the herpes simplex 1 TK promoter, at position −110 of the pBL2TK-CAT reporter vector. The C2 reporter had no detectable basal activity in COS-1 cells, indicating that deletion of the previously described negative element contained in the D2R promoter does not lead to an increased promoter activity in these cells (Fig. 2B, lanes 1–3). However, cotransfection of the C2 reporter with a RARα expression vector leads to a 35-fold increase of CAT activity after induction with t-RA (Fig. 2B, lane 5). Cotransfection of RXRγ with C2 and induction with 9-cis-RA led to 21-fold increase in CAT activity (Fig. 2B, lane 7). Similarly to the C1 reporter, coexpression of RARα/RXRγ resulted in a 66-fold increase of CAT activity in the presence of t-RA and 9-cis-RA (Fig. 2B, lane 9), as compared with control (Fig. 2B, lane 3). As for the C1 reporter, CAT activity observed in the absence of ligand in Fig. 2B, lanes 4, 6, and 8, is most probably due to endogenous RA.
To demonstrate that the putative RARE was involved in the increase of reporter activity by liganded RAR–RXR, we used the reporter C3. This construct has a constitutive basal activity when transfected in COS-1 cells probably due to the TK promoter (Fig. 2C, lane 1). Cotransfection of the C3 reporter with either RARα or RXRγ, both in the presence or in the absence of the related ligands, did not result in detectable induction (Fig. 2C, lanes 2–7). This suggests that the D2R RARE is sensitive to its promoter-flanking sequences that might be required for its full activity. In contrast, cotransfection of the C3 reporter with RARα–RXRγ expression vectors, in the presence of t-RA and 9-cis-RA, resulted in an 11-fold increase of CAT activity as compared with the basal TK activity in the absence of ligands (Fig. 2C, lanes 1 and 9).
These results indicate that the RARE-like site present in the D2R promoter responds to the presence of RA, through the binding of RAR–RXR heterodimers, with no apparent significant preference for particular RAR isotypes. In agreement with this conclusion, mutations in the D2R RARE sequence in the reporter construct lead to the absence of induction of CAT activity by RAR–RXR. Indeed, when the C4 reporter was used, in which the D2R RARE was replaced by the mutant D2 m1, which carries 6-bp conversions within the site (Fig. 3A), no detectable CAT activity was observed upon cotransfection with RAR–RXR in the presence of t-RA and 9-cis-RA (Fig. 2C, lanes 10 and 11).
The D2R Promoter Contains a RARE.
Experiments were performed to establish whether the putative RARE present in the D2R promoter could bind RARs and RXRs in vitro. A 32P-labeled oligonucleotide corresponding to the RARE-like site (RARE D2) present in the D2R promoter was synthesized (Fig. 3A). The binding ability of this oligonucleotide was assessed in gel shift assays using either recombinant RAR proteins or nuclear extracts from MMQ cells. Fig. 3B shows that RARα and RXRα individually bind poorly to the D2R RARE, whereas the presence of both receptors greatly enhances D2R RARE binding because of the higher affinity of RAR–RXR heterodimers to a RARE (17, 26). Furthermore, the D2R RARE was able to form a complex with endogenous proteins present in MMQ cell nuclear extracts, similarly to the RAREβ2 (Fig. 3C, lanes 1 and 2). Analysis of mRNA extracted from these cells revealed the presence of transcripts for RARα, RXRα, and RXRβ (data not shown). To confirm the presence of the retinoid receptors in the retarded complex, we performed gel shift analysis in the presence of a monoclonal antibody directed against RARα. The results clearly show that RARα antibody supershifted the binding complex (Fig. 3C, lane 3). When antibodies against RARα and RXRs were used in the same reaction, a similar supershift pattern of the retarded complex was observed (data not shown). The complex observed with MMQ extracts is specific as demonstrated by the absence of the retarded complex when the mutant D2 m1 oligonucleotide was used instead of D2R RARE (Fig. 3C, lanes 4 and 5).
