Abstract
Chromium(VI) compounds (e.g. chromates) are cytotoxic, mutagenic, and potentially carcinogenic. The reduction of Cr(VI) can yield reactive intermediates such as Cr(V) and reactive oxygen species. Bronchial epithelial cells are the primary site of pulmonary exposure to inhaled Cr(VI) and are the primary cells from which Cr(VI)-associated human cancers arise. BEAS-2B cells were used here as a model of normal human bronchial epithelium for studies on the reductive activation of Cr(VI). Cells incubated with Na2CrO4 exhibited two Cr(V) ESR signals, g = 1.979 and 1.985, which persisted for at least one hour. The g = 1.979 signal is similar to that generated in vitro by human microsomes and by proteoliposomes containing P450 reductase and cytochrome b5. Unlike many cells in culture, these cells continued to express P450 reductase and cytochrome b5. Studies with the non-selective thiol oxidant diamide indicated that the g = 1.985 signal was thiol-dependent whereas the g = 1.979 signal was not. Pretreatment with phenazine methosulfate eliminated both Cr(V) signals suggesting that Cr(V) generation is largely NAD(P)H-dependent. ESR spectra indicated that a portion of the Cr(VI) was rapidly reduced to Cr(III). Cells incubated with an insoluble chromate, ZnCrO4, also generated both Cr(V) signals, whereas Cr(V) was not detected with insoluble PbCrO4. In clonogenic assays, the cells were very sensitive to Na2CrO4 and ZnCrO4, but considerably less sensitive to PbCrO4.
Keywords: chromate, chromium(V), BEAS-2B cells, thiol, diamide
1. Introduction
While chromium (Cr) is naturally present in a variety of environments in small amounts, the major human exposures are from industrial use and release, including the production and use of chromate pigments, stainless steel machining and welding, zinc chromate primer paints, chrome plating, corrosion inhibition, leather tanning, and others. Contact with industrial sources has provided valuable information on the numerous toxic effects associated with exposure to hexavalent chromium, Cr(VI) [1–8]. Since greater than 105 tons of Cr are released annually into the environment, and since Cr is a significant contaminant at many sites (Superfund and other) across the U.S.A. [9–11], environmental exposure is of increasing concern.
The predominant stable oxidation states are Cr(VI) and Cr(III). Cr(III) compounds are generally insoluble and do not easily cross cell membranes [12], whereas several Cr(VI) compounds are soluble and can be transported into cells via an anion carrier [13]. Some industrially used chromates are water-insoluble but are still implicated in toxicity. In vitro studies have implicated Cr(VI) as a predisposing factor to Cr-induced genotoxicity [14–18]. Electron donors that transfer three electrons in a single step have not been identified in biological systems, so the reduction of Cr(VI) to Cr(III) must proceed stepwise through Cr(V) and/or Cr(IV) which are reactive Cr intermediates. These reactive Cr intermediates, and their potential to generate reactive oxygen species, are likely important components in the toxicity resulting from Cr(VI) exposure [17, 19–23].
Different intracellular Cr(VI) reductants could result in the generation of distinct types of reactive species, each mediating particular types of damage. Therefore, it is important to examine the types of reactive species generated in appropriate model systems. Several possible Cr(VI) reductants have been identified, including ascorbate, cysteine, glutathione, glutathione reductase, rat microsomal enzymes, and others [24–29]. In human systems, lung and liver microsomes generate Cr(V) during Cr(VI) reduction [30]. Purified human microsomal enzymes (P450 reductase plus cytochrome b5) similarly generate Cr(V) [31, 32].
Animal models have been used for several studies on the reductive activation of Cr(VI), but some key differences between humans and rodents have been reported [33, 34], underscoring the need for more studies with human systems. Human Jurkat cells (derived from human T cell leukemia) exposed to 2 mM dichromate generate Cr(V) [35]. However, these cells are not targets of primary exposure, which is often the inhalation of Cr-containing dusts, particles, and fumes. Therefore, respiratory effects of Cr (pulmonary fibrosis, chronic bronchitis, and lung cancer) are of particular concern [3, 7, 36–39]. In the lung, bronchial epithelial cells line the airways and are therefore directly exposed to inhaled chromium; these epithelial cells are major sites of the formation of associated pulmonary tumors. Because the intracellular reduction of Cr(VI) is directly linked to its toxic effects, a complete understanding of the reductive activation of Cr(VI) in these epithelial cells is essential.
The goal of these studies was to determine if normal human bronchial epithelial cells generate Cr(V) when exposed to various chromate compounds, and to provide insights into the nature of the Cr(V) species generated and their potential links to possible cellular reducing systems.
2. Experimental
2.1 Chemicals and reagents
LHC-9 medium was obtained from Biosource (Rockville, MD) or Invitrogen (Carlsbad, CA). Hanks' Balanced Salt Solution (HBSS) was from Invitrogen. BEAS-2B cells were obtained from the American Type Culture Collection. Sodium chromate (99+%) was the highest purity available from Aldrich Chemical (Milwaukee, WI). Lead chromate was from Alfa Aesar (Ward Hill, MA) and zinc chromate was from Pfaltz & Bauer (Waterbury, CT). Na253CrO4 was graciously provided by Dr. David H. Petering (University of Wisconsin–Milwaukee). Chromates are known carcinogens and should be handled accordingly. γ-Glutamyl glutamate (γ-Glu-Glu) was obtained from MP Biochemicals (Solon, OH). Trichloroacetic acid (TCA), K2CO3, and methanol (HPLC grade) were from Fisher Scientific (Pittsburgh, PA). Sodium acetate trihydrate was from JT Baker (Phillipsburg, NJ) and glacial acetic acid was from EMD Chemicals (Gibbstown, NJ). All other chemicals and reagents were purchased from Sigma Chemical (St. Louis, MO) or from sources indicated below.
2.2 Cell culture
BEAS-2B cells were grown at 37°C in humidified air containing 5% CO2 in serum-free LHC-9 medium. Flasks were pre-coated for 3–48 h with LHC-9 medium containing bovine fibronectin (Sigma F4759) (final conc. of 9.1 µg per ml), collagen (Vitrogen, Cohesion Technologies, Palo Alto, CA) (final conc. of 27.3 µg per ml), and bovine serum albumin (Biosource cat. no. 118–500) (final conc. of 90.9 µg per ml). The cells were fed every 48 h, and were subcultured prior to reaching confluence using the Reagent Pak system (Clonetics, CC-5034). Normal plating density was 3000 to 5000 cells/cm2. Alternatively, BEAS-2B cells were grown in Dulbecco's Modified Eagle's Medium (DMEM) with 25 mM HEPES and 4.5 g/L glucose (BioWhittaker 12-709F, Cambrex BioScience), 10% fetal bovine serum (FBS Optima, Atlanta Biologicals; or Valley Biomedical, Winchester, VA), penicillin (100 U/ml), and streptomycin (100 µg/ml).
2.3 Clonogenic assay
Clonogenic survival was determined using the method of Blankenship et al. [40] adapted for the growth requirements of the BEAS-2B cells. Cells were grown for 3 days in LHC-9 medium in 6-well dishes, exposed to Cr(VI) for 24 h, and then subcultured using the Reagent Pak system, and plated in 60-mm dishes (200 cells per dish). An aliquot of the cell suspension was examined for trypan blue staining; positively stained cells were classified as nonviable. After 11 days incubation, they were stained with crystal violet, and the number of colonies (>50 cells each) was determined. Stock suspensions of PbCrO4 and ZnCrO4 were washed in acetone and prepared as described [41], whereas an aqueous stock solution of Na2CrO4 was used.
2.4 Electron Spin Resonance
Cr(V) generation was examined using direct detection by ESR. Cells were harvested by scraping into the cell culture medium. The cells were pelleted for 5 min at 800 × g, the pellets were washed in pre-warmed LHC-9 medium, and the washed pellets were resuspended in a small volume of LHC-9. A small aliquot was used to determine the cell count. The cell suspension was pre-incubated for 3 min at 37°C, and Cr(VI) was added followed by incubation for the indicated times. Cr(VI) was added from aqueous stock solutions; those for ZnCrO4 and PbCrO4 were sonicated 15 sec prior to use to create a more uniform suspension of these insoluble particles. The samples, in quartz ESR tubes, were then immersed in liquid nitrogen (77 K) and stored, typically for less than one week, until analysis by ESR. For some samples, spectra were first obtained at liquid helium temperature (6.3 K) using a Bruker E500 ELEXSYS spectrometer (Silberstreifen, Germany) with an Oxford Instruments ESR-9 helium flow cryostat (Oxfordshire, UK) and a Bruker DM0101 cavity. For Cr(V) ESR, samples were quickly thawed, placed in a quartz flat cell and ESR spectra were obtained without delay at room temperature using a Bruker EMX spectrometer. The spectra were stable during the ESR analysis time, with no noticeable changes in spectral intensity or pattern. Instrument settings are indicated in the results. ESR spectra were confirmed in replicate experiments. The g values were determined by comparison to the 2,2,-diphenyl-1-picrylhydrazyl radical (DPPH) which has a g value of 2.0036.
2.5 HPLC Analysis of Glutathione
Adherent cells (equivalent to one T150 flask) were washed twice in pre-warmed HBSS and scraped into ice-cold 5% TCA. The samples were iced and TCA-insoluble proteins were removed by centrifugation at 4°C (9900 × g for 5 min). The pellets were saved for total protein analysis (see below). The acid supernatants were stored frozen until derivatization and analysis. Samples were derivatized and analyzed by HPLC using a method kindly provided by Dr. Tak Yee Aw [42]. Briefly, γ-Glu-Glu was added to 0.5 ml sample as an internal standard. Then, 0.05 ml 80 mM iodoacetic acid was added followed by 0.34 ml of 1 M K2CO3 to raise the pH to ≥8.0. After 60 min incubation at room temperature, 0.125 ml of 6% 1-fluoro-2,4-dinitrobenzene (in ethanol) was added, followed by 0.35 ml 1 M K2CO3 (to raise the pH to 10). Samples were incubated overnight in the dark at 4°C. The samples were centrifuged (9900 × g) at 4°C for 20 min at 11,000 rpm. An aliquot of each supernatant was filtered through a 0.45 µm Acrodisc syringe filter, and analyzed by HPLC. HPLC conditions were: Sphereclone NH2 5 µ column (250 × 4.6 mm) plus a 4.6 mm guard column; absorbance detected at 365 nm; flow rate of 1.5 ml per min. The mobile phase was 90% solvent A (80% methanol) and 10% solvent B (2% sodium acetate in 64% methanol) for the first five min. During the next 10 min, a linear gradient was applied ending at 1% solvent A and 99% solvent B. 1% A/90% B was maintained for 10 min, and then a gradient was applied to return and equilibrate the column to the starting conditions. The retention times of the peaks for GSH and GSSG in the samples were compared to those of GSH and GSSG standards that were derivatized in the same manner. GSH eluted at 11.8 to 12 min, whereas GSSG eluted 2 min later. Concentrations were determined from peak area relative to the standard curves, and were normalized per µg of protein.
