Abstract
The production of melanin is a complex biochemical process in which several enzymes may play a role. Although phenoloxidase and serine proteases are clearly key components, the activity of other enzymes, including dopa decarboxylase and dopachrome conversion enzyme may also be required. We tested the effect of knockdown of gene expression for these two enzymes on melanization of abiotic targets in the mosquito, Anopheles gambiae. Knockdown of dopa decarboxylase and dopachrome conversion enzyme resulted in a significant reduction of melanization of Sephadex beads at 24h after injection. Knockdown of a third enzyme, phenylalanine hydroxylase, which is involved in endogenous production of tyrosine, had no effect on bead melanization. Quantitative analysis of gene expression demonstrated significant upregulation of phenylalanine hydroxylase, but not the other two genes, following injection.
Keywords: Anopheles gambiae, insect immunity, melanization, mosquito
Introduction
The pigment melanin is widespread in the animal kingdom, with varied functional roles ranging from formation of surface color patterns and protection against ultraviolet radiation to internal defense against pathogens. The latter role is apparent in many insects where melanized capsules have been observed around a wide variety of foreign objects including abiotic targets, bacteria, nematode worms, parasitoids, and protists (Salt 1963; 1970). In mosquitoes, melanization functions as a natural mechanism of resistance against human malaria parasites in Anopheles culicifacies (Adak et al., 2006) and against the nematode Brugia malayi in Armigeres subalbatus (Infanger et al., 2004). Melanization of malaria parasites has also been artificially selected in a laboratory colony of An. gambiae (Collins et al., 1986; Paskewitz et al., 1988) and a significant body of work has developed to identify the molecular determinants of this phenotype (Blandin et al., 2004; Volz et al., 2006; Kumar et al., 2003).
The biochemical pathways leading to melanin formation are complex and can involve multiple branches (True, 2003; Christensen et al., 2005; Nappi and Christensen 2005). Central to melanin biosynthesis in insects are the phenoloxidase enzymes (PO). Tyrosinase-type POs exist in zymogen forms (ProPO) which can be activated by serine proteases (termed proPO activating protease (PAP) or proPO activating enzyme (PPAE)). Other serine proteases are involved in activating PPAE and in initiating the activation cascade (Ji et al., 2004). Once PO is activated, a series of enzyme-catalyzed and spontaneous reactions may occur. As a first step, the amino acid tyrosine must be hydroxylated to produce dopa by the action of PO or tyrosine hydroxylase. Tyrosine can be dietary or can be derived endogenously by the action of phenylalanine hydroxylase (PAH) on its substrate, phenylalanine. Once dopa is formed, it can then be oxidized to dopaquinone. PO also catalyzes this reaction. Dopaquinone spontaneously converts to dopachrome, which can be decarboxylated by dopachrome conversion enzyme (DCE; also known as dopachrome isomerase) to form 5,6-dihyroxyindole which is then oxidized by PO (Christensen et al., 2005). The resulting compound will polymerize to form melanin. Dopamine, derived from dopa via the action of dopa decarboxylase (DDC), can also serve as the catecholamine substrate for melanin production. Identifying the enzymes and accessory proteins that contribute to melanin formation during different physiological processes remains an active area of research (Huang et al., 2005; Jiang et al., 2005; Volz et al., 2006; Tang et al., 2006; Warr et al., 2006).
The molecular basis of melanogenesis in mosquitoes has been studied through gene silencing experiments which have provided clear demonstrations of the functional roles of some of the enzymes noted above. In culicine mosquitoes, knockdown of PO I (Shiao et al., 2001), Ddc (Huang et al., 2005a), Dce (Huang et al., 2005b), or Pah (Infanger et al., 2004) reduces melanization of nematode worms. However, the extent to which these observations hold true for different targets and for anopheline mosquitoes has not been determined. Genetic studies have demonstrated that melanization of different targets are often controlled by different pathways even within a species (Zheng et al., 2003; Tang et al., 2006). Melanization in Anopheles gambiae can be easily induced in response to the injection of an abiotic target, Sephadex beads (Paskewitz and Riehle, 1994). Thus, in this study, we used the bead system and RNAi-mediated gene silencing to investigate the role of three enzymes, DDC, DCE and PAH, in the melanization response.
Materials and Methods
Mosquitoes
The G3 strain of A. gambiae was used for all experiments. The G3 strain originated from mosquitoes collected from The Gambia. The G3 strain has the ability to melanize malaria parasites and was used in genetic selection of a line that fully melanizes a wide range of Plasmodium species and strains (Collins et al. 1986). Mosquitoes were reared as described previously (Paskewitz et al., 1999).
Identification of A. gambiae DDC, PAH, and DCE genes
Sequences for all three genes were identified by database screening. Accession numbers are: dopa decarboxylase AF063021, dopachrome conversion enzyme AJ459959, and phenylalanine hydroxylase AF283273 (Oduol et al., 2000).
RNA interference
An in vitro transcription template was produced using a two-step PCR protocol (Dudley et al., 2002). First, a pair of 35 bp primers, each designed to include 15 bp of T7 promoter sequence (in bold) plus 20 bp of the target gene sequence, were used to amplify a product from mosquito cDNA. The gene-specific primers were: Dopa decarboxylase 5′CGACTCACTATAGGGACACCGGGCCAGCCTTCGAG3′ and 5′CGACTCACTATAGGGCTGCTGCTGCTGGCGTTCAT; Phenylalanine hydroxylase 5′CGACTCACTATAGGGGTGCGTCTTCTGCGGTGTGA3′ and 5′CGACTCACTATAGGGGAAGCTGACCAACCCGATCA3′; Dopachrome conversion enzyme 5′CGACTCACTATAGGGCTGGGACAGGATTGCGTAAA3′ and 5′CGACTCACTATAGGGTCGACAGGCCCGCGTGCTTC3′.