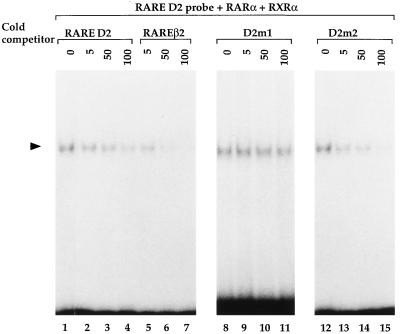
To characterize the specificity of binding, competition assays were performed using recombinant RARα and RXRα receptors in the presence of 5-, 50-, and 100-fold excess of D2R RARE oligonucleotides (Fig. 4). Binding was efficiently blocked by competition with a 50-fold excess of unlabeled D2R RARE oligonucleotide and almost disappeared in the presence of 100-fold excess (Fig. 4, lanes 1–4). When the RAREβ2 oligonucleotide (Fig. 3A) was used as a competitor, a drastic decrease of the labeled complex was already observed when a 5-fold excess of unlabeled RAREβ2 was used and was complete at 50-fold excess (Fig. 4, lanes 1, 5–7). The higher competing capacity of the RARE β2 oligonucleotide is in keeping with the presence of a direct repeat spaced by five nucleotides (DR5) in contrast to the D2R RARE, which, instead, contains a direct repeat spaced by three nucleotides (DR3) (23, 25, 27).
Figure 4.
DNA binding competition assays. Gel shift analysis using labeled RARE D2 and recombinant RARα and RXRα as indicated. As competitors, RARE D2, RAREβ2, and the mutant D2 m1 and D2 m2 cold oligonucleotides were used as indicated at 5-, 50-, and 100-fold molar excess. The black arrowhead indicates the shifted complex.
To further characterize the D2R RARE element and its binding affinity to retinoid receptors, mutations were introduced into this sequence, generating D2 m1 and D2 m2 (Fig. 3A). These mutated oligonucleotides were used for competition in gel shift assays (Fig. 4). The unlabeled D2 m1 oligonucleotide, in which the D2R RARE was drastically altered, failed to compete with the labeled oligonucleotide to bind the RAR–RXR heterodimer even after addition of 100-fold excess of the competitor (Fig. 4, lanes 8–11). In contrast, the mutant D2 m2 (Fig. 3A), which converts the D2R RARE into a perfect DR3 element (GGGTCANNNTGGTCA), was more efficient at competing than the native site (compare, in Fig. 4, lanes 12–15 to lanes 1–4).
The above data further support the conclusion that the retarded complex formed by the binding of the heterodimer RARα–RXRα on the D2R RARE is specific.
Altered Expression of the D2R Gene in Mice Mutants for RARs and RXRs.
Our results strongly suggest that RARs regulate D2R gene transcription. Thus, the absence of one of these transcription factors in vivo may alter the levels of D2R messenger RNA. We thus performed in vivo studies by analyzing the level of expression of D2R in mice in which the expression of one or more RARs has been knocked out by homologous recombination.
Interestingly, dopaminergic neurons of the nigrostriatal pathway have been found to synthesize RA (28). In addition, whereas RARα is uniformly expressed in the mouse brain, RARβ, RXRβ, and RXRγ receptors are more abundantly expressed in certain brain regions, such as caudate putamen and nucleus accumbens (W. Krezel, personal communication), both striatal regions that highly express D2R.
D2R mRNA expression was analyzed in RXRβ−/− and RXRγ−/− single mutants (29, 30). Ten micrograms of total RNA from striatal tissues of these mice was analyzed by Northern blotting, using a mouse D2R-specific probe. D2R expression in mutants was compared with the expression in wild-type littermates. In support of our in vitro data, we found a 40% reduction of D2R mRNA expression in the striatum of RXRγ−/− mice (Fig. 5), whereas no reduction of D2R mRNA was observed in the same tissue of RXRβ-deficient mice (Fig. 5).
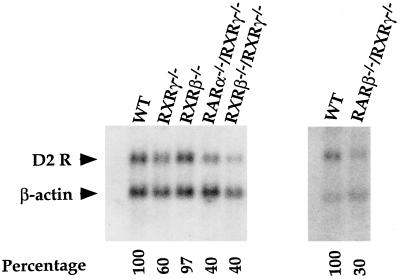
Figure 5.