For protein analysis, the TCA-insoluble pellets (from above) were dissolved in 0.1 M NaOH for ≥5 days at room temperature and then protein was determined by a modified Lowry method, with bovine serum albumin as the standard [43].
2.6 Immunofluorescence
Cells were grown on coverslips in LHC-9 medium to approximately 70% confluence. The adherent cells were washed with pre-warmed phosphate-buffered saline (PBS) and then fixed in 1% paraformaldehyde in PBS for 8 min. They were rinsed four times with PBS, permeabilized with 0.2% Triton X-100 in PBS for 5 min, and then rinsed again four times. They were then incubated with a primary antibody (0.04 mg per ml) in 1% bovine serum albumin in PBS for 30 min at room temperature. The primary antibodies against human P450 reductase, human cytochrome b5, and human b5 reductase were previously described [31]. After five washes with PBS, they were incubated in the dark with the secondary antibody (0.01 mg per ml; Alexa Fluor goat anti-rabbit IgG, Molecular Probes) for 30 min at room temperature. After four washes with PBS, Slow Fade reagent (MolecularProbes) was applied per manufacturer's directions and the cells were examined using a Nikon Eclipse 600 epifluorescence microscope using the FITC filter set.
3. Results
3.1 Cr(V) generated by medium
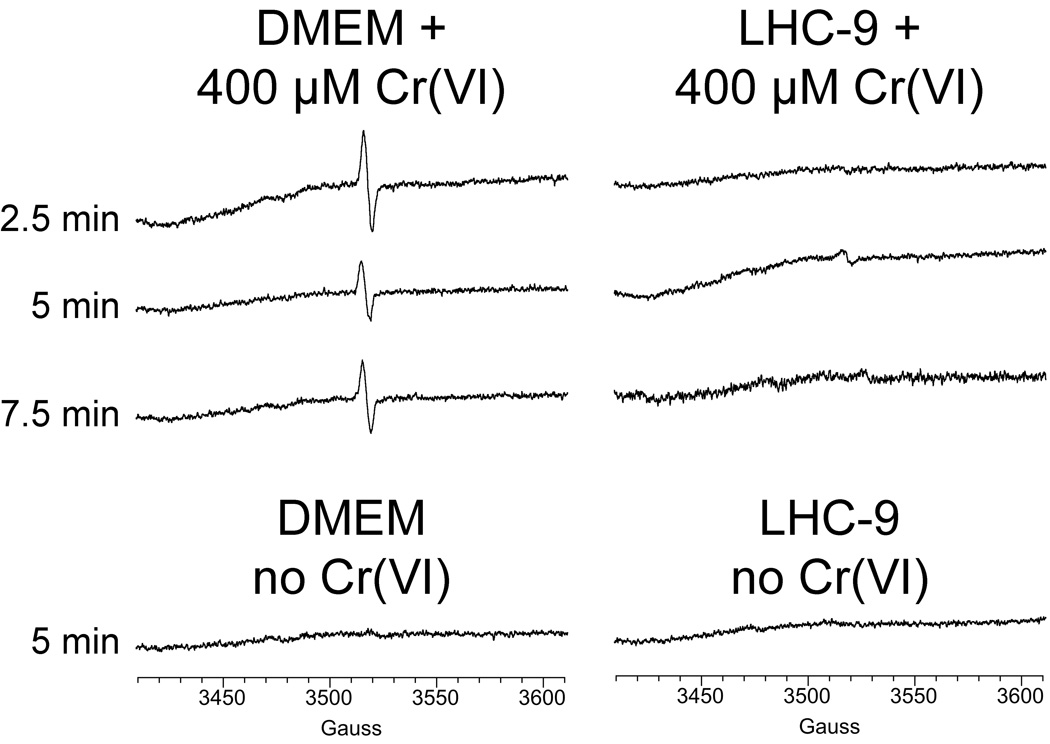
Cr(V) (e.g. as Cr(V)O43−) is a d1 paramagnetic species [44, 45] that can be directly detected by ESR spectroscopy by its sharp line spectrum (g = 1.979) at conventional X-band frequency. As initial controls, Cr(VI) (as Na2CrO4) was incubated with cell culture media in the absence of cells (Fig. 1). A definitive signal consistent with Cr(V) (g = 1.979) was seen with the DMEM medium, but the signal was not evident in LHC-9 medium (Fig. 1). In DMEM without Cr(VI), the Cr(V) signal was absent (Fig. 1). A Cr(V) signal was also observed when DMEM was incubated with 0.2 mM Na2CrO4 (not shown). These data indicate that DMEM medium reduces some of the Cr(VI) to Cr(V). This could potentially interfere with the interpretation of data on the generation of Cr(V) by cells. LHC-9 medium was therefore used for ESR experiments with cells because of its lack of significant Cr(V) generation.
Fig. 1.
DMEM-HEPES medium incubated with 0.4 mM Na2CrO4 at 37°C under room air generates Cr(V) (left), whereas LHC-9 medium does not (right). The times of incubation with Na2CrO4 are indicated. Representative ESR spectra obtained at room temperature are shown. Instrument settings were: 5 G modulation amplitude, 50 mW microwave power, 6.32 × 104 receiver gain, 40.96 msec time constant, 9.76 GHz microwave frequency, sweep width = 200 G, field set = 3510 G, modulation frequency = 100 kHz, scan time = 42 sec; number of scans, 9.
3.2 Cr(V) generated by cells
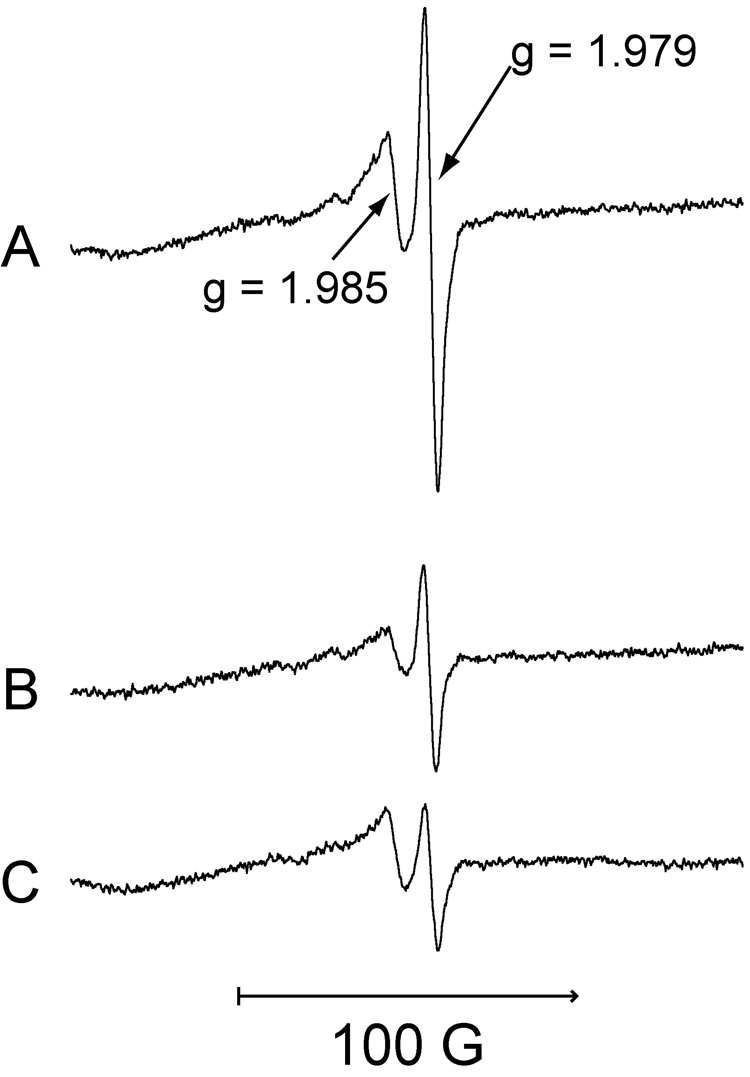
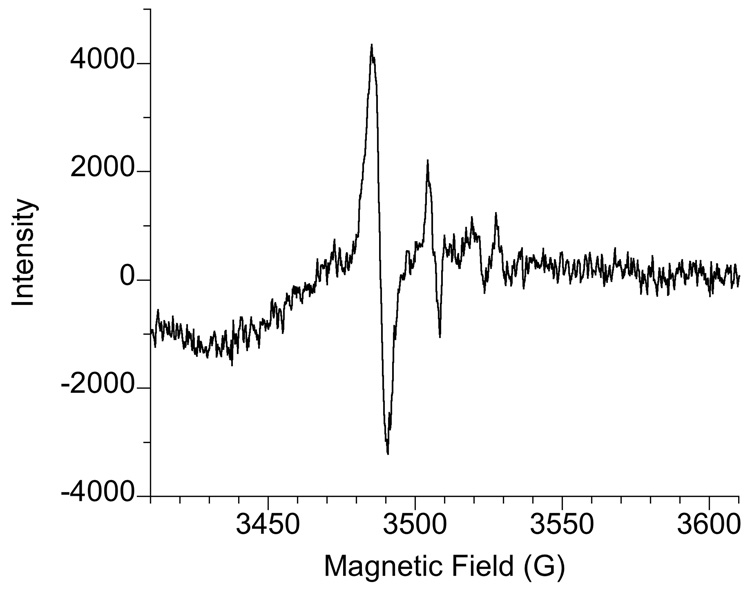
When freshly harvested cells (grown and resuspended in LHC-9 medium) were incubated for 5 min with Na2CrO4, two ESR signals were observed: a prominent signal at g = 1.979 and a signal of lesser intensity at g = 1.985 (Fig. 2). The g = 1.979 signal is similar to that for Cr(V) generated in vitro by human microsomes [30] and by proteoliposomes containing purified human P450 reductase and cytochrome b5 [31, 46]. The signal at g = 1.985 suggests a Cr(V) species that is dependent on thiols. While the g = 1.985 signal might theoretically represent something other than Cr(V), both the g = 1.979 and g = 1.985 signals were dependent on Cr(VI) because neither were seen in cells incubated without Cr(VI) (see below).
