Abstract
Dystrobrevin, a dystrophin-related and -associated protein, has been proposed to be important in the formation and maintenance of the neuromuscular junction. Dystrobrevin coprecipitates with both the acetylcholine receptor complex as well as the dystrophin glycoprotein complex. Although the nature of dystrobrevin’s association with the dystrophin glycoprotein complex remains unclear, it is known that dystrobrevin binds directly to the syntrophins, a heterologous group of dystrophin-associated proteins. Using the yeast two-hybrid system to identify protein–protein interactions, we present evidence for the heterodimerization of dystrobrevin directly with dystrophin. The C terminus of dystrobrevin binds specifically to the C terminus of dystrophin. We further refined this site of interaction to these proteins’ homologous coiled-coil motifs that flank their respective syntrophin-binding sites. We also show that the interaction between the dystrobrevin and dystrophin coiled-coil domains is specific and is not due to a nonspecific coiled-coil domain interaction. From the accumulated evidence of protein–protein interactions presented here and elsewhere, we propose a partially revised model of the organization of the dystrophin-associated glycoprotein complex.
Dystrobrevin, a member of the dystrophin family of proteins, was originally identified from the Torpedo californica electric organ as an 87-kDa phosphoprotein associated with the cytoplasmic face of the postsynaptic membrane (1, 2). It has been postulated that the 87-kDa protein plays a role in synapse formation or stability because it copurifies with acetylcholine receptors from the electric organ membranes. Antibodies raised against 87 kDa show that it is concentrated with acetylcholine receptors at the synaptic region but is also found extrasynaptically at the sarcolemma of both Torpedo electric organ and vertebrate skeletal muscle (2). Furthermore, the 87-kDa protein is also found in association with dystrophin and the 58-kDa syntrophins in the Torpedo electric organ (3).
In mammalian skeletal muscle, dystrophin is found in association with several integral and peripheral membrane proteins, forming a complex known as the dystrophin glycoprotein complex (DGC) (4–6). The syntrophins, as well as a minor component at 94 kDa (A0), copurify with the DGC in rabbit skeletal muscle membrane (4, 6). Based upon the amino acid sequence of the rabbit 94-kDa dystrophin associated protein, A0 appears to be the mammalian homologue of the Torpedo 87-kDa protein (7). The syntrophins are a biochemically heterogeneous group of 58-kDa intracellular membrane-associated proteins (8–10), which have distinct patterns of expression and distribution along the cell membranes and are encoded by a family of three separate genes.
Dystrophin is a member of the spectrin superfamily of actin-binding proteins (11) and has been divided into four general structural domains: a N-terminal actin-binding domain, a central rod region, a cysteine-rich region (CR), and a C-terminal domain (CT) (for a review, see ref. 12). The CR and CT are the domains that interact with the sarcolemmal glycoprotein complex (13, 14). By analogy to the spectrins, dystrophin had been thought to function as a homodimer (15). However, electron microscopy analyses of rotary-shadowed images of dystrophin suggest that dystrophin can exist either as a monomer or a dimer (16, 17). More recent biochemical experiments by Chan and Kunkel (18), as well as observations by Kahana et al. (19), provide further evidence that dystrophin exists as a monomer, based on its inability to homodimerize in vitro and the physical properties of the rod domain, respectively. The CR and CT regions contain several potential binding domains: the WW domain (20, 21), a motif found in signaling molecules and thought to mediate the dystrophin/β-dystroglycan interaction, two EF-hands that potentially bind calcium (11), a ZZ domain thought to be the binding site of calmodulin (22), the syntrophin-binding domain (23–25), as well as a coiled-coil motif of unknown function (26). Such coiled-coil domains are well-characterized mediators of protein–protein interactions (27). Coiled-coil structures consist of two right-handed α-helices wrapped around one another and are found in many proteins including tropomyosin, the keratins, and leucine zipper containing proteins such as the yeast transcription factor GCN4 (27, 28). The sequences are characterized by heptad repeats that form a hydrophobic interface to the helices (27). In addition to the hydrophobic interface, overall electrostatic interactions between the helices are very important (29, 30).
Based on regional homology to dystrophin, three general structural domains comprise dystrobrevin: the CR and CT domains, homologous to the cysteine-rich and C-terminal domains of dystrophin, respectively; and a C-terminal domain unique to dystrobrevin (DUR) (1, 31, 32). In contrast to the single known Torpedo dystrobrevin molecule, mammals have five different splice forms of this protein (31, 32). These various forms include different domains of dystrobrevin and have different tissue distributions.
The syntrophin-binding site on dystrophin has been delineated to exon 74, directly upstream from the coiled-coil motif (23–25). A similar site has also been documented on dystrobrevin, specifically dystrobrevin exons 13 and 14 (amino acids 427–480) (23, 33). In comparison to other dystrophin-associated proteins, the syntrophin content in biochemically purified dystrophin complex appears to be 2-fold higher (4, 6). Two different theories have accounted for this observation: a second syntrophin-binding site on dystrophin or syntrophin homodimerization (24, 25). The existence of a second syntrophin-binding site on dystrophin is suggested by the direct binding of a dystrophin peptide to an ≈60-kDa protein copurifying with the dystrophin complex (24). Likewise, syntrophin binds to a ≈60-kDa protein in blot overlay experiments, suggesting possible homodimerization of the syntrophins (25). An alternative hypothesis has been proposed from direct immunoprecipitation of syntrophins from tissues where dystrobrevin may interact with dystrophin, thus accounting for two molecules of syntrophin for each dystrophin (34).