Northern blot analysis of D2R mRNA expression in the striatum of wild-type and mutant mice. Genotypes are indicated on the top of each lane. Arrowheads indicate the bands corresponding to D2R and β-actin mRNAs. The expression level of D2R mRNA in wild-type mice was arbitrarily set at 100%.
We next analyzed the following double mutants: RARα−/−–RXRγ−/−, RXRγ−/−–RXRβ−/−, and RARβ−/−–RXRγ−/−. Interestingly, these mice showed a more severe decrease in the striatal D2R gene expression (Fig. 5). The D2R mRNA level in the striatum of RARα−/−–RXRγ−/− and RXRγ−/−–RXRβ−/− mutant mice was decreased by 60% as compared with wild-type littermates and by 70% in the RARβ–RXRγ-deficient mice.
These data indicate that RARα, RARβ, RXRβ, and RXRγ are likely to be involved in the regulation of the expression of the D2R gene in vivo. Note that the expression pattern of two retinoid receptors, RARβ and RXRγ, correlates with the reduction of the striatal D2R mRNA levels in the mice lacking these receptors.
DISCUSSION
Dopamine controls a wide variety of physiological functions through the activation of membrane receptors. Alterations in the dopaminergic system in humans result in mental diseases such as schizophrenia and Parkinson’s disease. We have shown that deletion of the D2R gene in knockout mice results in an impaired locomotor phenotype with analogies with Parkinson’s disease and in pituitary tumors (8, 9). These observations suggest that gene regulation of dopamine receptors, and in particular of the D2R, is a key element in the normal function of the dopaminergic system. In this respect, it is interesting to note that the two major dopamine receptors, D1 and D2, are thought to possess promoters with housekeeping characteristics (10, 11). Thus, specificity in the expression pattern of these receptors must be temporally and spatially dictated by cell-specific transcription factors.
We have characterized here a RARE that is located in the 5′ flanking region of the dopamine D2 receptor gene at position −68. The described D2R RARE is made up of repeated motifs closely related to the core motif 5′-PuG(G/T)TCA-3′. The two motifs are spaced by 3 bp, a spacing that has been described to favor the binding of vitamin D3 receptor (23, 25, 27). In this paper, we demonstrate that RAR–RXR heterodimers bind to this motif and activate transcription from the D2R promoter. At present, we cannot exclude that this element might also be a target for the vitamin D3 receptor in a class of dopaminergic or target cell neurons or in cells of the pituitary gland. Interestingly, a canonical Sp1 binding site is contiguous to the D2R RARE. It could be speculated that in particular cells expressing the D2R, an interaction between members of the retinoid receptor family and the Sp1 factor might affect the D2R RARE binding specificities.
The nonconsensus response elements present in D2R and some other natural promoters may result in lower binding affinities for retinoid receptors, as observed here in gel shift competition assays using D2R RARE, D2 m2, and RAREβ2 oligonucleotides (Fig. 4). This might correspond to a regulatory mechanism aimed at attenuating the response to specific ligands in vivo. It is also possible that transcription factors recognizing nonconsensus motifs may be subject to cooperative interactions with other factors binding to nearby or adjacent sites (31, 32). The presence of these cooperative interactions could determine the strength and cellular specificity of the transcriptional response. In the case of the D2R promoter, the close vicinity of the D2R RARE and the Sp1 sites might represent an example of such cooperativity.
It should be pointed out that the D2R is expressed in different regions of the brain and in particular by the dopaminergic cells in mesencephalic neurons and by the dopaminergic neuron targets, such as the medium spiny neurons in the striatum, but also by cortical and hypothalamic cells. In addition, outside the brain the D2R is highly expressed in the pituitary gland by two cell types, melanotrophs and lactotrophs. It is thus conceivable that the D2R promoter might be controlled by different combinations of members of the thyroid hormone/RA and vitamin D3 receptor families in different cell types.
However, our results point to a crucial role of members of RAR and RXR families in the control of D2R expression in the striatum. This control is dependent on the binding of RAR–RXR heterodimers as demonstrated by the stronger decrease of D2R expression in RAR–RXR double mutants as compared with single mutants. This suggests that absence of one retinoid receptor might be partially compensated by other members of the RAR–RXR family in simple mutants. These results strongly support our in vitro data and identify these receptors as specific transcription factors required for full expression of the D2R gene in the striatum.