Fig. 2.
LHC-9-grown BEAS-2B cells incubated with Na2CrO4 for 5 min at 37°C in LHC-9 medium under room air generate Cr(V). Representative ESR spectra (obtained at room temperature) for Cr(V) are shown for: (A) 400 µM Na2CrO4 plus 1 × 107 cells.; (B) 400 µM Na2CrO4 plus 4.1 × 106 cells; (C) 200 µM Na2CrO4 plus 1 × 107 cells. For each, the total reaction volume was 0.25 ml. Instrument settings were the same as for Fig. 1.
The relative intensity of these two signals varied with the initial Cr(VI) concentration and the cell density (Fig. 2). When comparing two cell densities with a constant amount of Cr(VI) (0.4 mM), the intensity of both signals declined 2.3-to 2.4-fold with a 2.4-fold decrease in cell number (Fig. 2A,B). When comparing a two-fold decrease in Cr(VI) concentration at a constant cell number, the g = 1.979 signal decreased to a greater extent (3.3-fold) relative to the g = 1.985 signal (1.4-fold) (Fig. 2A,C). The ratio of Cr(VI) to cell number ratio can therefore impact the relative intensity of these signals.
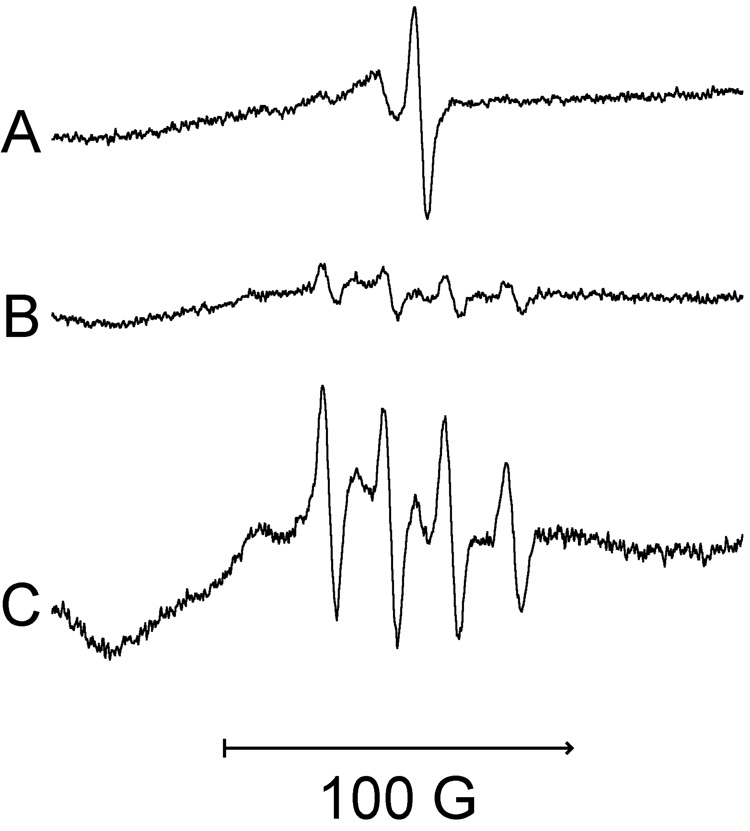
To authenticate if one or both signals in Fig. 2 are due to Cr(V), analogous studies were conducted with isotopically pure 53Cr (stable isotope) as Na253CrO4. Because 53Cr has a nuclear spin of I = 3/2, Cr(V) signals will be split into four lines (separation of 17.4 G), typical for the hyperfine coupling for 53Cr(V) [44]. With 53Cr, both the g = 1.979 and g = 1.985 components of the Cr(V) signal (Fig. 3A) were altered by 53Cr (Fig. 3B,C), confirming that Cr(V) is responsible for both components. The 53Cr signals are best seen in Fig. 3C in which the increased number of scans enhanced the signal-to-noise ratio. The four splittings derived from the original g = 1.979 component are readily observed in Fig. 3B and 3C. Note that the single g = 1.985 signal from Fig. 3A is not evident with 53Cr in Figs. 3A,B, supporting its identity as a Cr(V) species. Some of the features consistent with the 53Cr splitting of the original g = 1.985 component can be seen in Fig. 3C.
Fig. 3.
Representative Cr(V) signals from LHC-9-grown BEAS-2B cells exposed under room air to Na2CrO4 for 5 min at 37°C in LHC-9 medium. Spectra were recorded at room temperature. (A) 400 µM Na2CrO4 plus 4.1 × 106 cells, 9 scans. (B) 400 µM isotopically pure Na253CrO4 plus 4.9 × 106 cells, 9 scans. (C) The same sample as B except 36 scans were collected. The reaction volume and instrument settings were the same as those in Fig. 1.
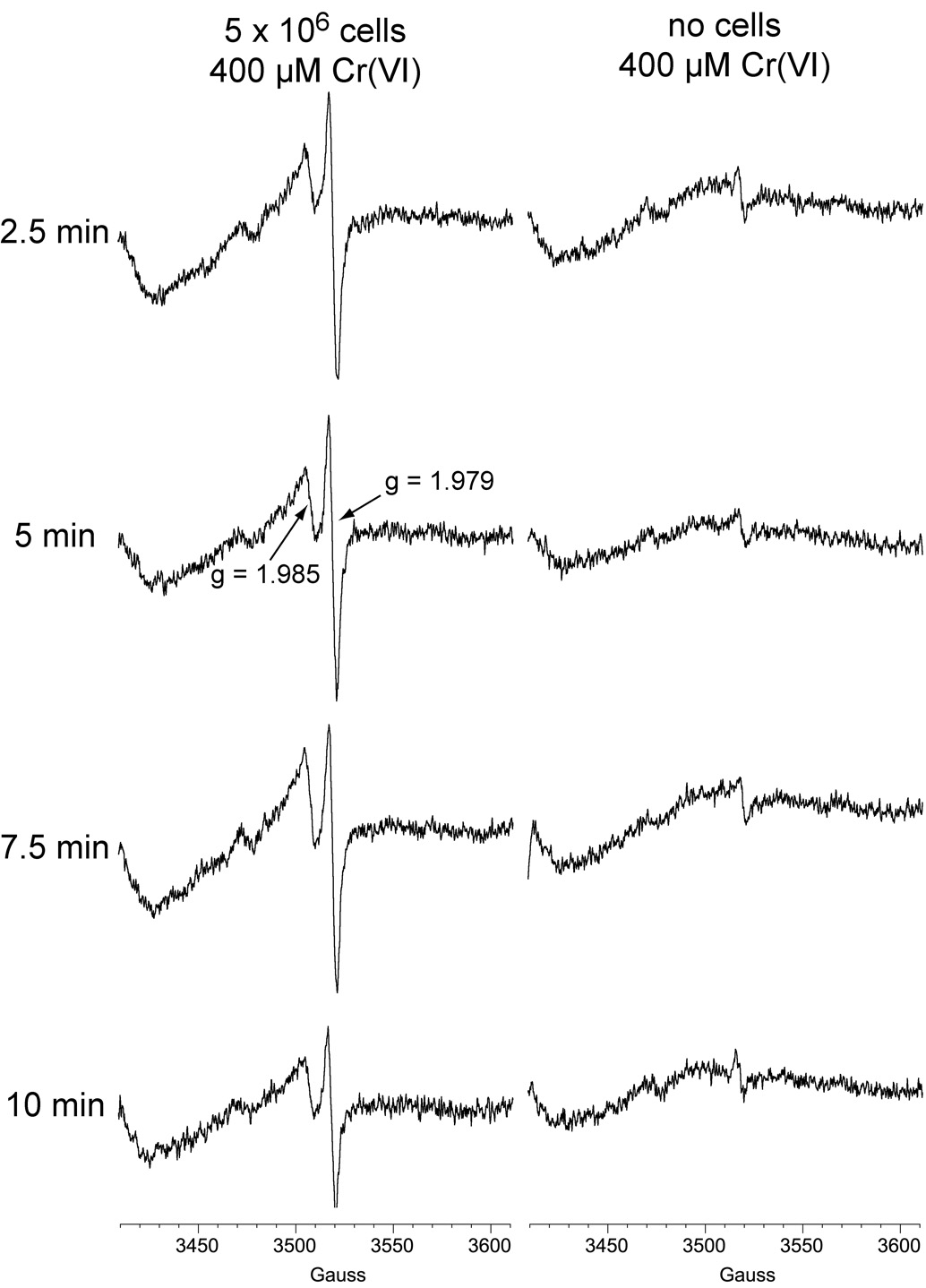
3.3 Cr(V) time course analyses
Experiments were conducted to follow the intensity of the Cr(V) signals over time. LHC-9-grown cells generated pronounced Cr(V) signals 2.5 min after the Cr(VI) was added, which was 10 the earliest timepoint examined (Fig. 4). The Cr(V) signals declined somewhat over time, although both were still prominent after 10 min (Fig. 4). In in vitro studies, Cr(V) is a short-lived transient intermediate [30] and its signal intensity does not represent the total amount produced over time but rather the relative level at that point in time. The data with cells therefore indicate a rapid generation of Cr(V), but also a continual generation over time. Both Cr(V) signals are still detected 1 h after sodium chromate was added (see below). The rapid onset and extended timeframe of Cr(V) generation matches that for in vitro experiments with human liver and lung microsomes [30]. Cr(V) generation in these lung cells is also consistent with findings that room air only inhibits a minority of Cr(VI) reduction by human microsomes [33, 47], and suggests that significant Cr(V) generation is likely in human bronchial epithelial cells in vivo.
Fig. 4.
Representative ESR spectra of Cr(V) obtained following incubation of LHC-9-grown BEAS-2B cells with Na2CrO4. Cells were grown in LHC-9 medium, harvested by scraping, washed in pre-warmed LHC-9 medium, and resuspended in a small volume of LHC-9. Aliquots of the cell suspension containing the indicated number of cells were incubated with Na2CrO4 for the indicated times at 37°C. For each, the total reaction volume was 0.25 ml. Instrument settings were the same as for Fig. 1.
While the LHC-9 medium does not create significant background for Cr(V) ESR, it can be cost-prohibitive for extensive use with experiments requiring large numbers of cells. We therefore explored the possibility of growing cells in DMEM, and then washing and resuspending the cells in LHC-9 medium for the Cr(V) ESR experiments. Similar to LHC-9 grown cells, these DMEM-grown cells generated both the g = 1.979 and g = 1.985 Cr(V) signals over time (Fig. 5). The cell-free controls showed that the washing process largely eliminated the Cr(VI)-reducing properties of the DMEM. The Cr(V) ESR signals of these DMEM-grown cells are therefore similar to those for cells grown in LHC-9 medium. This indicates that DMEM-grown cells are suitable for these studies as long as they are washed and resuspended in LHC-9 prior to incubation with Cr(VI).