To gain a better understanding of the interaction of dystrobrevin with the DGC, we searched for as yet unknown binding partners of dystrobrevin by using the yeast two-hybrid system. We focused our attention on the CT domain of dystrobrevin because this domain is known to interact with the syntrophins and contains a coiled-coil motif. This coiled-coil domain is located downstream of the syntrophin-binding site, and hence it is likely to be fully available for a potentially novel protein–protein interaction. With this assay, we isolated several cDNAs from a human skeletal muscle cDNA library that specifically interacted with the dystrobrevin CT domain [exons 11A to 17B (33)]: these included the expected α-syntrophin as well as cDNAs coding for dystrophin. The respective binding sites for dystrobrevin and dystrophin were characterized and the interaction was confirmed by an in vitro coimmunoprecipitation assay. These studies revealed that the proteins interact via their coiled-coil motifs thus confirming a predicted interaction of the coiled-coil motif of dystrophin with A0 (24), the rabbit orthologue of dystrobrevin (7).
EXPERIMENTAL PROCEDURES
Constructs.
Constructs were prepared by PCR amplification of specific regions corresponding to different portions of dystrobrevin and dystrophin with the following primer sets: DTN e11A-15 (cgcgaattcGCGGCCGCGTCGACG; TCTCTGcaGTGGGCTGAGAAGCTTG), DTN e14–16 (CCTGAatTCTCTTTCACCATCGATG; CTCCTGcAGAGCAGACATTCTCTG), DTN e15–17B (CAGGAatTCCAGAGACTTCGGCTAG; ATACATTAGAGCCAAAGCAATGAG), Dys e64–70 (GCTGAatTcAATAATGTCAGATTC; ACAGTCgaCACTGGCAGGTAGCCC), Dys e68–75 (CCCGaatTcCACAGAGTGGCTGC; TTGgTCgAcCAGCTGCCTTAGCC), Dys e74–78 (TTAGAattcGAGGAAAGAGGGGAG; ACTgtCgACATTGTGTCCTCTCTC), Dys e74.5 (AATGAattCATAGATGATGAAC; AATgtcGAcCTGGGCAGGACTACG), Dys e74.5–75.5 (AATGAattCATAGATGATGAAC; GAGtcGACGGCAGTGGGGACAGGCC), Dys e75.5 (CCTGAAtTcATGCCCACCTCTCCC; TTGgTCgAcCAGCTGCCTTAGCC), Dys e75.5–76.5 (CCTGAAtTcATGCCCACCTCTCCC; GGAGtcGACACCGTTGTGCCATTCAC). In each PCR, 100 ng of the plasmid encoding the region of interest was used as template with 50 ng of each primer. To maintain the fidelity of the amplified DNA, we used the Taq extender DNA polymerase (Stratagene) in all amplification reactions. Amplified DNA was cloned into either the pGBT9 and pGAD vectors for the two-hybrid constructs (CLONTECH) or the pMGT and pFHR vectors for expression (23). The dystrophin C-terminal construct C2979, as well as the β1-syntrophin constructs Tβ1S, were as described (23).
Yeast Two-Hybrid Assay.
The HF7c yeast strain, containing two Gal4-inducible reporter genes HIS3 and LacZ, was transformed simultaneously with both a DNA-binding domain plasmid encoding dystrobrevin exons 11A-17B (pGBT9-DTN-e11A-17B) (trp−) and a transactivating domain library encoding human adult skeletal muscle cDNAs (leu−) (CLONTECH). Six and a half million independent double transformants were plated onto selection plates lacking tryptophan, leucine, and histidine (TDO plates) and supplemented with 5 mM 3-amino-1,2,4-triazole (Sigma). After 8 days at 30°C, colonies grown on this selective medium were replicated on a TDO plate and incubated overnight at 30°C. The His+ colonies were then lifted onto 3-mm Whatman paper that was then immersed in liquid nitrogen for a few seconds and layered onto a second piece of Whatman paper, humidified with 3 ml of Z buffer [60 mM Na2HPO4/40 mM NaH2PO4/10 mM KCl/1 mM MgSO4/30 mM 2-mercapthoethanol/5-bromo-4 chloroindolyl-β-d-galactoside (X-Gal) at a final concentration of 2 mg/ml] for 8 h at room temperature. Positive clones were rescued and tested for specificity by retransformation into HF7c either with dystrobrevin or with a extraneous target. We verified that all hybrid DNA-binding domain fusion constructs lacked transcriptional self-activation by cotransforming the HF7c yeast strain with each construct and a control vector encoding the transactivating domain.
In Vitro Translation and Immunoprecipitation Assay.
Dystrobrevin, dystrophin, and syntrophin protein fragments were produced by in vitro transcription/translation of the expression vector pMGT by the TnT T7 coupled reticulocyte lysate system (Promega) in a reaction volume of 50 μl as per the manufacturer’s protocol. Some reactions were carried out in a reaction buffer containing 0.2 μCi (4 μl) of l-[U-14C]leucine (>300 mCi/mmol; 1 Ci = 37 GBq; Amersham).