Because injection of dsRNA can suppress the mosquito immune response (Blandin et al., 2002), we used an exogenous gene for control injections. A cloned gene for green fluorescent protein (GFP) was used to produce control dsRNA as described above. Primers included 15 bp of T7 promoter sequence plus 20 bp GFP sequence were used to amplify a product using the phMFGP vector (Promega, Madison, WI, USA) as the template DNA. The gene specific primers were (5′CGACTCACTATAGGGCGTGATCAAGCCCGACA-3′ and 5′-CGACTCACTATAGGGTGGGCTTCGGCGTGCT-3′).
Amplification parameters were an initial denaturation step of 92°C for 1 min followed by 35 cycles of 92°C for 30 s, 58°C for 45 s, 72°C for 45 s. After a final 10 min extension at 72°C, reactions were held at.4°C until they were frozen. Each PCR product was purified using a Qiagen gel extraction kit (Qiagen, Valencia, CA, USA). Purified products then were used in a second PCR reaction with a primer containing a full T7 site: 5′TAATACGACTCACTATAGGG3′. The resulting product was again purified using the Qiagen Gel Extraction kit and 1-2 μg of the product were used as template for transcription. The Megascript RNAi kit (Ambion, Austin, TX, USA) was used for transcription and the production of dsRNA following the manufacturer’s directions. All dsRNA preparations were quantified by measuring absorbance at 260 nm, checked for integrity on an agarose gel, and stored at -20°C.
For most experiments, mosquitoes (0-36 h after eclosion) were injected with 0.1 μg dsRNA (in 0.2 μL dH2O) using pulled glass needles. To increase the efficiency of knockdown, some cohorts were injected with up to 1.2 μg of dsRNA.
Four days after dsRNA for GFP or for DDC, DCE and PAH were injected, we injected one CM Sephadex bead into each mosquito as described in the next section. After an incubation period of 24 h, we collected the carcasses for analysis of the efficacy of knockdown and also removed beads for scoring melanization. Each experiment was independently replicated three or more times and at least 75 mosquitoes each were assessed for control and knockdown treatments.
Sephadex bead inoculation
Four days after dsRNA injections, mosquitoes were injected with CM-25 Sephadex beads (40-120 μm; Sigma-Aldrich, St. Louis, MO, USA) as previously described (Chun et al., 1995; Gorman et al., 1998). Briefly, beads were hydrated in mosquito saline prior to inoculation. Each bead was aspirated with less than 0.5 μL of saline into a pulled glass needle and then injected into the hemocoel of a female that had been anesthetized on ice. One bead was injected per mosquito at the junction between the thoracic and abdomen using a mouth aspirator. For all experiments, mosquitoes were placed into small humidified plastic cages supplied with 10% sucrose and allowed to recover in an incubator at 70-80% relative humidity and 25-26°C
Semiquantitative RT-PCR
Total RNA from carcasses of at least 10 female mosquitoes was extracted and used to verify that the knockdown was successful. Samples were extracted with the AquaPure RNA isolation kit (BioRad, Hercules, CA,, USA) and then treated with amplification grade DNaseI (Invitrogen, San Jose, CA, USA). First strand cDNA was synthesized using oligo dT and reverse transcriptase Superscript III (Invitrogen) and used for subsequent PCR reactions.
Samples were subjected to semi-quantitative RT-PCR after first using the ribosomal S7 gene (5′-TGCTGCAAACTTCGGCTAT-3′ and 5′-CGCTATGGTGTTCGGTTCC-3′) to normalize the samples (30 sec at 92°C, 56°C, and 1.0 min at 72°C for 21 cycles). To amplify enzymes genes, gene-specific primers (10 pmol) were used in 20 μL reactions with Amplitaq (1 unit, Promega, Madison, WI, USA), 200 μM dNTP, 2.0 mM MgCl2. Conditions for the PCR reactions were: 30 s at 92°C, 56°C, and 1.0 min at 72°C for 35 cycles for CLIPB9. The same conditions and 30 cycles were used for all others.
Primers used for each gene were: DDC 5′-GTGCGTCTTCTGCGGTGTGA-3′ and 5′-GAAGCTGACCAACCCGATCA-3′; PAH 5′-CAAACAGTGCTGGCGCTAGA-3′ and 5′-TGCGGCCCGAAGCTAACCAC-3′; DCE 5′-CATCCGGTCCGAGATTGTCC-3′ and 5′-TGTGCGACATCCCGAACGAG-3′.
Real-Time PCR
Whole mosquitoes were used for RNA extraction following bead injection. Total RNA was isolated as described above and was treated with RQ1 DNAse (Promega) to remove contaminating genomic DNA. RNA concentration was measured with micro-spectrophotometry (Nanodrop NT1000, Thermofisher, Waltham, MA, USA). Next, removal of DNA from the RNA samples was confirmed by real-time PCR using housekeeping gene primer sets (no cDNA control). Samples that yielded threshold cycle (Ct) values larger than 33 were deemed acceptable (Rotenberg et al. 2006). One hundred ng of high-quality total RNA was reverse transcribed using the High Capacity cDNA Archive kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions. The resulting cDNA was diluted 1:5 and stored at 4°C. In our experience, freeze/thaw cycles result in degraded cDNA samples.