Fig. 5.
Representative ESR spectra of Cr(V) obtained following incubation of DMEM-grown BEAS-2B cells with Na2CrO4. Cells were grown in DMEM-HEPES medium with 10% FBS, harvested by scraping, washed in pre-warmed LHC-9 medium, and resuspended in a small volume of LHC-9. Aliquots of the cell suspension containing the indicated number of cells were incubated with Na2CrO4 for the indicated times at 37°C. Controls without cells are shown at right. For each, the total reaction volume was 0.25 ml. Instrument settings were the same as for Fig. 1.
3.4 Role of thiols
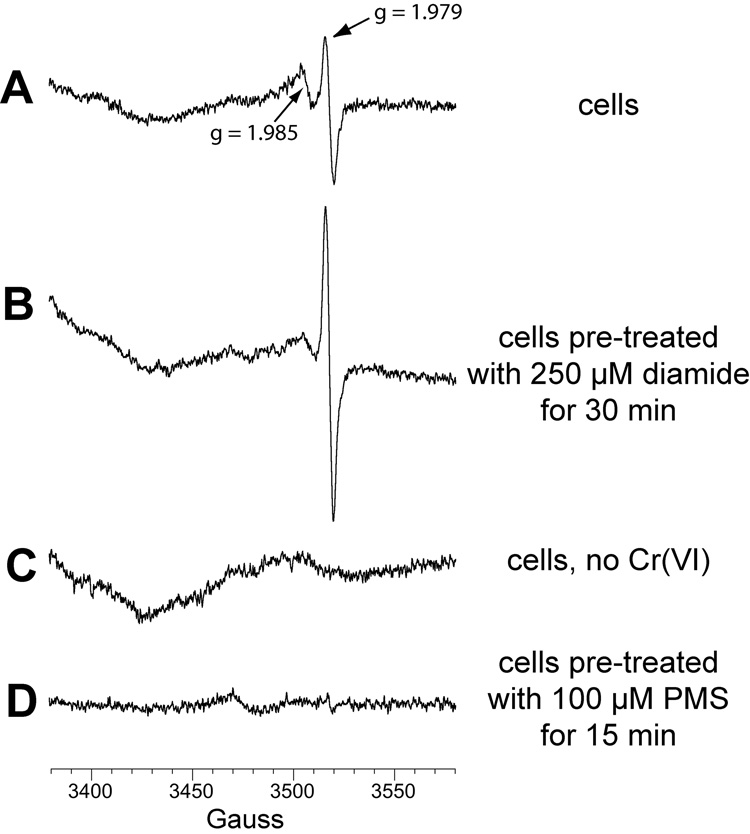
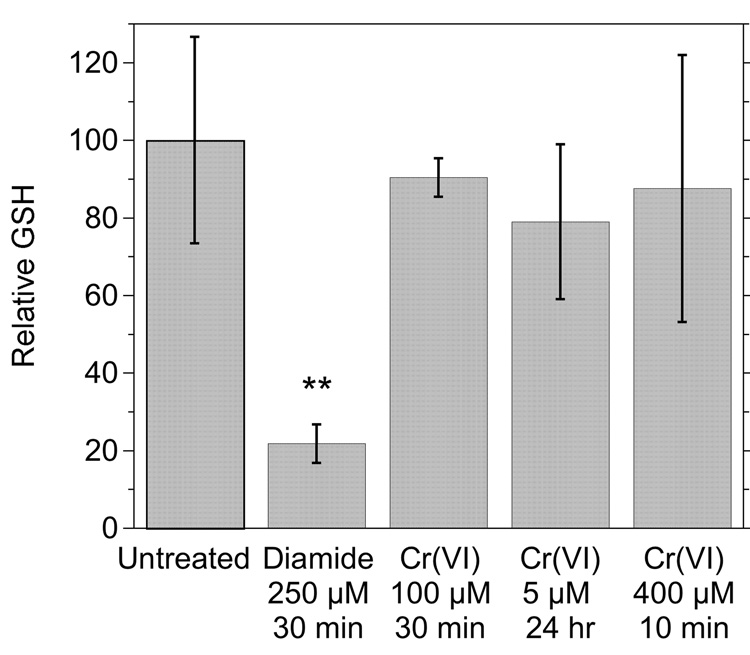
The g = 1.985 Cr(V) signal might be dependent on one or more thiol species. This was further explored by examining cells pre-treated with diamide [(CH3)2NCON=NCON(CH3)2] which is a potent and non-specific thiol oxidant. In diamide-treated cells (Fig. 6B), the g = 1.985 signal intensity was significantly diminished (Fig. 6A), consistent with a thiol dependence. In contrast, the g = 1.979 signal intensity increased (2.1- to 2.2-fold to that of control cells) in diamide-treated cells (Fig. 6B). This suggests that thiols are not necessary for the reduction of Cr(VI) to Cr(V) in these cells. Neither of the Cr(V) ESR signals were seen in cells incubated without chromate (Fig. 6C). The ability of diamide to oxidize cellular thiols was confirmed by its ability to deplete GSH by an average of 78% in these cells (Fig. 7). Diamide did not generate significant GSSG, consistent with the known ability of diamide to yield GS-protein adducts in cells [48]. That GSSG was below detection limits in cells is consistent with GSSG normally comprising <2% of the total glutathione pool [49]. However, the in vitro oxidation of GSH by H2O2 resulted in GSSG as expected (not shown), demonstrating the capability of the method to detect GSSG. While GSH is a cytosolic antioxidant and cytosolic Cr(VI) reductant, incubation of cells with Na2CrO4 for 10 min (400 µM), 30 min (100 µM) or 24 h (5 µM) did not result in significant changes in GSH (Fig. 7). The cells retained normal morphology after these Cr(VI) treatments (not shown). Some GSH likely contributed to Cr(VI) reduction in the cells, but it was not to an extent that decreased GSH levels.
Fig. 6.
Representative Cr(V) signals from BEAS-2B cells exposed to 0.4 mM Na2CrO4 for 5 min at 37°C in LHC-9 medium under room air. Cells were grown in DMEM-HEPES medium with 10% FBS, harvested by scraping, washed in pre-warmed LHC-9 medium, and resuspended in a small volume of LHC-9 for incubation with Na2CrO4. Reactions contained 3 × 106 cells (A, B, C), or 5 × 106 cells (D). Prior to harvesting and washing in LHC-9, the cells in B were pretreated with 0.25 mM diamide for 30 min, and those in D with 0.1 mM PMS. For each, the total reaction volume was 0.25 ml and instrument settings were the same as in Fig. 1 except that the field set was 3480 G.
Fig. 7.
Relative levels of GSH in cells exposed to various chemicals. Cells were either untreated or pre-exposed as indicated to: diamide (0.25 mM for 30 min), or Cr(VI) as Na2CrO4 (100 µM for 30 min, 5 µM for 24 h, or 400 µM for 10 min). **, P <0.01 vs. the other treatments.
3.5 Dependence on NADPH
The presence of the g = 1.979 Cr(V) signal and the absence of the g = 1.985 signal in diamide-treated cells (Fig. 6) suggests that non-thiol reductants contribute significantly to Cr(VI) reduction. The effect of pretreating cells with 0.1 mM phenazine methosulfate (PMS), a nonenzymatic oxidant of NAD(P)H, was therefore also examined. Neither of the Cr(V) signals were seen in PMS-treated cells (Fig. 6D), suggesting that the generation of Cr(V) is largely the result of NAD(P)H-dependent enzymes. The Cr(V) signals were similarly absent in cells pretreated with a higher concentration (0.4 mM) of PMS (not shown).
3.6 Hyperfine structure of the Cr(V) signal
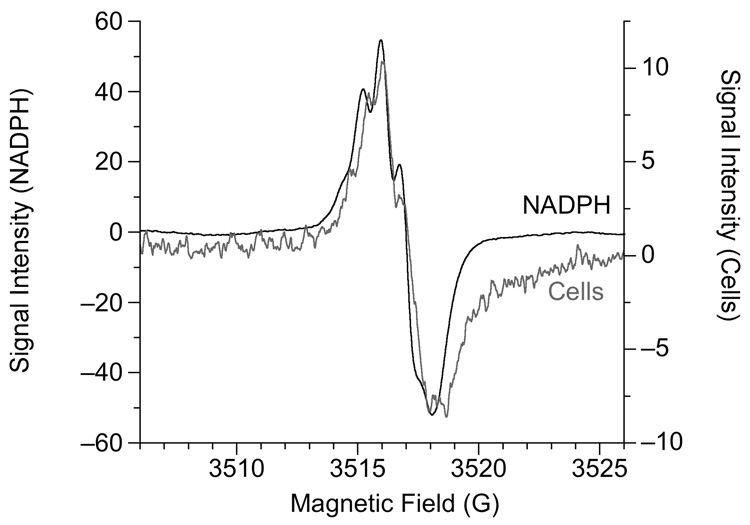
To examine the hyperfine structure of the Cr(V) signal, experiments with cells were repeated in which the modulation amplitude was reduced to 0.1 G. While this greatly diminished the intensity of the Cr(V) signal, doubling the Cr(VI) concentration from 400 to 800 µM allowed for sufficient signal intensity to observe the hyperfine structure of the g = 1.979 signal which contained 5 or 6 splittings (Fig. 8). This structure was similar, but not identical, to that for Cr(V)-NADPH (Fig. 8) and suggests the influence of nearby hydrogens in Cr(V)-diol complexes [50]. In fact, several other Cr(V)-diol complexes yield similar spectra including those due to NADH, FAD, and various sugars (fructose, ribose, cellobiose, and others) [50]. The similarity of these spectra precludes identification of the identity of the Cr(V) species, but the cellular data suggest Cr(V)-diol type species. Given the variety of carbohydrates, flavins, pyridine nucleotides, and other components in cells, it is likely that the g = 1.979 hyperfine structure represents a variety of Cr(V)-diol species. At 0.1 G modulation amplitude, the intensity of the g = 1.985 component of the Cr(V) signal was too small to attempt to resolve any hyperfine structure.
Fig. 8.