Our data indicate an involvement of RA in adult CNS functions, as the absence of retinoid receptors results in a reduced expression of the DA D2 receptor. We have previously shown that, in mice, altered expression of this receptor results in a parkinsonian phenotype (8) and pituitary tumors (9). It is well established that Parkinson’s disease is generated by a strong reduction of DA levels. Our data raise the possibility that aberrant control of dopaminergic receptor expression might also lead to neurological disorders.
Acknowledgments
We greatly acknowledge Philippe Vernier for the gift of the rat D2R promoter, M. Sporn for 9-cis-RA, B. Boulay and J. M. Lafontaine for artwork, N. Ferandel and M. Acker for cell cultures, and D. Bonnier for recombinant proteins. We thank Ph. Kastner and N. Ghyselinck for RAR and RXR mouse mutants and P. Sassone-Corsi for discussions. This work was supported by funds from the Centre National de la Recherche Scientifique, the Institut National de la Santé et de la Recherche Médicale, the Collège de France, the Centre Hospitalier Universitaire Régional, the Association pour la Recherche sur la Cancer, the Human Frontier Science Program, and Bristol-Myers Squibb. T.A-S. is a recipient of a fellowship from the Centre National de la Recherche Scientifique (Bourse Docteur Ingenieur), and W.K. is a recipient of a fellowship from the Ministère de la Recherche and the Association pour la Recherche sur la Cancer.
ABBREVIATIONS
- RA
retinoic acid
- RAR
RA receptor
- RXR
retinoid X receptor
- DA
dopamine
- D2R
dopamine D2 receptor
- D1R
dopamine D1 receptor
- RARE
RA response element
- t-RA and 9-cis-RA
all-trans- and 9-cis-retinoic acid
- CAT
chloramphenicol acetyltransferase
- TK
thymidine kinase
- CNS
central nervous system
References
- 1.Gingrich J A, Caron M G. Annu Rev Neurosci. 1993;16:299–321. doi: 10.1146/annurev.ne.16.030193.001503. [DOI] [PubMed] [Google Scholar]
- 2.Dal Toso R, Sommer B, Ewert M, Herb A, Pritchett D B, Bach A, Shivers B D, Seeburg P H. EMBO J. 1989;8:4025–4034. doi: 10.1002/j.1460-2075.1989.tb08585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jackson D M, Westlind-Danielsson A. Pharmacol Ther. 1994;64:291–369. doi: 10.1016/0163-7258(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 4.Montmayeur J-P, Borrelli E. Proc Natl Acad Sci USA. 1991;88:3135–3139. doi: 10.1073/pnas.88.8.3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Montmayeur J-P, Guiramand J, Borrelli E. Mol Endocrinol. 1993;7:161–170. doi: 10.1210/mend.7.2.7682286. [DOI] [PubMed] [Google Scholar]
- 6.Guiramand J, Montmayeur J-P, Ceraline J, Bhatia M, Borrelli E. J Biol Chem. 1995;270:7354–7358. doi: 10.1074/jbc.270.13.7354. [DOI] [PubMed] [Google Scholar]
- 7.Civelli O, Bunzow J R, Grandy D K, Zhou Q Y, Van Tol H H. Eur J Pharmacol. 1991;19:277–286. doi: 10.1016/0922-4106(91)90001-x. [DOI] [PubMed] [Google Scholar]
- 8.Baik J-H, Picetti R, Saiardi A, Thiriet G, Dierich A, Depaulis A, Lemeur M, Borrelli E. Nature (London) 1995;377:424–428. doi: 10.1038/377424a0. [DOI] [PubMed] [Google Scholar]
- 9.Saiardi A, Bozzi Y, Baik J-H, Borrelli E. Neuron. 1997;19:115–126. doi: 10.1016/s0896-6273(00)80352-9. [DOI] [PubMed] [Google Scholar]
- 10.Minowa T, Minowa M T, Mouradian D M. Biochemistry. 1992;31:8389–8396. doi: 10.1021/bi00151a001. [DOI] [PubMed] [Google Scholar]