Primers for internal reference genes (ribosomal protein S7, RPS3, RPS31, tropomyosin, and actin) and the three target genes (Ddc, Dce, and Pah) were designed using the Beacon designer software (Premier Biosoft International, Palo Alto, CA, USA). The program setting “avoid template structure” was chosen to limit primer sequences to regions of little secondary template structure. Primers chosen were: actin-forward CAGTCCAAGCGTGGTATC actin-reverse GTTAGCCTTCGGGTTCAG; RPS7-forward CCTATGGTGTTCGGTTCC, RPS7-reverse GATCGCCTTCTTGTTGTTG; Tropomyosin-forward GAACGGATTCAGCAGGTG, Tropomyosin-reverse TTCTCAGCATCTTCAAGCC; RPS31-forward GACAGCATTTGGGACGATTC, RPS31-reverse GTTGATTAGGACAGACTTTACCG; RPS3-forward CTATGAAGGTTATGCTCTGC, RPS3-reverse CATCAGGTAGTCGGTCAG; DDC-forward GTGGGAAGAGGTGATGGC, DDC-reverse GTAGGCGTGGAACTTGGG; DCE-forward GCGGTCCACCATAAGAAC, DCE-reverse AGTTAGGATAGGGCTTCAGG; PAH-forward CGACTACAAGGATAACGAAGC, PAH-reverse GTACGACAGGATCTGGTTGG.
The primers were synthesized by IDT Technologies (Coralville, IA, USA). The performance of these primers (PCR efficiency and standard curves) was tested on five dilutions of cDNA prepared as described above. All standard curves were generated from triplicate reactions of the five-fold dilution series of cDNA. Suitable internal reference gene primer sets were chosen on the basis of i) primer efficiencies that were close to 100% and ii) threshold cycles were within 5 cycles of those seen for the target genes. Both reference and target primers exhibited similar efficiencies as determined using a dilution series of cDNA derived from A. gambiae total RNA (uninoculated adult female control).
Real-time PCR was carried out using an iCycler machine (BioRad) and analyzed using the iQ software package (BioRad). All reactions were performed in triplicate 25 μL volumes using iQ SYBRGreen Supermix (BioRad). A master mix was prepared for each primer set containing SYBRGreen and an appropriate volume of each primer to yield a final primer concentration of 200 nM. The reaction conditions were enzyme activation and well factor determination at 95°C for 3 min followed by 40 cycles of 95°C for 10 s (denaturation) and 58°C for 45 s (annealing and elongation); the melt curve protocol began immediately after amplification and consisted of 95°C for 1 min, followed by 55°C for 1 min and then 80-10s steps of 0.5°C increases at each step. Threshold values for threshold cycle (Ct) determination were generated automatically by the iCycler software. The absence of primer artifacts was determined from the melt curve profile of the PCR products.
The stability of the reference genes in comparison to each other during the treatments was analyzed using the BestKeeper program (Pfaffl et al., 2004; MS-Excel based Q-PCR data analysis program).
Results
Effect of bead injection on transcript levels
We examined whether transcript abundance for the three enzymes changed during activation of the melanization pathway. Three independent cDNA samples were prepared from mosquitoes that were harvested following injection with one CM Sephadex bead. Each experiment included four time points (preinjection (0h), and 3h, 6h and 24h after bead injection).
Because large variation in housekeeping gene expression occurs in vertebrate systems (Vandesompele et al., 2002; Brunner et al., 2004; Radonic et al., 2004), we tested five genes for stability following bead injection. For this initial analysis, we used total RNA input as the reference (Sindelka et al., 2006). Actin exhibited large increases in threshold levels when comparing between time 0 and 24h, suggesting that gene expression may be suppressed during the response to wounding and injection. The other four genes exhibited a minimal increase of <2 fold change in expression at all times. Although gene expression was stable, RPS3 and S7 transcripts were much more abundant than the target genes (a difference greater than 5 cycles) which is undesirable for quantitative PCR. Tropomyosin and RPS31 were expressed at levels similar to the target genes but tropomyosin did not produce consistent results at the 24 h timepoint. Thus, RPS31 was chosen as the best reference gene. Interestingly, three of the stable housekeeping genes do not vary much in terms of functionality; they are ribosomal proteins involved in protein biosynthesis. Similar results have recently been reported in an evidence-based approach to housekeeping gene identification for vertebrates (De Jong et al., 2007)
Figure 1 provides the results of all three trials for each target gene because the relative fold change in expression varied somewhat between experiments (interassay variation). This was apparent especially at the 24 h timepoint, which demonstrates the risk of relying on RNA from a single experiment for Q-PCR. Earlier timepoints were more consistent. Compared with the preinoculation controls (0 h), Pah was upregulated at 3 and 6 h postinoculation by an average of 3 fold. Dce was upregulated by approximately 2.5 fold at 3 and 6 h. Ddc transcript levels were not significantly altered during these trials (1.8 fold increase or less). These experiments do not discriminate between changes due to the injection alone (wounding) or to introduction of the bead. However, melanin formation is induced in either case.