Coprecipitation assays were performed on proteins taken directly from the translation reactions, combining 5 μl of each of the proteins of interest and incubating them together in 20 μl of TBST buffer (10 mM Tris, pH 8.0/0.1% Tween-20/150 mM NaCl). After 1.5–2 h of incubation on ice, 20 μl of antibody (1:10 dilution in TBST) directed against one of the proteins (the cognate protein) was added. Four different antibodies were used in the immunoprecipitation experiments: antibodies raised against the N terminus or the C terminus of dystrophin (Dys1 and Dys2, respectively; NovoCastra, Newcastle, U.K.), a mAb raised against Torpedo syntrophin (SYN1351) which has been described (35) (gift from Sealock Laboratory, Chapel Hill, NC), and an antidystrobrevin antibody (DB433) prepared against a peptide corresponding to mouse dystrobrevin as described (31) (gift from the Froehner Laboratory, Chapel Hill, NC). After a 1-h incubation on ice, interacting proteins were precipitated by the addition of 50 μl of a 50% suspension of protein G-Sepharose (Sigma) for 30 min on ice. The protein–antibody–bead complexes were pelleted at 10,000 × g for 2 min at room temperature, and the supernatant was removed. The beads were then washed three times with 1 ml of TBST buffer, and the pellet was resuspended in 10 μl of 2× loading buffer and stored at −20°C.
All samples were separated by electrophoresis on SDS/PAGE gels, and protein sizes were compared with 14C-methylated high molecular weight standards (10–50 μCi/mg) (Amersham). Gels were dried onto 3-mm Whatman paper and exposed to a storage phosphor plate for 1–7 days, which was then scanned by a PhosphorImager (Molecular Dynamics) and analyzed with imagequant software (Molecular Dynamics).
RESULTS
To search for cDNA clones encoding proteins that interact with dystrobrevin, we used the two-hybrid system, an assay that detects physical interactions between protein domains that are expressed as fusion proteins in yeast. Interacting fusion proteins combine to form a DNA-binding and a transcriptional activation dimer that induces synthesis from a reporter gene (36). Two different C-terminal dystrobrevin-2 constructs were employed, both encoding exons 11A through 17B, but one (construct B) excluding exon 12, a normally occurring splice form of dystrobrevin (33). The dystrobrevin constructs were fused to the DNA-binding domain of Gal4 in the pGBT9 plasmid and used as a target in the two-hybrid screen. The yeast strain HF7c, containing the two Gal4-responsive reporter genes HIS3 and LacZ, was cotransformed with the dystrobrevin hybrid and a human skeletal muscle cDNA fused to the activating domain of Gal4 in the pGAD plasmid. Six and a half million independent yeast transformants were tested for activation of the histidine marker by selection on His− medium. Fifty-six yeast colonies were able to grow on deprived medium, and were therefore tested for β-galactosidase activity; 20 of the 56 yeast colonies showed β-galactosidase activity. The DNA from each of the 20 colonies was isolated and sequenced at both ends to determine clone identity. Both dystrobrevin constructs gave equivalent resulting clones. Ten cDNAs showed identity to human α-syntrophin, three cDNAs showed identity to human dystrophin. An additional seven cDNAs including several novel expressed tag sequences were also identified and are currently being characterized.
The majority of the cDNAs isolated encoded α-syntrophin, thus confirming previous results and serving as an internal control for the yeast two-hybrid system. The three dystrophin clones each encoded a different, but overlapping portion of dystrophin. The three dystrophin cDNAs (DYS e61–79, DYS e56–79, and DYS e51–79) were completely sequenced to ensure that there was no chimeric sequence that might account for the transcriptional activation of the histidine and LacZ genes. DYS e61–79 was found to contain dystrophin nucleotides 9,250–13,972, encoding the last 14 amino acids of dystrophin exon 60, exons 61–79 and the entire 3′ untranslated region (UTR). DYS e56–79 contains dystrophin nucleotides 8,431–12,994 encoding exons 56–79, and a portion 3′ UTR, whereas DYS e51–79 encodes dystrophin nucleotides 7,513–13,852, corresponding to exons 51–79, and a portion of the 3′ UTR. These cDNAs are three independent clones which have a common region of overlap of the dystrophin protein, encoded by exons 61–79. This region corresponds to a portion of the last central rod repeat, as well as the CR and the CT domains of dystrophin.