Hyperfine structure of the g = 1.979 Cr(V) ESR signal. The "Cells" spectrum was obtained following 5 min incubation of 107 cells at 37°C in 0.25 ml of LHC-medium containing 800 µM Na2CrO4. The "NADPH" spectrum was obtained following 30 min incubation of 5 mM NADPH plus 5 mM Na2CrO4 at 37°C in 0.3 ml 43.8 mM potassium phosphate buffer (pH 7.4). Instrument settings were: 0.1 G modulation amplitude, 50 mW microwave power, 6.32 × 104 receiver gain, 327.68 msec time constant, 9.764 GHz microwave frequency, sweep width = 20 G, field set = 3516 G, modulation frequency = 100 kHz, scan time = 83.89 sec; conversion time = 81.92 msec. The number of scans was 5 and 50 for the "NADPH" and "Cells" samples, respectively. Note the different y-axis intensity scales for the two samples. The signals intensities were divided by 1000 to obtain the values on the graphs.
3.7 ESR for Cr(III)
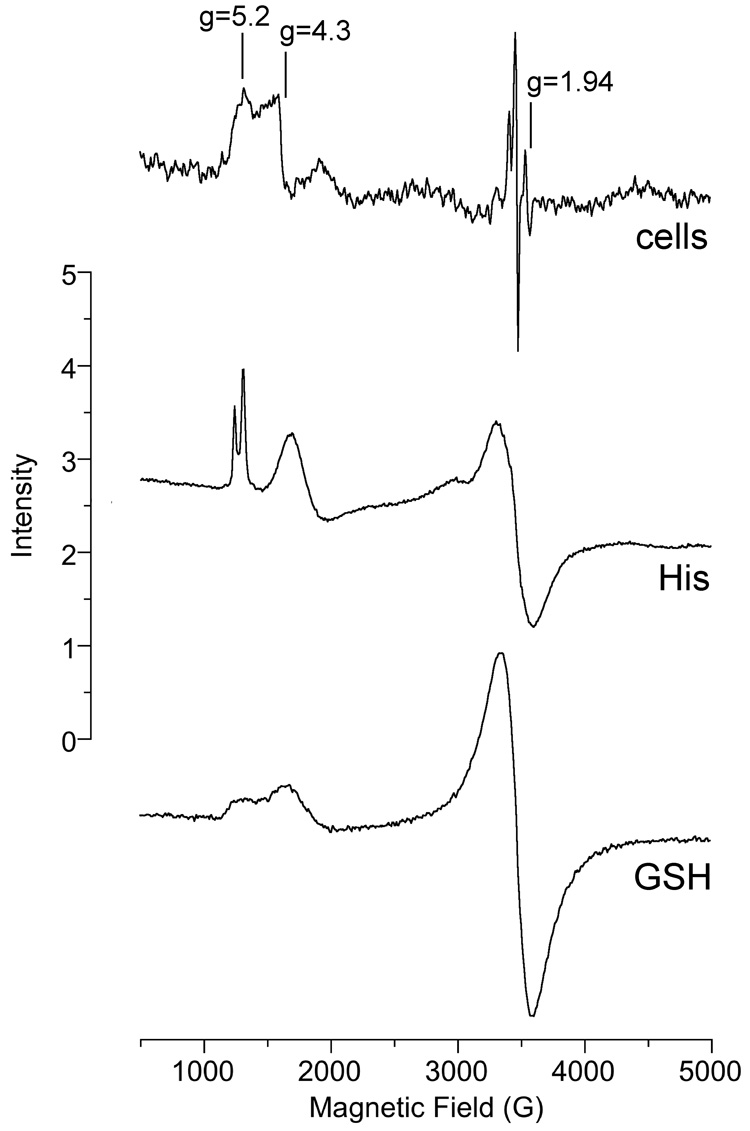
To assess the possible further reduction of chromium to Cr(III) in cells incubated with Na2CrO4, ESR spectra at 6.3 K were also obtained on some of the samples. ESR lines for Cr(III) were evident in cells incubated with Na2CrO4 for 5 min, indicating the rapid conversion of at least some of the Cr(VI) to Cr(III) (Fig. 9, cells). The lines in the g = 5 region are characteristic of a Cr(III) complex with a large ZFS (zero field splitting), signifying a distorted geometry. The line with geff(z) = 5.2 should have geff(y) = 2.3 and a broad line at about 1.6 for geff(x) as detemined from a rhombogram [51]. E/D (the ratio of E to D, which are the axial and rhombic zero field splitting parameters, respectively) for the cells spectrum is 0.25 signifying a large rhombic component. However, neither the lines for geff(y) and geff(x) were detected with the cells, probably because they are too broad. Thus there are two Cr(III) signals in cells: the geff = 5.2 just described and the second with geff = 4.3 which has an E/D of about 0.04. These cellular signals are similar to the signals for the Co(II) sites (S = 3/2) in horse liver alcohol dehydrogenase [52].
Fig. 9.
ESR spectra of samples that were incubated and then frozen and analyzed by ESR at 6.3 K. "Cells" (top): BEAS-2B cells were incubated with 400 µM Na2CrO4 for 5 min at 37°C prior to freezing. "His" (middle): 10 mM histidine plus 1.25 mM Cr(III) (as CrCl3) were incubated in 0.5 M HEPES buffer (pH 7.4) at room temperature for 96 h prior to freezing. "GSH" (bottom): 100 mM GSH plus 1.25 mM Cr(III) were incubated in 0.5 M HEPES buffer (pH 7.4) at room temperature for 72 h prior to freezing. Instrument settings were: temperature = 6.3 K, 5 G modulation amplitude, 0.63 mW microwave power, 60 dB receiver gain, 82 msec time constant, 9.633 GHz microwave frequency, scan time = 83 sec; number of scans, 9.
Similar lines in the g = 5 region have been reported following the reduction of Cr(VI) by GSH [53]. We observed broader lines in this region following extended incubation of Cr(III) with GSH (Fig. 9, GSH). The broadness of these lines may be due to strain for the ZFS parameters. Sharper lines in the g = 5 region were obtained upon extended incubation of Cr(III) with histidine (Fig. 9, His). The Cr(III)-histidine complex may be similar in structure to the Cr(III)-GSH in which non-thiol residues (e.g. γ-Glu of GSH or GSSG) provide the ligands for Cr(III) [53]. The cellular signal at g = 1.94 is likely due to iron-sulfur complexes (usually oxidized) and can be seen in cells and tissues incubated without Cr(VI) [54]. The sharp signals to the left of g = 1.94 (Fig. 9) are Cr(V) species. Overall, the 6.3 K ESR spectra indicate that at least two Cr(III) species are formed quickly in these cells; while their exact composition is unknown, the spectral features suggest complex(es) with ligands from cellular components.
3.8 Soluble vs. insoluble chromates
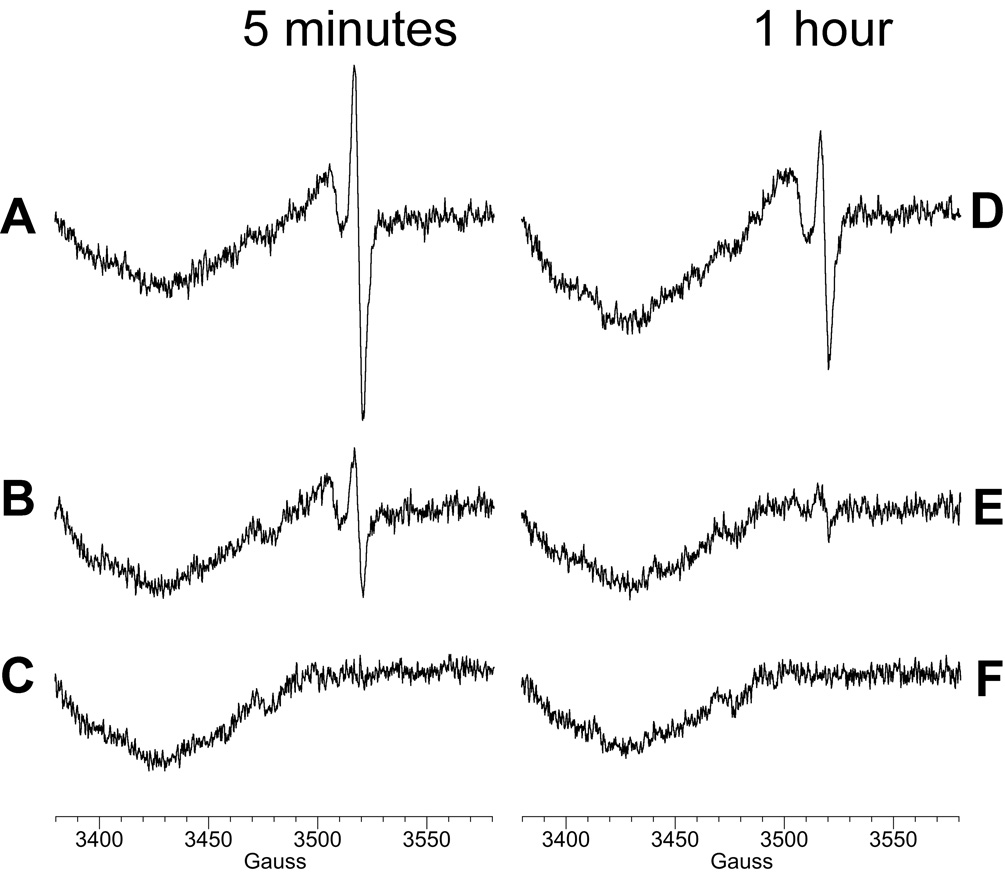
The data to this point were generated with Na2CrO4, which is highly water soluble. Insoluble forms of chromate (e.g. PbCrO4, ZnCrO4) are of interest from the perspective of industrial use and occupational exposure, and the potential of increased carcinogenicity. The ability of the BEAS-2B cells to generate a detectable Cr(V) signal with these insoluble chromates was therefore compared to Na2CrO4. Both the g = 1.979 and g = 1.985 Cr(V) signals were seen following 5 and 60 min incubation with Na2CrO4 (Fig. 10A,D). Both signals were also seen after 5 min with ZnCrO4 (Fig. 10B) although the intensity of the g = 1.979 signal in particular was much lower than with Na2CrO4. After 1 h with ZnCrO4, only a small g = 1.979 signal was seen (Fig. 9E). Cr(V) signals were not seen at either timepoint with PbCrO4 (Fig. 10C,F).
Fig. 10.
ESR spectra of BEAS-2B cells incubated with Na2CrO4 (A, D), ZnCrO4 (B, E), or PbCrO4 (C, F) for 5 min (A, B, C) or 1 h (D, E, F) at 37°C in LHC-9 medium under room air. For each, the initial chromate concentration was 0.4 mM, and the cell density was 5 × 106 cells in a total reaction volume of 0.25 ml. Instrument settings were as for Fig. 1 except that the field set was 3480 G.