Figure 1.
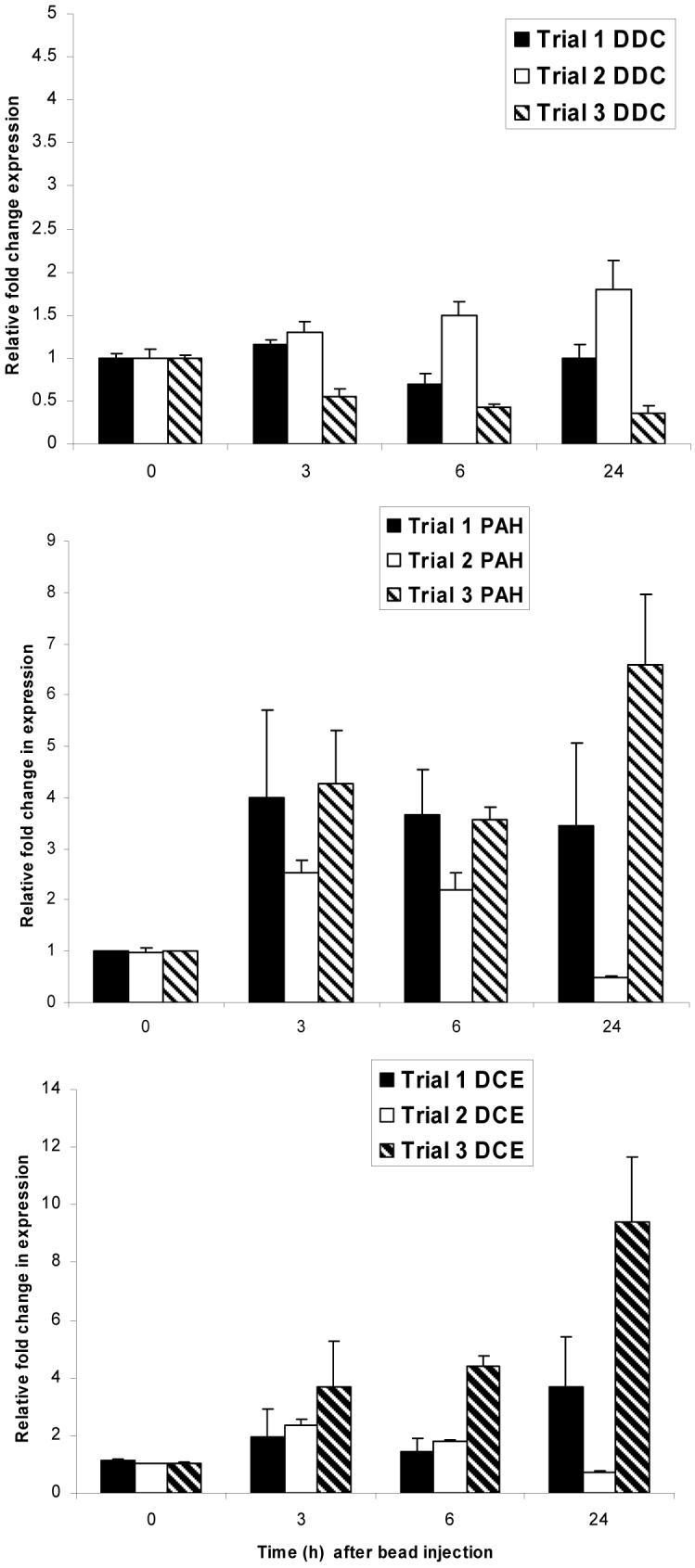
Real time PCR analysis of expression profiles for three genes, dopa decarboxylase (DDC), dopachrome conversion enzyme (DCE) and phenylalanine hydroxylase (PAH) following bead injection. Results of three replicate assays are provided for each gene. Ribosomal protein S31 was used to normalize the samples. Expression at times 3, 6 and 24 h was determined relative to time 0 (arbitrarily set at 1); time 0 results are included to illustrate variation between replicate experiments. Note that the relative expression scale differs for each gene.
Gene silencing
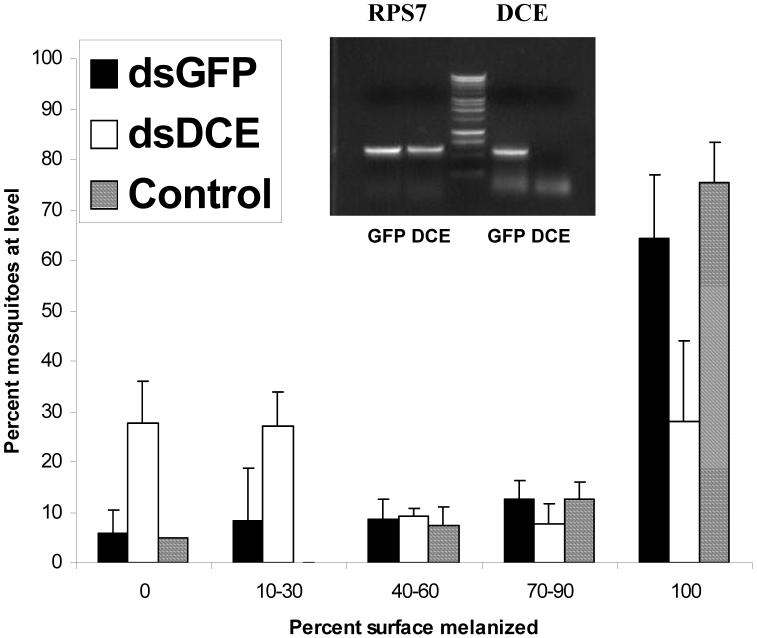
The RT-PCR results demonstrate that significant knockdown of all three enzymes occurred (Fig. 2, 3, 4). Knockdown of Dce resulted in reduced melanization of CM Sephadex beads (Fig. 2). Over three experiments, the average proportion of mosquitoes that completely melanized beads was 28% compared with 64% when the control dsGFP was injected (Student’s t-test, p<0.05). While we found unmelanized beads in only 6% of the GFP controls, 28% of the Dce knockdowns failed to initiate melanization on the bead surface. In other experiments, the amount of dsRNA for Dce was increased from 0.1 to 0.66 μg, resulting in further reduction in the expression of this gene but this did not significantly alter the bead melanization outcomes.