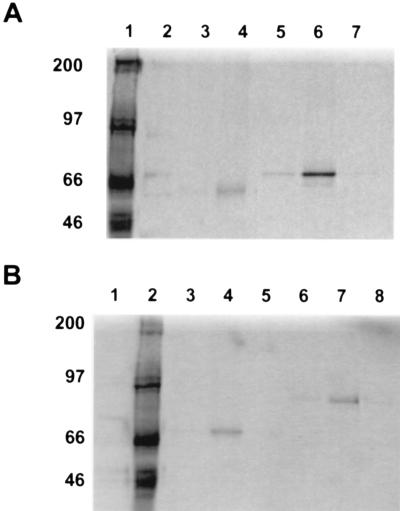
To confirm the apparent dystrobrevin-dystrophin interaction, the C terminus of dystrophin, β1-syntrophin, and a polypeptide corresponding to the full ORF of dystrobrevin-2 (DTN-2) were translated in an in vitro transcription/translation assay, and each was assayed in various combinations with the others for interactions by immunoprecipitation. The dystrophin construct that encompasses nucleotides 9,148–11,263 (amino acids 2,980–3,685) (C2979) gives rise to an 84-kDa protein product (23). This corresponds to dystrophin exons 60–79 producing a polypeptide encoding an equivalent region to the two-hybrid DYS e61–79 construct; the β1-syntrophin construct encodes the full-length β1-syntrophin (Tβ1-S-41, amino acids 1–538), and produces a 58-kDa translated product (23); and the full-length dystrobrevin-2 construct encodes exons 1–17B (amino acids 1–567) and gives rise to a 68-kDa translated product. The conditions of the coprecipitation were optimized by using the known interactions of β1-syntrophin and the dystrophin polypeptide C2979 (data not shown) as well as the interaction of β1-syntrophin and the dystrobrevin-2 polypeptide (Fig. 1). Following immunoprecipitation of the complex with protein-G Sepharose and an antibody specific to either dystrobrevin (DB433) or syntrophin (SYN1351), the samples were subjected to SDS/PAGE (Fig. 1A, lanes 4 and 6, respectively). The antibody directed against dystrobrevin coprecipitated syntrophin in the presence of dystrobrevin-2 (lane 4). Conversely, the antibody directed against syntrophin was capable of precipitating dystrobrevin-2 in the presence of syntrophin (lane 6). Specificity was documented by withholding from the reaction either the probe protein (lanes 3 and 7) or the antibody directed against the cognate protein (lanes 2 and 5).
Figure 1.
Dystrobrevin–syntrophin and dystrobrevin–dystrophin interactions by coimmunoprecipitation. In vitro translated proteins were incubated together and precipitated with a specific antibody in the presence of G-Sepharose. The pellet was washed three times and resuspended in sample buffer for analysis on SDS/PAGE. The radiolabeled translated products were detected by autoradiography (see Experimental Procedures). (A) Lanes: 1, [14C]methylated molecular weight standards; 2, translated radiolabeled β1-syntrophin incubated with dystrobrevin-2 alone; 3, translated radiolabeled β1-syntrophin incubated with an antidystrobrevin antibody (AB433) alone; 4, coprecipitation of dystrobrevin-2 (DTN-2) with translated radiolabeled β1-syntrophin (Tβ1-S), with an antidystrobrevin antibody (AB433); 5, translated radiolabeled dystrobrevin-2 incubated with an antisyntrophin antibody (SYN1351) alone; 6, coprecipitation of β1-syntrophin (Tβ1-S) with translated radiolabeled dystrobrevin-2 (DTN-2), with an antisyntrophin antibody (SYN1351); 7, translated radiolabeled dystrobrevin-2 incubated with β1-syntrophin alone. A background of nonspecific aggregation of β1-syntrophin, dystrobrevin, or dystrophin was seen regardless of whether a specific antibody was used as has been previously reported. This background is variable from different experiments but is never higher than the specific coprecipitation reactions (37). The dystrobrevin–dystrophin coprecipitation was accomplished a minimum of 20 times, and no significant differences were observed. (B) Lanes: 1, translated radiolabeled dystrobrevin-2 incubated with C-terminal dystrophin polypeptide (C2979) alone; 2, [14C]methylated molecular weight standards; 3, translated C-terminal dystrophin polypeptide (C2979) and radiolabeled dystrobrevin-2 incubated with a nonspecific antibody to the N terminus of dystrophin (Dys1); 4, coprecipitation of C-terminal dystrophin polypeptide (C2979) with translated radiolabeled dystrobrevin-2 (DTN-2), by using an antibody against dystrophin C-terminus (Dys2); 5, translated radiolabeled dystrobrevin-2 incubated with Dys2 antibody alone; 6, translated radiolabeled dystrophin polypeptide (C2979) incubated with dystrobrevin-2 alone; 7, coprecipitation of dystrobrevin-2 (DTN-2) with translated radiolabeled dystrophin polypeptide(C2979), by using an antidystrobrevin antibody (AB433); 8, translated radiolabeled dystrophin polypeptide (C2979) incubated with AB433 alone.
By using the same conditions under which known interactions were documented, the dystrobrevin–dystrophin interaction was verified. An antibody directed against the C-terminal region of dystrophin (Dys2) coprecipitated dystrobrevin-2 in the presence of dystrophin (Fig. 1B, lane 4). An antibody directed against dystrobrevin (DB433) was alternatively able to coprecipitate dystrophin in the presence of dystrobrevin (Fig. 1B, lane 7). Again, by withholding from the reaction either respective cognate proteins (lanes 5 and 8) or antibodies (lanes 1 and 6), or by adding an antibody directed against the amino terminus of dystrophin (lane 3), the complex failed to precipitate, thus confirming the interaction between dystrobrevin and dystrophin uncovered by the two-hybrid screen.