3.9 Clonogenic survival
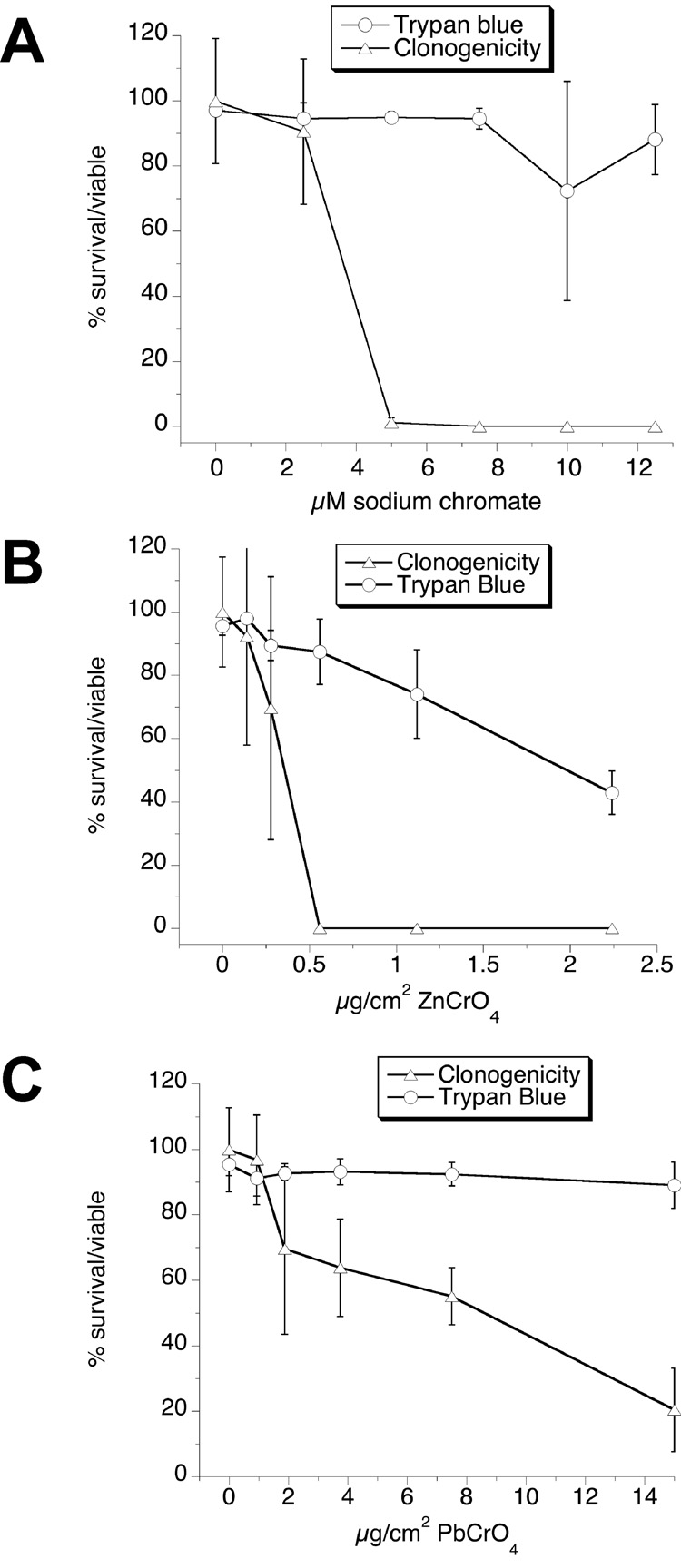
The effects of 24-h exposure to these three chromates were examined by clonogenic survival or by exclusion of the dye trypan blue. In all cases, trypan blue exclusion was a much less sensitive indicator of viability (Fig. 11). Trypan blue exclusion should not therefore be considered a reliable indicator of the ability of BEAS-2B cells to survive exposure to chromates. For Na2CrO4 and ZnCrO4 the cells showed a steep decline in clonogenic survival over narrow concentration ranges, with estimated 50% survival at 3.6 µM Na2CrO4 and 0.36 µg ZnCrO4 per cm2. Since ZnCrO4 is water-insoluble, its dose is expressed in mass per culture surface area rather than in units of concentration. If ZnCrO4 were soluble, however, the 0.36 µg per cm2 would correspond to 9.9 µM. The cells tolerated much higher levels of PbCrO4 (50% survival estimated at 8.6 µg/cm2) and the slope of the PbCrO4 clonogenic survival curve was much more gradual. Since PbCrO4 is also insoluble, its dose is not expressed in µM.
Fig. 11.
Relative survival/viability of BEAS-2B cells exposed for 24 h in LHC-9 medium to different concentrations of Na2CrO4 (A), ZnCrO4 (B), or PbCrO4 (C), determined by a clonogenic assay and trypan blue exclusion. Results are the means ± SD, n = 3 experiments.
3.10 Microsomal protein expression
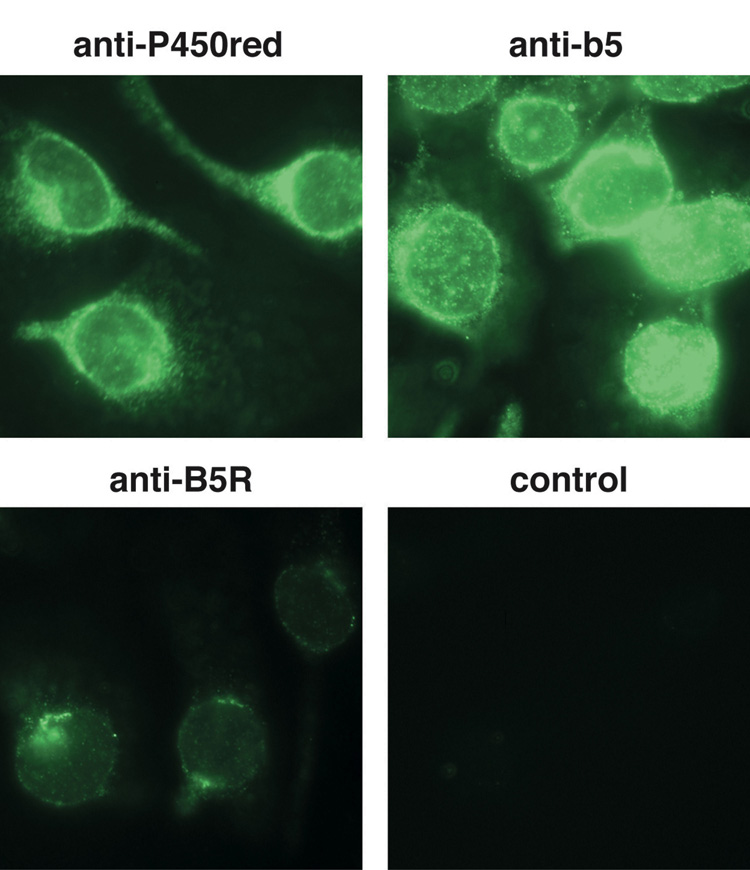
Human microsomal enzymes have been implicated in Cr(VI) reduction including the generation of Cr(V) [32]. While the expression of several microsomal enzymes typically declines markedly in cell culture, immunofluorescence demonstrated that BEAS-2B cells continue to express the microsomal proteins cytochrome b5, b5 reductase, and P450 reductase (Fig. 12). Their subcellular localization in the nuclear membrane and perinuclear region is as expected for smooth endoplasmic reticular proteins. BEAS-2B cells therefore retain these enzymes which could contribute to Cr(VI) reduction and Cr(V) generation.
Fig. 12.
Immunofluorescent detection of the proteins P450 reductase (P450red), cytochrome b5 (b5), and b5 reductase (B5R) using antibodies specific for each protein, and a fluorescein conjugated secondary antibody. The control was treated identically except that only a secondary antibody was applied (no primary antibody).
4. Discussion
4.1 Cr(V) generated by medium
In cellular studies on reductive Cr(VI) activation, the potential contribution of the cell culture medium has received little attention. However, this report demonstrates that DMEM, a commonly used medium, can generate Cr(V) in the absence of cells (Fig. 1). This could potentially confound interpretation of the cellular contribution to Cr(VI) reduction. In these studies, the use of cells grown in LHC-9 medium, or grown in DMEM and washed and resuspended in LHC-9, eliminated Cr(VI) reduction by the medium and therefore provided for a direct assessment of the cellular role in Cr(V) generation. The DMEM findings also indicate the importance of considering cell culture media for assessing Cr(VI) toxicity. It is possible that extracellular Cr(VI) reduction by the medium would decrease toxicity by decreasing Cr(VI) uptake and therefore the resulting intracellular generation of reactive Cr species. Consistent with this concept, co-treatment of clonal human bronchial fibroblasts with ascorbate as an extracellular Cr(VI) reductant prevents Cr uptake and clastogenic effects [55]. By decreasing Cr(VI) available for uptake, extracellular Cr(VI) reduction by the medium might similarly diminish Cr-associated ROS generation associated with intracellular reductants such as cytochrome b5 and P450 reductase [32]. The potential impact of medium-generated extracellular Cr(V) on cells has not been explored, but the transient nature of Cr(V) suggests that it would not accummulate in the medium. This is consistent with the decline in the DMEM-generated Cr(V) signal intensity over time (Fig. 1).
4.2 Generation of the g = 1.979 Cr(V) signal
Bronchial epithelial cells are a major site of tumor formation in the lung [3, 7, 37, 39, 56, 57] and are among the first targets of inhaled toxins, including Cr-containing fumes and particles. Primary lung epithelial cells, however, have a limited lifespan in culture and would be difficult to routinely expand to the large cell numbers required for ESR. BEAS-2B cells were therefore chosen as a model of normal human bronchial epithelium. They originate from normal tissue and are nontumorigenic even after extensive passage numbers [58], they can routinely be grown to the cell numbers needed for ESR studies, and they have been used as a model for cytotoxicity associated with airborne particulates [59–62]. Unlike many other cells, BEAS-2B cells maintain expression of key microsomal enzymes when grown in culture [59, 63] including cytochrome b5 and P450 reductase (Fig. 12) which can generate Cr(V) and hydroxyl radical by Cr-mediated redox cycling [32]. In human lungs, P450 reductase is localized primarily in the bronchial and bronchiolar epithelial cells [64].