Figure 2.
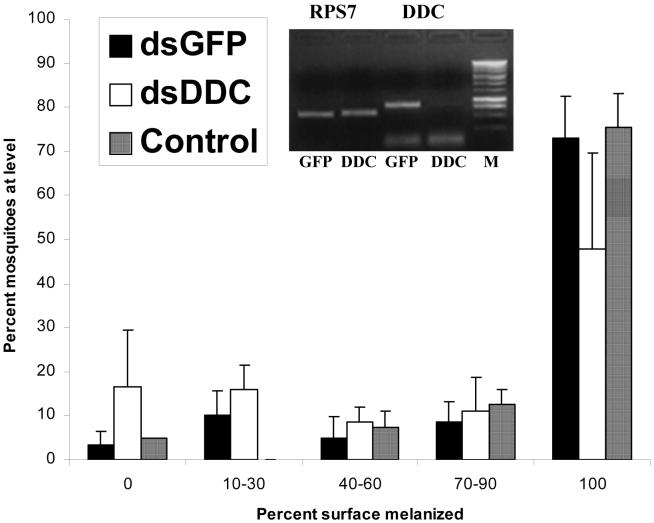
Effect of RNAi mediated silencing of dopa decarboxylase on melanization of CM-Sephadex beads in Anopheles gambiae. A frequency distribution for 5 categories of increasing strength of melanization is presented. Control mosquitoes were not injected with double stranded RNA. GFP mosquitoes were injected with dsGFP. Inset: ethidium bromide stained gel demonstrating the degree of knockdown as measured by RT-PCR. RPS7 is used as a calibration control, resulting in approximately equal transcript levels of this nontarget gene in both dsDDC and dsGFP treatments.
Figure 3.
Effect of RNAi-mediated silencing of dopachrome conversion enzyme on melanization of CM-Sephadex beads in Anopheles gambiae. A frequency distribution for 5 categories of increasing strength of melanization is presented. Control mosquitoes were not injected with double stranded RNA. GFP mosquitoes were injected with dsGFP. Inset: ethidium bromide stained gel demonstrating the degree of knockdown as measured by RT-PCR. RPS7 is used as a calibration control, resulting in approximately equal transcript levels of this nontarget gene in both dsDCE and dsGFP treatments.
Figure 4.
Effect of RNAi-mediated silencing of phenylalanine hydroxylase on melanization of CM-Sephadex beads in Anopheles gambiae. A frequency distribution for 5 categories of increasing strength of melanization is presented. Control mosquitoes were not injected with double stranded RNA. GFP mosquitoes were injected with dsGFP. Inset: ethidium bromide stained gel demonstrating the degree of knockdown as measured by semiquantitative RT-PCR. RPS7 is used as a calibration control, resulting in approximately equal transcript levels of this nontarget gene in both dsPAH and dsGFP treatments.
Silencing of Ddc also resulted in significantly reduced melanization of CM Sephadex beads (Figure 3). When we used mosquitoes for dsRNA injection two days after eclosion, the average proportion of mosquitoes that completely melanized beads was 38% in Ddc knockdowns compared with 73% for GFP controls (Student’s t-test, p<0.05). Two additional experiments were carried out using mosquitoes that were injected with dsRNA on the first day after eclosion but here there were no differences between Ddc knockdowns and controls. RT-PCR indicated that knockdown of Ddc was successful in all replicates, suggesting that newly eclosed mosquitoes may have additional resources for bead melanization that render the activity of Ddc redundant.
Silencing of Pah did not reduce melanization of CM-Sephadex beads in comparison with GFP controls (Fig. 4).
Discussion
Melanization can play a key role in containing and killing foreign organisms in mosquitoes. Inactivation of this immune mechanism can result in enhanced survival of malaria parasites (Volz et al., 2006) and filarial worms (Shiao et al., 2001). Melanization can also be elicited by bacteria (Hillyer et al., 2003a, b) and abiotic particles like Sephadex beads (Paskewitz and Riehle, 1994). In Anopheles gambiae, there is clear evidence that several components of the melanization pathway are essential for melanization of both abiotic targets and Plasmodium berghei (Blandin et al., 2004; Paskewitz et al., 2006; Volz et al., 2006; Warr et al., 2006). These studies identified specific serine proteases and a recognition protein as critical upstream regulators of the process. Silencing of serine proteases CLIPB4 and CLIPB8 (Paskewitz et al., 2006; Volz et al. 2006) and TEP1 (Warr et al., 2006) strongly reduce bead and parasite melanization. Other proteins of unknown function (leucine rich repeat immune protein) also regulate the bead and parasite melanization response (Warr et al., 2006; Osta et al., 2004).