- 11.Valdenaire O, Vernier P, Maus M, Dumas Milne Edwards J B, Mallet J. Eur J Biochem. 1994;220:577–584. doi: 10.1111/j.1432-1033.1994.tb18658.x. [DOI] [PubMed] [Google Scholar]
- 12.Luckow B, Schütz G. Nucleic Acids Res. 1987;15:5490. doi: 10.1093/nar/15.13.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Green S, Issemann I, Sheer E. Nucleic Acids Res. 1988;16:369. doi: 10.1093/nar/16.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Judd A M, Login I S, Kovacs K, Ross P C, Spangelo B L, Jarvis W D, Macleod R M. Endocrinology. 1988;123:2341–2350. doi: 10.1210/endo-123-5-2341. [DOI] [PubMed] [Google Scholar]
- 15.Forget H, Painson J-C, Drews R T, Lagacé G, Collu R. Mol Cell Endocrinol. 1993;93:125–133. doi: 10.1016/0303-7207(93)90115-z. [DOI] [PubMed] [Google Scholar]
- 16.Petkovich M, Brand N J, Krust A, Chambon P. Nature (London) 1987;330:444–450. doi: 10.1038/330444a0. [DOI] [PubMed] [Google Scholar]
- 17.Mader S, Leroy P, Chen J-Y, Chambon P. J Biol Chem. 1993;268:591–600. [PubMed] [Google Scholar]
- 18.Kumar V, Chambon P. Cell. 1988;55:145–156. doi: 10.1016/0092-8674(88)90017-7. [DOI] [PubMed] [Google Scholar]
- 19.Nicholson R C, Mader S, Nagpal S, Rochette-Egly C, Chambon P. EMBO J. 1990;9:1986–1989. doi: 10.1002/j.1460-2075.1990.tb07895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaub M P, Rochette-Egly C, Lutz Y, Ali S, Matthes H, Scheuer I, Chambon P. Exp Cell Res. 1992;201:335–346. doi: 10.1016/0014-4827(92)90282-d. [DOI] [PubMed] [Google Scholar]
- 21.Auffray C, Rougeon F. Eur J Biochem. 1980;107:303–314. doi: 10.1111/j.1432-1033.1980.tb06030.x. [DOI] [PubMed] [Google Scholar]
- 22.Minowa M T, Minowa T, Mouradian M M. J Biol Chem. 1994;269:11656–11662. [PubMed] [Google Scholar]
- 23.Leid M, Kastner P, Chambon P. Trends Biochem Sci. 1992;17:427–433. doi: 10.1016/0968-0004(92)90014-z. [DOI] [PubMed] [Google Scholar]
- 24.Mangelsdorf D J, Borgmeyer U, Heyman R A, Zhou J, Y, Ong E S, Oro A E, Kakizuka A, Evans R M. Genes Dev. 1992;6:329–344. doi: 10.1101/gad.6.3.329. [DOI] [PubMed] [Google Scholar]
- 25.Chambon P. FASEB J. 1996;10:940–954. [PubMed] [Google Scholar]
- 26.Kliewer S A, Umesono K, Mangelsdorf D J, Evans R M. Nature (London) 1992;355:446–449. doi: 10.1038/355446a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schütz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans R M. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCaffery P, Dräger U C. Proc Natl Acad Sci USA. 1994;91:7772–7776. doi: 10.1073/pnas.91.16.7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kastner P, Mark M, Leid M, Gansmuller A, Grondona J-M, Décimo D, Krezel W, Dierich A, Chambon P. Genes Dev. 1996;10:80–92. doi: 10.1101/gad.10.1.80. [DOI] [PubMed] [Google Scholar]
- 30.Krezel W, Dupé V, Mark M, Dierich A, Kastner P, Chambon P. Proc Natl Acad Sci USA. 1996;93:9010–9014. doi: 10.1073/pnas.93.17.9010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Day R N, Koike S, Sakai M, Muramatsu M, Maurer M A. Mol Endocrinol. 1990;4:1964–1971. doi: 10.1210/mend-4-12-1964. [DOI] [PubMed] [Google Scholar]
- 32.Rhodes S J, Chen R, Dimattia G E, Scully K M, Kalla K A, Lin S C, Yu V C, Rosenfeld M G. Genes Dev. 1993;7:913–932. doi: 10.1101/gad.7.6.913. [DOI] [PubMed] [Google Scholar]