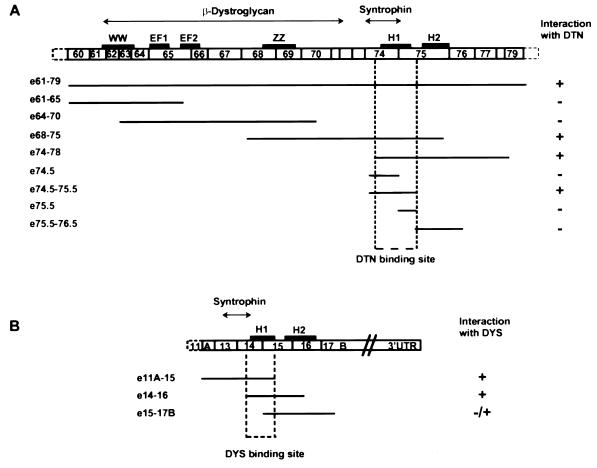
To further characterize the structural requirements of the dystrobrevin–dystrophin interaction, we created a set of progressively smaller constructs for dystrophin and dystrobrevin and tested these for interactions with each larger construct in the yeast two-hybrid assay (Fig. 2). The minimum region of overlap for binding between the two proteins was found to be between amino acids 3,501 and 3,596 of dystrophin, encoding the last 17 amino acids of exon 74, and all the amino acids of exon 75 (Fig. 2A). This region corresponds to the coiled-coil domain of dystrophin (26). To further delineate the binding site of dystrobrevin on dystrophin, we created additional smaller constructs to divide the helical domain of dystrophin in half and found that dystrobrevin binds dystrophin at the first set of heptad repeats (Figs. 2A and 3), particularly at amino acids 3,501–3,541. The analogous dystrophin-binding site on dystrobrevin was located via the same strategy between amino acids 457 and 497, corresponding to the last 24 amino acids of exon 14, and the first 17 amino acids of exon 15 (Figs. 2B and 3). This region contains the first five heptad repeats of dystrobrevin’s coiled-coil domain (32).
Figure 2.
Schematic diagram of dystrophin and dystrobrevin overlapping peptide fusion constructs used in the two hybrid assay to map the narrowest region of interaction. (A) The open boxes represent a schematic of dystrophin C-terminal exons showing the domain organization [WW domain, calcium-binding EF hand (EF1 and EF2), ZZ domain, and the coiled-coil motif (H1 and H2)]. The regions for β-dystroglycan and syntrophin binding are indicated above (double ended arrow horizontal lines). Each fusion construct tested in the two-hybrid assay for interaction with dystrobrevin (DTN) is schematically represented below. The status of interaction with dystrobrevin (+ or −) as determined by the transcriptional activity of both Gal4-inducible histidine and LacZ is indicated (see Experimental Procedures). The dystrobrevin-binding site on dystrophin was delineated to the first helix of the coiled-coil motif (H1). (B) The open boxes represent a schematic of dystrobrevin-2 C-terminal exons showing the domain organization and the coiled-coil motif (H1 and H2). The syntrophin-binding site is indicated above (double-ended arrow horizontal line). Each fusion construct tested in the two-hybrid assay for interaction with dystrophin is schematically represented below. The status of interaction with dystrophin (+ or −) as determined by the transcriptional activity of both Gal4-inducible histidine and LacZ is indicated. The dystrophin-binding site on dystrobrevin was delineated to the first helix of the coiled-coil motif (H1).
Figure 3.
Sequence alignment of part of the C terminus of the dystrophin (amino acids 3,474–3,603) and dystrobrevin (amino acids 436–551) proteins. The letters a–g designate the positions of residues within the heptad of the coiled-coil domain. The dystrobrevin-binding site on dystrophin is represented by a single underline, whereas the dystrophin-binding site on dystrobrevin is represented by a double underline. The respective pIs of the coiled-coil domains are given below.
Dystrobrevin and dystrophin are related proteins. Therefore, the specificity of the coiled-coil heterodimerization was determined by testing the possibility of homodimerization of both dystrophin and dystrobrevin in the two-hybrid system. Cotransformation of the HF7c strain with a Gal4 DNA-binding domain–DYS e61–79 fusion protein and a Gal4 activation domain–DYS e61–79 fusion protein lacked Gal4-induced transcriptional activation. Similarly, cotransformation of the HF7c strain with a Gal4 DNA-binding domain–DTN e11A–17B fusion protein and a Gal4 activation domain–DTN e11A–17B fusion protein yielded no transcriptional activation. Therefore, neither the C terminus of dystrophin nor the C terminus of dystrobrevin homodimerize in the yeast two-hybrid system. Furthermore, the respective pI of the coiled-coil domains of dystrophin and dystrobrevin are 5.44 and 8.56 (Fig. 3). Therefore, destabilizing electrostatic interactions in the respective homodimers would favor the heterodimerization of the two proteins (29, 30).
DISCUSSION
There are several lines of evidence which suggest that dystrobrevin and dystrophin interact in vivo. Dystrobrevin and dystrophin not only colocalize at the sarcolemma and neuromuscular junction in skeletal muscle (2, 31), but also copurify biochemically (1). Dystrobrevin has been shown to interact directly with the dystrophin-associated syntrophins, a multigene family of modular adapter proteins thought to be involved in signaling (23). By blot overlay, Ozawa and colleagues (7, 24) have shown that a peptide corresponding to the coiled-coil motif of dystrophin binds to a protein A0, which has subsequently been shown to be the rabbit orthologue of dystrobrevin. Furthermore, Froehner and colleagues (34) observed that complexes immunoprecipitated with an antibody directed against the linker sequence between the coiled-coils of dystrobrevin has a reduced content of dystrophin as compared with preparations from other dystrobrevin antibodies. These results and the evidence that dystrophin binds A0, lead Froehner and colleagues (34) to hypothesize that dystrobrevin might directly interact with dystrophin.