To our knowledge, Cr(V) formation in noncancerous human bronchial epithelial cells has not been previously reported. We observed two Cr(V) signals (g = 1.979 and 1.985) in BEAS-2B cells exposed to chromates. The most prominent signal (g = 1.979) is similar to that seen with NADPH-dependent Cr(VI)-reducing enzymes such as glutathione reductase and lipoyl dehydrogenase [28, 65] and human cytochrome b5 plus P450 reductase [32]. The hyperfine structure of this g = 1.979 signal (Fig. 8) is similar to that observed in vitro with various Cr(V)-diol complexes [50]. A Cr(V) signal similar to g = 1.979 has been reported in other biological systems exposed to Cr(VI), including live mice injected with dichromate (g = 1.980) [66], and CHO cells incubated with Na2CrO4 (g = 1.978) [67]. A g = 1.979 signal was not however seen in chick embryo liver [68].
Since this signal can be generated by various Cr(VI) reductants, its formation cannot be linked to a specific reductant, and it may in fact be dependent on several cellular reductants. The elimination of this signal by pre-treatment with the NAD(P)H oxidant PMS (Fig. 6D) implies that Cr(VI) reduction and the generation of the g = 1.979 signal largely depends on NAD(P)H-dependent enzymes. Studies with dichromate-injected mice similarly suggested a role for NAD(P)H-dependent enzymes [35]. Since the intensity of this signal was not decreased (and was actually increased) by pre-treatment with the thiol oxidant diamide (Fig. 6), thiols are not necessary for the generation of this signal. This is consistent with observations in other studies, including: (a) thiols such as GSH may not contribute significantly to Cr(V) formation in Chinese hamster V-79 cells, [69]; (b) thiol depletion of chick embryos did not affect the level of hepatic Cr(V) after 70 min [68]; and (c) GSH depletion increased the Cr(V) signal in the kidney (but not the liver) of rats injected with Cr(VI) [70].
In BEAS-2B cells exposed to Na2CrO4, the major Cr(V) signal (g = 1.979) was persistent. It was maximal early and declined only 35% over 1 hour (Fig. 10). The g = 1.979 Cr(V) signal in dichromate-injected mice declined to a greater extent (75%) after 1 hour [66]. In CHO cells incubated with Na2CrO4, the intensity of the g = 1.978 Cr(V) signal increased over time from 2 to 14.5 min [67], although longer timepoints were not reported. Since Cr(V) has been implicated in various types of cellular damage, the persistence of Cr(V) over time, which implies continual generation, could indicate the potential for continued cellular damage over longer intervals.
4.3 Generation of the g = 1.985 Cr(V) signal
While g = 1.985 was the smaller Cr(V) signal in the BEAS-2B cells, the possible precursor to this signal (g = 1.996) has been noted in some systems. The reduction of Cr(VI) by GSH in Tris buffer (pH 8) yielded two Cr(V) signals, g = 1.996 and g = 1.986, with the latter predominant [71]. In phosphate buffer, the g = 1.995 signal was predominant [72]. After 5 min of GSH-mediated Cr(VI) reduction in phosphate buffer, we similarly observed a predominant g = 1.996 signal and a lesser signal at g = 1.986 (Fig. 13). Levina et al. [53] reported a similar spectrum for GSH plus Cr(VI) at pH 7.0, and for a Cr(V)-GSH complex dissolved in water for two min. Their signal for the Cr(V)-GSH complex (g = 1.996) diminished by 7 min as the GSH-Cr(V) complex decomposed, whereas the intensity of the g = 1.986 signal increased [53]. They reported that all ESR signals from this Cr(V)-GSH complex disappeared after 10 min, indicating instability in aqueous solution. With lower reactant concentrations (5 mM chromate plus 20 mM GSH), their g = 1.986 signal became predominant with a lesser signal at g = 1.996 [53]. In the BEAS-2B cells, we never observed a g = 1.996 component, even at early timepoints. It is possible that a Cr(V)-GSH complex is generated in BEAS-2B cells, but that its levels are below the sensitivity of detection by EPR. It is also possible that Cr(V)-GSH is even less stable in these cells than in the in vitro experiments conducted by Levina et al. [53]. A Cr(V) signal indicative of g = 1.996 was, however, reported for human A549 cells [73]. Since these spectra were run on frozen samples, the signal was seen at g = 1.987 which is equivalent to a g = 1.996 signal at room temperature where giso = ⅓[2.014 + 2(1.987)]. However, these A549 cells are different in many ways from BEAS-2B in that A549 cells: (a) are not normal epithelial cells, but rather a hypotriploid lung adenocarcinoma [74]; (b) consist of at least 4 different cell types [75]; and (c) are 10-fold more resistant to H2O2 than are BEAS-2B cells [76]. Given these marked differences, the differences in Cr(V) spectra are not surprising.
Fig. 13.
Representative Cr(V) signals following incubation of 400 µM Na2CrO4 with 1 mM GSH for 5 min in 10 mM potassium phosphate pH 7.4 under room air. The reaction volume was 0.3 ml and the instrument settings were the same as those in Fig. 1.
Levina et al. proposed that the g = 1.986 signal represents a Cr(V) intermediate resulting from the decomposition of the g = 1.996 GSH-Cr(V) complex [53]. If so, the g = 1.985 signal in BEAS-2B cells could represent indirect evidence for the formation of a GSH-Cr(V) complex. This signal in BEAS-2B cells is dependent on thiols, i.e. it was greatly diminished in diamide-treated cells. In vitro, Cr(V)-GSH appears to largely decompose by ligand oxidation (intramolecular redox reactions) and may therefore not react as readily with cellular macromolecules [53]. In cells, Cr(V)-GSH species might participate in ligand exchange reactions with carbohydrates [53]. Cr(V)-carbohydrate species exhibit ESR signals at g = 1.979 [77, 78], and it is possible that cellular carbohydrates could extend the half-life of the Cr(V) species [79]. Since thiols are depleted in diamide-treated cells, and since the g = 1.979 signal is not diminished in diamide-treated cells, the g = 1.979 signal is not dependent on Cr(V)-GSH/carbohydrate ligand exchanges. Given the abundance of carbohydrates in cells, however, the g = 1.979 signal in cells could include Cr(V)-carbohydrate species which would be consistent with the hyperfine structure noted in Fig. 8. The g = 1.979 signals are not necessarily dependent on carbohydrates because other Cr(V)-diols generate similar spectra and g = 1.979 is predominant in a variety of carbohydrate-free in vitro systems with enzymatic Cr(VI) reductants [28, 32, 65]. Lastly, with lower reactant concentrations in vitro (5 mM chromate plus 20 mM GSH), the g = 1.986 signal declined by 90% after 50 min [53]. In contrast, in the BEAS-2B cells with Na2CrO4, the g = 1.985 signal did not decline after 1 hour (Fig. 10). This signal, could represent an intermediate Cr(V)-GSH complex with non-thiol donor atoms, and its persistence in BEAS-2B cells could either result from a greater stability or a continued generation over time.
From the discussion above, it is apparent that the relative ratio of the Cr(V) signals (g = 1.996, 1.985, 1.979) generated by different systems can vary dependent on the specific conditions. Even within BEAS-2B cells, the ratio of the g = 1.985 and g = 1.979 components varied with the ratio of Cr(VI) to cell number and with time.
4.4 Soluble versus insoluble chromates
Both soluble and insoluble chromates are used industrially. BEAS-2B cells generated detectable Cr(V) signals from both Na2CrO4 (highly soluble) and ZnCrO4 (water insoluble), although the signals were considerably more intense for Na2CrO4 (Fig. 10). While PbCrO4 treated RAW 264.7 cells generate hydroxyl radical [80], to our knowledge this is the first report of Cr(V) detected in cells exposed to an insoluble chromate. ZnCrO4 is widely used as a corrosion inhibitor in primer paints and other industrial coatings so the generation of Cr(V) has potential occupational health considerations. Signals were not, however, detected for PbCrO4 which is also water-insoluble. Cr(V) is a short-lived intermediate and its signal does not accummulate over time. In vitro studies with human microsomes demonstrated a turnover time for Cr(V) of approximately 1.2 min [30]. Similarly, Cr(V) generated by glutathione reductase is short-lived [28]. Therefore, each Cr(V) signal represents the Cr(V) at that point in time. Insoluble chromates would be expected to penetrate cells more slowly, so it is not surprising that their Cr(V) signals are smaller. Since a Cr(V) signal was observed for ZnCrO4 even after just 5 min with the cells (Fig. 10), a portion of this chromate must enter cells within this timeframe. The manner in which ZnCrO4 enters the cells is not yet known. Among the insoluble chromates, ZnCrO4 proved the most soluble in complete medium (DMEM plus FBS) when followed over 7 days [81]. While the serum-free LHC-9 medium used in these studies is different from DMEM, it is still possible that the medium somehow facilitated the dissolution of at least a small portion of the ZnCrO4. Alternatively, the size or surface structure of the ZnCrO4 particles may make them more amenable to cellular components that facilitate dissolution or entry. As noted in the methods, both the ZnCrO4 and PbCrO4 particle suspensions were sonicated just before use to attempt to create as fine a dispersion as possible, and this may have been more effective for ZnCrO4. The absence of Cr(V) signals with PbCrO4 at either 5 or 60 min might indicate either a slower or a considerably delayed uptake of Cr(VI). Given the sensitivity limits of ESR, slow uptake of PbCrO4 may result in Cr(V) levels that are below the detection limit. With Syrian hamster embryo cells, PbCrO4 uptake occurs gradually over 7 days and PbCrO4 solubilization is facilitated by cells [81]. Studies with human bronchial fibroblasts similarly indicate that the cells facilitate the extracellular dissolution of PbCrO4 [82] but that dissolution was not complete [83]. With CHO cells, intracellular Cr started to accummulate 3 hours after PbCrO4 exposure and was maximal at 24 h [84].
In addition to the lack of a Cr(V) signal with PbCrO4, the BEAS-2B cells tolerated much higher levels of PbCrO4 than ZnCrO4 (Fig. 11). The BEAS-2B cells were less sensitive to PbCrO4 when compared with reports for primary human bronchial fibroblasts, e.g. BEAS-2B cells exhibited clonogenic survivals of 70 and 55% for 2 and 7.5 µg PbCrO4/cm2, respectively, whereas fibroblast survival was 16 and 1.9% for 1 and 5 µg/cm2, respectively [83]. The BEAS-2B cells were also less sensitive to PbCrO4 than were human foreskin fibroblasts [41] and WTHBF-6 cells [85]. The decreased sensitivity to PbCrO4 does not reflect a decreased sensitivity of BEAS-2B to all chromates, however, because BEAS-2B cells were somewhat more sensitive to Na2CrO4 than were human bronchial fibroblasts [83] or CHO-AA8 cells [40].