Pah, which functions to produce tyrosine from phenylalanine, could be an additional upstream regulator of melanization if stores of this essential substrate are limited. Some evidence supports a role for this enzyme in mosquito immunity. Pah transcript abundance increases following immune challenge with the dog heartworm, Dirofilaria immitis or bacteria in Aedes aegypti (Johnson et al., 2003) and following lipopolysaccharide injection of An. gambiae (Oduol et al., 2000). We also found convincing evidence of upregulation of this gene following bead injection. Knockdown of Pah in Ar. subalbatus or Ae. aegypti was reported to reduce melanization of injected D. immitis, although a dsRNA control was not performed (Infanger et al., 2004). Knockdown of Pah had no effect on bead melanization in An. gambiae. However, examination of bead melanization at times earlier than 24 h might reveal a delay in the process. It is also possible that the PAH protein in An. gambiae was not significantly reduced in spite of strong reduction of the transcript. In Ar. subalbatus subjected to RNAi, the PAH protein appeared unusually stable and did not decrease until after injection of D. immitis (Infanger et al., 2004). Finally, it is possible that the silencing of PAH had no impact on bead melanization but would have a significant impact on melanization of another challenge type. Further investigation will be needed to resolve this issue in An. gambiae.
Enzymes that function downstream of PO have also been investigated in mosquitoes. Biochemical analyses demonstrated the presence of dopachrome conversion enzyme in Ae. aegypti hemolymph and its role in accelerating melanization in vitro (Li et al., 1994). Johnson and colleagues (2001) observed upregulation of transcripts following inoculation of D. immitis in Ae. aegypti. Silencing of Dce in Ar. subalbatus by RNAi resulted in reduction of melanization of D. immitis (Huang et al., 2005b). In An. gambiae, Dce transcripts were slightly upregulated following infection by the human malaria parasite, Plasmodium falciparum (Dong et al., 2006) and following bead injection (this study). A significant reduction of bead melanization occurred after knockdown of Dce. This effect is small relative to that seen for TEP1 and serine proteases. In accordance with the biochemical data, we conclude that Dce accelerates but is not an essential factor for melanization of these abiotic targets in An. gambiae.
For dopa decarboxylase, silencing was accomplished through engineering of Sindbis virus and introduction of antisense virus into Ar. subalbatus (Huang et al., 2005a). Silencing resulted in an initial reduction of melanization of D. immitis microfilariae that had been injected into the hemocoel. However, longer incubation times revealed a progressive increase in melanization. Ddc-silenced Armigeres mosquitoes also exhibited high mortality, abnormal movement and overfeeding, conditions that are attributable to the role of DDC in producing dopamine, a neurotransmitter. We monitored for these effects but, surprisingly, did not observe them for An. gambiae. Quantitative PCR demonstrated a significant rise in the relative expression of Ddc in comparison with actin in Ar. subalbatus mosquitoes 48 h after blood feeding or injection of D. immitis (Huang et al., 2005a). Ddc transcripts increased in An. gambiae following exposure to oxidative stress (Dimopoulos et al., 2002) but not following bead injection (this study). Silencing of Ddc resulted in a small but significant decrease in the percentage of beads that were fully melanized by 24 h.
Our findings, together with those of previous studies, suggest that melanization pathways in mosquito immunity usually use DDC and DCE to accelerate formation of the end products. Since melanization clearly limits the ability of some mosquito species to transmit pathogens (Adak et al., 2006; Christensen et al. 2005; Infanger et al., 2004), further investigation of this complex phenomenon may provide avenues for development of novel control strategies for these parasites.