We present evidence from two independent in vitro methods for the direct association between dystrobrevin and dystrophin. Initially, an interaction between the C-terminal domain of dystrophin (exons 61–79) and the homologous C-terminal domain of dystrobrevin (exons 11A–17B) was detected by using the two-hybrid assay. This was substantiated by showing that an antibody specific to dystrophin is able to coprecipitate dystrobrevin in the presence of dystrophin (Fig. 1B, lane 4). Analogously, an antibody specific to dystrobrevin is able to coprecipitate dystrophin in the presence of dystrobrevin (Fig. 1B, lane 7). The structural requirements of the dystrobrevin–dystrophin interaction were further mapped by two-hybrid analysis to the proteins’ respective coiled-coil motifs, regions flanking the syntrophin-binding sites. This binding domain was further delineated to the first helix of the coiled-coil motif, specifically to dystrophin amino acids 3,501–3,541 and dystrobrevin amino acids 457–497.
Coiled-coil interactions occur in both homodimerization and heterodimerization binding (27). In previous studies with blot overlay and coprecipitation experiments, we had detected no evidence of dystrophin homodimerization (18). To be sure, we tested the possibility of homodimerization of both dystrophin and dystrobrevin through their coiled-coil motif in the two-hybrid system. No transcriptional activation of the reporter gene was detected, confirming that neither the C terminus of dystrophin nor the C terminus of dystrobrevin homodimerize. This is consistent with the heterologous dimerization of dystrophin and dystrobrevin via their homologous coiled-coil domains. This result signifies that this interaction is specific for dystrobrevin’s and dystrophin’s coiled-coil motifs, rather than simply due to a nonspecific coiled-coil interaction. Furthermore, the favorable electrostatic interactions in the heterodimer as predicted by the respective sequences are likely helping to determine the pairing specificity (29, 30).
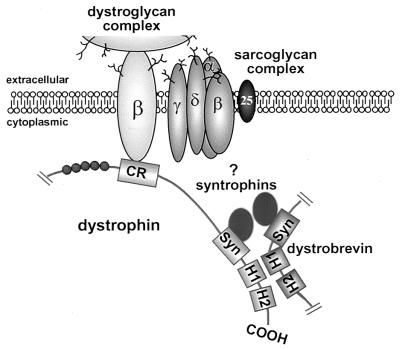
These results provide a working model for the DGC (Fig. 4), consistent with that presented by Peters et al. (34). The DGC provides a link from the intracellular actin cytoskeleton to the extracellular matrix (4–6), and through this connection, dystrophin is thought to contribute to the maintenance of the membrane during cycles of muscle contraction and relaxation. Based on analogy to the spectrin molecules, dystrophin had been initially suggested to functions as a homodimer (15). Recent biochemical experiments, however, have provided evidence that dystrophin is unable to dimerize in vitro (18). In addition, we were unable to detect any dystrophin–dystrophin interactions in the yeast two-hybrid system. Dystrobrevin’s direct interaction with dystrophin further reinforces the model of dystrophin working as a monomer. In this partially new model, a single dystrophin molecule binds actin at its N terminus, interacts with the dystroglycan complex via its CR, binds a syntrophin molecule through exon 74, and finally heterodimerizes with dystrobrevin via the first helix of its coiled-coil motif (Fig. 4).
Figure 4.
Partial schematic model of the dystrophin glycoprotein complex. Schematic diagram of some of the proposed intermolecular interactions of dystrophin. Dystrophin as a monomer associates at its N terminus with actin (data not shown), and the dystrophin-associated glycoprotein complex near the C terminus. Dystrophin interacts with the dystroglycan complex via its CR, binds a syntrophin molecule through exon 74, and finally heterodimerizes with dystrobrevin via the first helix of its coiled-coil motif. Together, dystrobrevin and dystrophin are proposed to recruit two syntrophins per complex. The pair of syntrophins (α-, β1-, or β2-syntrophin) present in the complex is unclear (as indicated by ?) as no intrinsic binding specificity can be allocated to either dystrophin or dystrobrevin (see Discussion).
The syntrophin content in purified dystrophin complexes has been shown to be twice the amount of other dystrophin associated proteins by stoichiometric analysis (4, 6). Two different theories have previously explained this observation: the presence of a second syntrophin-binding site on dystrophin as well as the possible dimerization of the syntrophins (24, 25). In agreement with the observation made by Froehner and colleagues (34), we believe that the presence of two syntrophins could be explained by interactions with dystrobrevin. Furthermore, the simultaneous binding of dystrobrevin to syntrophin and dystrophin supports observations detected in Δ71–74 transgenic mdx mice (38, 39). Although these mice express a dystrophin transgene which lacks the syntrophin-binding site, syntrophin immunostaining was restored. However, the expressed dystrophin transgene includes the dystrobrevin-binding site, thus allowing dystrobrevin localization along the sarcolemma which would in turn bind syntrophin. Indeed, Peters et al. (34) find that dystrobrevin immunostaining is restored in the Δ71–74 transgenic mdx mice. We therefore believe that the straightforward explanation for the presence of two syntrophins per complex is through parallel syntrophin interactions with both dystrophin and dystrobrevin. Dystrobrevin and dystrophin, in turn, interact through their coiled-coil domains and together recruit two syntrophins per complex.
The syntrophins present different tissue distribution as well as expression specificity in skeletal muscle (25, 34, 40). It is tempting to suggest that one of dystrobrevin’s roles is to allocate this specificity by controlling which combination of syntrophins exist at a particular location in the cell, because the different dystrobrevin isoforms also exhibit different tissue specificity (32, 41). Our in vitro data, however, do not show any specificity of preferential binding of dystrobrevin for any of the three syntrophins. It is therefore probable that this intrinsic specificity arises in vivo from posttranslational modifications such as phosphorylation.
It is not clear which isoform of dystrobrevin associates with the dystrophin glycoprotein complex in vivo. The in vitro interaction studies presented here were performed with the dystrobrevin-2 isoform. It is clear, however, that dystrobrevin-3 is unlikely to interact with the DGC because it lacks both the syntrophin and the dystrophin-binding domains. Therefore, an interaction with the dystrophin glycoprotein complex cannot be the only function of the dystrobrevins. To elicit clues as to the other functions for this family of molecules, we are currently searching for proteins that interact with the CR, as well as the DUR domains of dystrobrevin by using the two-hybrid assay.
Acknowledgments
We are grateful to Yiu-Mo Chan for continual advice and with his help interpreting the coprecipitation data. We also wish to thank Dick Bennett, Julie Scarfo, and Ivan Guerrero for their assistance with the sequencing. Many thanks to the Kunkel Lab and Mary-Alice Abbott for reviewing the manuscript insightfully. The project described was supported by Grant NS23740 from the National Institute of Neurological Disorders and Stroke (to L.M.K.) L.M.K. is an Investigator of the Howard Hughes Medical Institute.
ABBREVIATIONS
- DGC
dystrophin glycoprotein complex
- CR
cysteine-rich region
- CT
C-terminal domain
- DUR
dystrobrevin unique region
- DTN
dystrobrevin
- Dys
dystrophin
References
- 1.Wagner K R, Cohen J B, Huganir R L. Neuron. 1993;10:511–522. doi: 10.1016/0896-6273(93)90338-r. [DOI] [PubMed] [Google Scholar]
- 2.Carr C, Fischbach G D, Cohen J B. J Cell Biol. 1989;109:1753–1764. doi: 10.1083/jcb.109.4.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butler M H, Douville K, Murnane A A, Kramarcy N R, Cohen J B, Sealock R, Froehner S C. J Biol Chem. 1992;267:6213–6218. [PubMed] [Google Scholar]
- 4.Ervasti J M, Campbell K P. Cell. 1991;66:1121–1131. doi: 10.1016/0092-8674(91)90035-w. [DOI] [PubMed] [Google Scholar]
- 5.Ibraghimov-Beskrovnaya O, Ervasti J M, Leveille C J, Slaughter C A, Sernett S W, Campbell K P. Nature (London) 1992;355:696–702. doi: 10.1038/355696a0. [DOI] [PubMed] [Google Scholar]
- 6.Yoshida M, Ozawa E. J Biochem (Tokyo) 1990;108:748–752. doi: 10.1093/oxfordjournals.jbchem.a123276. [DOI] [PubMed] [Google Scholar]
- 7.Yoshida M, Yamamoto H, Noguchi S, Mizuno Y, Hagiwara Y, Ozawa E. FEBS Lett. 1995;367:311–314. doi: 10.1016/0014-5793(95)00574-s. [DOI] [PubMed] [Google Scholar]
- 8.Adams M E, Butler M H, Dwyer T M, Peters M F, Murnane A A, Froehner S C. Neuron. 1993;11:531–540. doi: 10.1016/0896-6273(93)90157-m. [DOI] [PubMed] [Google Scholar]
- 9.Ahn A H, Yoshida M, Anderson M S, Feener C A, Selig S, Hagiwara Y, Ozawa E, Kunkel L M. Proc Natl Acad Sci USA. 1994;91:4446–4450. doi: 10.1073/pnas.91.10.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang B, Ibraghimov-Beskrovnaya O, Moomaw C R, Slaughter C A, Campbell K P. J Biol Chem. 1994;269:6040–6044. [PubMed] [Google Scholar]
- 11.Koenig M, Monaco A P, Kunkel L M. Cell. 1988;53:219–226. doi: 10.1016/0092-8674(88)90383-2. [DOI] [PubMed] [Google Scholar]
- 12.Sadoulet-Puccio H M, Kunkel L M. Brain Pathol. 1996;6:25–35. doi: 10.1111/j.1750-3639.1996.tb00780.x. [DOI] [PubMed] [Google Scholar]