Acknowledgements
We thank Professor Kyle Willis, University of Wisconsin, Department of Plant Pathology, for his help in establishing and interpreting quantitative PCR protocols. We thank Jun Wang for sharing her expertise in this area as well. Finally, we acknowledge the work of Beth Schadd, who reared all mosquitoes used in these experiments. This work was supported in part by a grant from the National Institutes of Health to SMP (AI46031).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adak T, Singh OP, Nanda N, Sharma VP, Subbarao SK. Isolation of a Plasmodium vivax refractory Anopheles culicifacies strain from India. Trop. Med. Int Health. 2006;11:197–203. doi: 10.1111/j.1365-3156.2005.01556.x. [DOI] [PubMed] [Google Scholar]
- Blandin S, Moita LF, Kocher T, Wilm M, Kafatos FC, Levashina EA. Reverse genetics in the mosquito Anopheles gambiae: targeted disruption of the Defensin gene. EMBO Reports. 2002;3:852–856. doi: 10.1093/embo-reports/kvf180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blandin S, Shiao SH, Moita LF, Janse CJ, Waters AP, Kafatos FC, Levashina EA. Complement-like protein TEP1 is a determinant of vectorial capacity in the malaria vector Anopheles gambiae. Cell. 2004;116:661–670. doi: 10.1016/s0092-8674(04)00173-4. [DOI] [PubMed] [Google Scholar]
- Brunner AM, Yakovlev IA, Strauss SH. Validating internal controls for quantitative plant gene expression studies. BMC Plant Biol. 2004;4:1–7. doi: 10.1186/1471-2229-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen BM, Li J, Chen CC, Nappi AJ. Melanization immune responses in mosquito vectors. Trends Parasitol. 2005;21:192–199. doi: 10.1016/j.pt.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Chun J, Riehle M, Paskewitz SM. Effect of mosquito age and reproductive status on melanization of Sephadex beads in Plasmodium-refractory and susceptible strains of Anopheles gambiae. J. Invert. Pathol. 1995;66:11–17. doi: 10.1006/jipa.1995.1054. [DOI] [PubMed] [Google Scholar]
- Collins FH, Sakai RK, Vernick KD, Paskewitz SM, Seeley DC, Miller L, Collins WE, Campbell CC, Gwadz RW. Genetic selection of a Plasmodium-refractory strain of the malaria vector Anopheles gambiae. Science. 1986;234:607–610. doi: 10.1126/science.3532325. [DOI] [PubMed] [Google Scholar]
- De Jonge HJM, Fehrmann RSN, de Bont ESJM, Hofstra RMW, Gerbens F, Kamps WA, de Vries EGE, van der Zee AGJ, Meerman GJ, Elst AT. Evidence-based selection of housekeeping genes. PLoS ONE. 2007;2:898–910. doi: 10.1371/journal.pone.0000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimopoulos G, Christophides GK, Meister S, Schultz J, White KP, Barillas-Mury C, Kafatos FC. Genome expression analysis of Anopheles gambiae: Responses to injury, bacterial challenge and malaria infection. Proc. Natl. Acad. Sci. USA. 2002;99:8814–8819. doi: 10.1073/pnas.092274999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Aguilar R, Xi Z, Warr E, Mongin E, Dimopoulos G. Anopheles gambiae immune responses to human and rodent Plasmodium parasite species. PLoS Pathogens. 2006;6:1–13. doi: 10.1371/journal.ppat.0020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley NR, Labbe JC, Goldstein B. Using RNA interference to identify genes required for RNA interference. Proc. Natl. Acad. Sci. USA. 2002;99:4191–4196. doi: 10.1073/pnas.062605199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman MJ, Schwartz AM, Paskewitz SM. The role of surface characteristics in eliciting humoral encapsulation of foreign bodies in Plasmodium-refractory and susceptible strains of Anopheles gambiae. J. Insect Physiol. 1998;44:947–954. doi: 10.1016/s0022-1910(98)00056-0. [DOI] [PubMed] [Google Scholar]
- Hillyer JF, Schmidt SL, Christensen BM. Rapid phagocytosis and melanization of bacteria and Plasmodium sporozoites by hemocytes of the mosquito Aedes aegypti. J. Parasitol. 2003a;89:62–69. doi: 10.1645/0022-3395(2003)089[0062:RPAMOB]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Hillyer JF, Schmidt SL, Christensen BM. Hemocyte-mediated phagocytosis and melanization in the mosquito Armigeres subalbatus following immune challenge by bacteria. Cell Tissue Res. 2003b;313:117–127. doi: 10.1007/s00441-003-0744-y. [DOI] [PubMed] [Google Scholar]
- Huang CY, Chou SY, Bartholomay LC, Christensen BM, Chen CC. The use of gene silencing to study the role of dopa decarboxylase in mosquito melanization reactions. Insect Mol. Biol. 2005a;14:237–244. doi: 10.1111/j.1365-2583.2004.00552.x. [DOI] [PubMed] [Google Scholar]
- Huang CY, Christensen BM, Chen CC. Role of dopachrome conversion enzyme in the melanization of filarial worms in mosquitoes. Insect Mol. Biol. 2005b;14:675–682. doi: 10.1111/j.1365-2583.2005.00597.x. [DOI] [PubMed] [Google Scholar]
- Infanger LC, Rocheleau TA, Bartholomay LC, Johnson JK, Fuchs J, Higgs S, Chen CC, Christensen BM. The role of phenylalanine hydroxylase in melanotic encapsulation of filarial worms in two species of mosquitoes. Insect Biochem. Mol. Biol. 2004;34:1329–1338. doi: 10.1016/j.ibmb.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Ji C, Wang Y, Guo X, Hartson S, Jiang H. A pattern recognition serine proteinase triggers the prophenoloxidase activation cascade in the tobacco hornworm, Manduca sexta. J. Biol. Chem. 2004;279:34101–34106. doi: 10.1074/jbc.M404584200. [DOI] [PubMed] [Google Scholar]
- Jiang H, Wang Y, Gu Y, Guo X, Zou Z, Scholz F, Trenczek TE, Kanost MR. Molecular identification of a bevy of serine proteases in Manduca sexta hemolymph. Insect Biochem Mol Biol. 2005;35:931–943. doi: 10.1016/j.ibmb.2005.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JK, Li J, Christensen BM. Cloning and characterization of a dopachrome conversion enzyme from the yellow fever mosquito, Aedes aegypti. Insect Biochem. Mol. Biol. 2001;31:1125–1135. doi: 10.1016/s0965-1748(01)00072-8. [DOI] [PubMed] [Google Scholar]
- Johnson JK, Rocheleau TA, Hillyer JF, Chen CC, Li J, Christensen BM. A potential role for phenylalanine hydroxylase in mosquito immune responses. Insect Biochem. Mol. Biol. 2003;33:345–354. doi: 10.1016/s0965-1748(02)00257-6. [DOI] [PubMed] [Google Scholar]
- Kumar S, Christophides GK, Cantera R, Charles B, Han YS, Meister S, Dimopoulos G, Kafatos FC, Barillas-Mury C. The role of reactive oxygen species on Plasmodium melanotic encapsulation in Anopheles gambiae. PNAS. 2003;100:14139–14144. doi: 10.1073/pnas.2036262100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Zhao X, Christensen BM. Dopachrome conversion activity in Aedes aegypti: significance during melanotic encapsulation of parasites and cuticular tanning. Insect Biochem. Mol. Biol. 1994;24:1043–1049. doi: 10.1016/0965-1748(94)90142-2. [DOI] [PubMed] [Google Scholar]
- Nappi AJ, Christensen BM. Melanogenesis and associated cytotoxic reactions: applications to insect innate immunity. Insect Biochem. Mol. Biol. 2005;35:443–459. doi: 10.1016/j.ibmb.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Oduol F, Xu J, Niare O, Natarajan R, Vernick KD. Genes identified by an expression screen of the vector mosquito Anopheles gambiae display differential molecular immune response to malaria parasites and bacteria. Proc. Natl. Acad. Sci. USA. 2000;97:11397–11402. doi: 10.1073/pnas.180060997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osta MA, Christophides GK, Kafatos FC. Effects of mosquito genes on Plasmodium development. Science. 2004;303:2030–2032. doi: 10.1126/science.1091789. [DOI] [PubMed] [Google Scholar]
- Paskewitz SM, Riehle MA. Response of Plasmodium-refractory and susceptible strains of Anopheles gambiae to inoculated Sephadex beads. Develop. Comp. Immunol. 1994;18:369–375. doi: 10.1016/0145-305x(94)90002-7. [DOI] [PubMed] [Google Scholar]
- Paskewitz SM, Brown MR, Lea AO, Collins FH. Ultrastructure of the encapsulation of Plasmodium cynomolgi (B strain) on the midgut of a refractory strain of Anopheles gambiae. J. Parasitol. 1988;74:432–439. [PubMed] [Google Scholar]
- Paskewitz SM, Reese-Stardy S, Gorman MJ. An easter-like serine protease from Anopheles gambiae exhibits changes in transcript levels following immune challenge. Insect Mol. Biol. 1999;8:329–338. doi: 10.1046/j.1365-2583.1999.83124.x. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004;26:509–515. doi: 10.1023/b:bile.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- Radonic A, Thulke S, Mackay IM, Landt O, Siegert W, Nitsche A. Guideline to reference gene selection for quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2004;313:856–862. doi: 10.1016/j.bbrc.2003.11.177. [DOI] [PubMed] [Google Scholar]
- Rotenberg D, Thompson TS, German TL, Willis DK. Methods for effective real time RT-PCR analysis of virus-induced gene silencing. J. Virol Meths. 2006;138:49–59. doi: 10.1016/j.jviromet.2006.07.017. [DOI] [PubMed] [Google Scholar]
- Salt G. The defence reactions of insects to metazoan parasites. Parasitol. 1963;53:527–642. doi: 10.1017/s0031182000073960. [DOI] [PubMed] [Google Scholar]
- Salt G. The cellular defence reactions of insects. Cambridge Monogr Exp Biol. 1970;16:1–118. [Google Scholar]
- Shiao SH, Higgs S, Adelman Z, Christensen BM, Liu SH, Chen CC. Effect of prophenoloxidase expression knockout on the melanization of microfilariae in the mosquito Armigeres subalbatus. Insect Mol. Biol. 2001;10:315–321. doi: 10.1046/j.0962-1075.2001.00268.x. [DOI] [PubMed] [Google Scholar]
- Sindelka R, Ferjentsik Z, Jonak J. Developmental expression profiles of Xenopus laevis reference genes. Dev Dyn. 2006;235:754–758. doi: 10.1002/dvdy.20665. [DOI] [PubMed] [Google Scholar]
- Tang H, Kambris Z, Lemaitre B, Hashimoto C. Two proteases defining a melanization cascade in the immune system of Drosophila. J. Biol. Chem. 2006;281:28097–28104. doi: 10.1074/jbc.M601642200. [DOI] [PubMed] [Google Scholar]
- True JR. Insect melanism: the molecules matter. Trends Ecol. Evol. 2003;18:640–647. [Google Scholar]
- Vandesompele K, De Preter F, Pattyn B, Poppe N, Van Roy A, De Paepe F, Speleman Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;34:1–11. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz J, Muller HM, Zdanowicz A, Kafatos FC, Osta MA. A genetic module regulates the melanization response of Anopheles to Plasmodium. Cell. Microbiol. 2006;8:1392–1405. doi: 10.1111/j.1462-5822.2006.00718.x. [DOI] [PubMed] [Google Scholar]
- Warr E, Lambrechts L, Koella JC, Bourgouin C, Dimopoulos G. Anopheles gambiae immune responses to Sephadex beads: involvement of anti-Plasmodium factors in regulating melanization. Insect Biochem. Mol. Biol. 2006;36:769–778. doi: 10.1016/j.ibmb.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Zheng L, Wang S, Romans P, Zhao H, Luna C, Benedict MQ. Quantitative trait loci in Anopheles gambiae controlling the encapsulation response against Plasmodium cynomolgi Ceylon. BMC Genetics. 2003;4:16–26. doi: 10.1186/1471-2156-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]