- 13.Suzuki A, Yoshida M, Yamamoto H, Ozawa E. FEBS Lett. 1992;308:154–160. doi: 10.1016/0014-5793(92)81265-n. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki A, Yoshida M, Hayashi K, Mizuno Y, Hagiwara Y, Ozawa E. Eur J Biochem. 1994;220:283–292. doi: 10.1111/j.1432-1033.1994.tb18624.x. [DOI] [PubMed] [Google Scholar]
- 15.Koenig M, Kunkel L M. J Biol Chem. 1990;265:4560–4566. [PubMed] [Google Scholar]
- 16.Fabbrizio E, Harricane M C, Pons F, Leger J, Mornet D. Biol Cell. 1992;76:167–174. doi: 10.1016/0248-4900(92)90209-j. [DOI] [PubMed] [Google Scholar]
- 17.Pons F, Augier N, Heilig R, Leger J, Mornet D, Leger J J. Proc Natl Acad Sci USA. 1990;87:7851–7855. doi: 10.1073/pnas.87.20.7851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan Y, Kunkel L M. FEBS Lett. 1997;410:153–159. doi: 10.1016/s0014-5793(97)00454-7. [DOI] [PubMed] [Google Scholar]
- 19.Kahana E, Flood G, Gratzer W B. Cell Motil Cytoskeleton. 1997;36:246–252. doi: 10.1002/(SICI)1097-0169(1997)36:3<246::AID-CM5>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 20.André B, Springael J Y. Biochem Biophys Res Commun. 1994;205:1201–1205. doi: 10.1006/bbrc.1994.2793. [DOI] [PubMed] [Google Scholar]
- 21.Bork P, Sudol M. Trends Biochem Sci. 1994;19:531–533. doi: 10.1016/0968-0004(94)90053-1. [DOI] [PubMed] [Google Scholar]
- 22.Ponting C P, Blake D J, Davies K E, Kendrick-Jones J, Winder S J. Trends Biochem Sci. 1996;21:11–13. [PubMed] [Google Scholar]
- 23.Ahn A H, Kunkel L M. J Cell Biol. 1995;128:363–371. doi: 10.1083/jcb.128.3.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki A, Yoshida M, Ozawa E. J Cell Biol. 1995;128:373–381. doi: 10.1083/jcb.128.3.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang B, Jung D, Rafael J A, Chamberlain J S, Campbell K P. J Biol Chem. 1995;270:4975–4978. doi: 10.1074/jbc.270.10.4975. [DOI] [PubMed] [Google Scholar]
- 26.Blake D J, Tinsley J M, Davies K E, Knight A E, Winder S J, Kendrick-Jones J. Trends Biochem Sci. 1995;20:133–135. doi: 10.1016/s0968-0004(00)88986-0. [DOI] [PubMed] [Google Scholar]
- 27.Lupas A. Trends Biol Sci. 1996;21:375–382. [PubMed] [Google Scholar]
- 28.Crick F H C. Acta Crystallogr. 1953;6:689–697. [Google Scholar]
- 29.Zhou N E, Kay C M, Hodges R S. J Mol Biol. 1994;7:500–512. doi: 10.1006/jmbi.1994.1250. [DOI] [PubMed] [Google Scholar]
- 30.O’Shea E K, Lumb K J, Kim P S. Curr Biol. 1993;3:658–667. doi: 10.1016/0960-9822(93)90063-t. [DOI] [PubMed] [Google Scholar]
- 31.Blake D J, Nawrotzki R, Peters M F, Froehner S C, Davies K E. J Biol Chem. 1996;271:7802–7810. doi: 10.1074/jbc.271.13.7802. [DOI] [PubMed] [Google Scholar]
- 32.Sadoulet-Puccio H M, Khurana T S, Cohen J B, Kunkel L M. Hum Mol Genet. 1996;5:489–496. doi: 10.1093/hmg/5.4.489. [DOI] [PubMed] [Google Scholar]
- 33.Sadoulet-Puccio H M, Feener C A, Schaid D J, Thibodeau S N, Michels V V, Kunkel L M. Neurogenetics. 1997;1:37–42. doi: 10.1007/s100480050006. [DOI] [PubMed] [Google Scholar]
- 34.Peters M F, Adams M E, Froehner S C. J Cell Biol. 1997;138:81–93. doi: 10.1083/jcb.138.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Froehner S C, Murnane A A, Tobler M, Peng H B, Sealock R. J Cell Biol. 1987;104:1633–1646. doi: 10.1083/jcb.104.6.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fields S, Song O. Nature (London) 1989;340:245–256. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 37.Ahn A H, Feener C A, Gussoni E, Yoshida M, Ozawa E, Kunkel L M. J Biol Chem. 1996;271:2724–2730. doi: 10.1074/jbc.271.5.2724. [DOI] [PubMed] [Google Scholar]
- 38.Rafael J A, Sunada Y, Cole N M, Campbell K P, Faulkner J A, Chamberlain J S. Hum Mol Genet. 1994;3:1725–1733. doi: 10.1093/hmg/3.10.1725. [DOI] [PubMed] [Google Scholar]
- 39.Rafael J A, Cox G A, Corrado K, Jung D, Campbell K P, Chamberlain J S. J Cell Biol. 1996;134:93–102. doi: 10.1083/jcb.134.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peters M F, Kramarcy N R, Sealock R, Froehner S C. NeuroReport. 1994;5:1577–1580. [PubMed] [Google Scholar]
- 41.Ambrose H J, Blake D J, Nawrotzki R A, Davies K E. Genomics. 1997;39:359–369. doi: 10.1006/geno.1996.4515. [DOI] [PubMed] [Google Scholar]