The differential sensitivity among the cell types could be due to differences in the cells themselves, but there could also be contributing factors from the different media. We used serum-free LHC-9 medium, which we demonstrated does not have significant inherent Cr(VI)-reducing properties. Nearly all other studies have used serum-supplemented media, some of which may reduce some Cr(VI) extracellularly as we observed for DMEM (see above). It has been noted that cells in salts-glucose medium are more sensitive to chromate than those in complete medium [40]. This would be consistent with the possibility of extracellular reduction by complete media and therefore decreased Cr(VI) uptake.
4.5 Summary
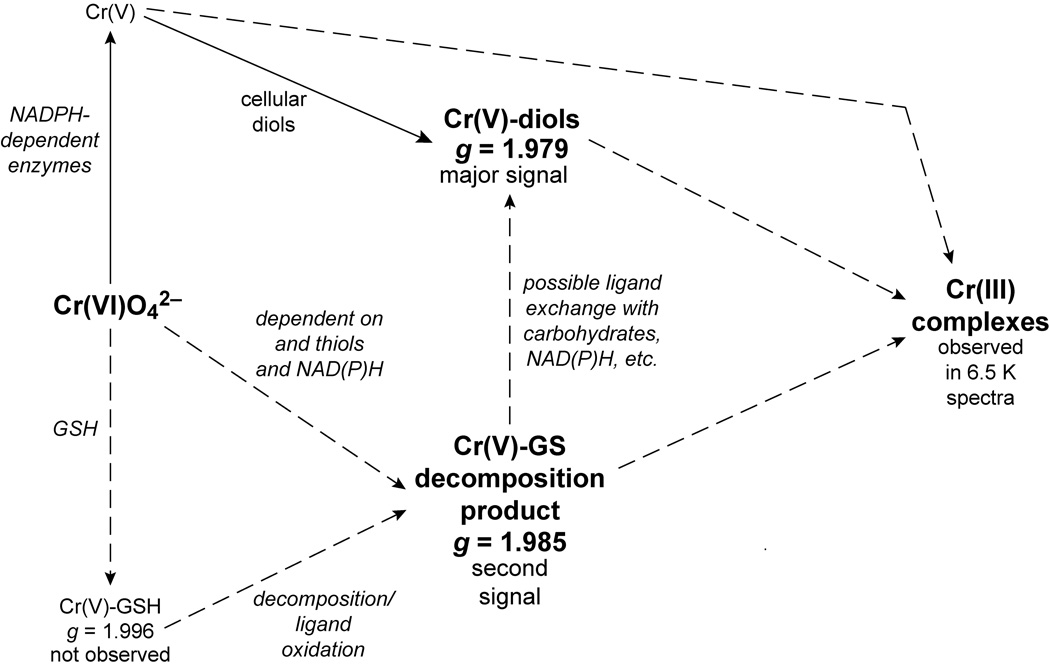
In summary, normal human bronchial epithelial cells generate Cr(V) from both Na2CrO4 (soluble) and ZnCrO4 (insoluble). Clonogenic survival demonstrated that the cells are very sensitive to both of these chromates, but much less sensitive to PbCrO4 for which Cr(V) generation was not detected. Two Cr(V) signals were generated in cells exposed to Na2CrO4 and ZnCrO4. The larger of the Cr(V) signals (g = 1.979) exhibits hyperfine structure consistent with Cr(V)-diol complexes (Fig. 14.) Only the smaller of the Cr(V) signals (g = 1.985) is dependent on thiols, but both are largely NAD(P)H-dependent suggesting a significant role for these enzymes in Cr(V) generation (Fig. 14). Unlike many cells in culture, BEAS-2B cells continue to express the microsomal Cr(VI)-reducing enzymes P450 reductase and cytochrome b5. Cr(V) is generated early after chromate exposure and persists over at least 1 hour which implies the potential for continued reactive species damage over time. At least a portion of the Cr(VI) is rapidly reduced to the next stable oxidation state, Cr(III) (Fig. 14), and at least two Cr(III) complexes were detected in cells. The importance of accounting for potential Cr(VI) reduction by the cell culture medium was also noted. Since bronchial epithelial cells are the primary target of inhaled chromium, BEAS-2B cells should be a suitable model to further study the cellular reductive activation of Cr(VI).
Fig. 14.
General scheme for the formation of the Cr species responsible for the ESR signals observed in BEAS-2B cells. The species associated with each signal are in boldface type. The dashed arrows indicate possible pathways to the formation of certain species, but the exact pathways are not known. The diagram is not meant to be inclusive of all possible pathways or Cr species, but rather is intended to relate the various species observed in these cells.
Acknowledgements
This project was supported by grant number ES012707 from the National Institute of Environmental Health Sciences (NIEHS), NIH. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIEHS, NIH.
The ESR facilities of the Department of Biophysics are supported by National Biomedical ESR Center Grant EB001980 from the NIH.
We are grateful to: Dr. Kasem Nithipatikom and Marilyn Isbell for kindly providing the HPLC instruments and expertise; to Rachel Forbes for technical assistance with the GSH analysis; to Dr. David H. Petering for kindly supplying the Na253CrO4; to Dr. Tak Yee Aw for providing the details of the glutathione analysis; and to Dr. Ronald Hines for his expertise on the culture conditions for BEAS-2B cells.
5. Abbreviations
- Cr
chromium
- DMEM
Dulbecco's Modified Eagle's Medium
- ESR
electron spin resonance
- FBS
fetal bovine serum
- γ-Glu-glu
γ-glutamyl glutamate
- GSH
glutathione, reduced
- GSSG
glutathione, oxidized
- HBSS
Hank's balanced salts solution
- PBS
phosphate-buffered saline
- PMS
phenazine methosulfate
- TCA
trichloroacetic acid
- ZFS
zero field splitting
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Taioli E, Zhitkovich A, Kinney P, Udasin I, Toniolo P, Costa M. Biol. Trace Elem. Res. 1995;50:175–180. doi: 10.1007/BF02785408. [DOI] [PubMed] [Google Scholar]
- 2.Costa M, Zhitkovich A, Toniolo P, Taioli E, Popov T, Lukanova A. Environ. Health Perspect. 1996;104:917–919. doi: 10.1289/ehp.96104s5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ishikawa Y, Nakagawa K, Satoh Y, Kitagawa T, Sugano H, Hirano T, Tsuchiya E. Br. J. Cancer. 1994;70:160–166. doi: 10.1038/bjc.1994.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ishikawa Y, Nakagawa K, Satoh Y, Kitagawa T, Sugano H, Hirano T, Tsuchiya E. Cancer Res. 1994;54:2343–2346. [PubMed] [Google Scholar]
- 5.Raithel H-J, Schaller K-H, Kraus T, Lehnert G. Int. Arch. Occup. Environ. Health. 1993;65:S197–S200. doi: 10.1007/BF00381340. [DOI] [PubMed] [Google Scholar]
- 6.Hayes RB. In: Biological and Environmental Aspects of Chromium. Langard S, editor. Amsterdam: Elsevier Biomedical Press; 1982. pp. 221–239. [Google Scholar]
- 7.Deschamps F, Moulin JJ, Wild P, Labriffe H, Haguenoer JM. Int. Arch. Occup. Environ. Health. 1995;67:147–152. doi: 10.1007/BF00626345. [DOI] [PubMed] [Google Scholar]
- 8.Langard S. Scand. J. Work Environ. Health. 1993;19 Suppl. 1:81–89. [PubMed] [Google Scholar]
- 9.Environmental Protection Agency. Chromium, Integrated Risk Information System. Washington, D.C.: Office of Health and Environmental Assessment, U.S. EPA; 1999. [Google Scholar]
- 10.Gadd GM, White C. TIBTECH. 1993 August11 [Google Scholar]
- 11.Zhitkovich A, Voitkun V, Kluz T, Costa M. Environ. Health Perspect. 1998;106 suppl. 4:696–974. doi: 10.1289/ehp.98106s4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kortenkamp A, Beyersmann D, O’Brien P. Toxicol. Environ. Chem. 1987;14:23–32. [Google Scholar]
- 13.Buttner B, Beyersmann D. Xenobiotica. 1985;15:735–741. doi: 10.3109/00498258509047435. [DOI] [PubMed] [Google Scholar]
- 14.Cantoni O, Costa M. Carcinogenesis. 1984;5:1207–1209. doi: 10.1093/carcin/5.9.1207. [DOI] [PubMed] [Google Scholar]
- 15.Christie NT, Cantoni O, Evans RM, Meyn RE, Costa M. Biochem. Pharmacol. 1984;33:1661–1670. doi: 10.1016/0006-2952(84)90289-2. [DOI] [PubMed] [Google Scholar]
- 16.Levy LS, Venitt S. Carcinogenesis. 1986;7:831–835. doi: 10.1093/carcin/7.5.831. [DOI] [PubMed] [Google Scholar]
- 17.Tsapakos MJ, Hampton TH, Wetterhahn KE. Cancer Res. 1983;43:5662–5667. [PubMed] [Google Scholar]
- 18.Whiting RF, Stich HF, Koropatnick DJ. Chem.-Biol. Interactions. 1979;26:267–280. doi: 10.1016/0009-2797(79)90030-9. [DOI] [PubMed] [Google Scholar]
- 19.Standeven AM, Wetterhahn KE. Chem. Res. Toxicol. 1991;4:616–625. doi: 10.1021/tx00024a003. [DOI] [PubMed] [Google Scholar]
- 20.Dillon CT, Lay PA, Bonin AM, Cholewa M, Legge GJF, Collins TJ, Kostka KL. Chem. Res. Toxicol. 1998;11:119–129. doi: 10.1021/tx9701541. [DOI] [PubMed] [Google Scholar]
- 21.Shi X, Chiu A, Chen CT, Halliwell B, Castranova V, Vallyathan J. Toxicol. Environ. Health. 1999;Pt. B 2:87–104. doi: 10.1080/109374099281241. [DOI] [PubMed] [Google Scholar]
- 22.Shi X, Dalal NS. Arch. Biochem. Biophys. 1992;292:323–327. doi: 10.1016/0003-9861(92)90085-b. [DOI] [PubMed] [Google Scholar]
- 23.Shi X, Ding M, Ye J, Wang S, Leonard SS, Zang L, Castranova V, Vallyathan V, Chiu A, Dalal N, Liu K. J. Inorg. Biochem. 1999;75:37–44. doi: 10.1016/S0162-0134(99)00030-6. [DOI] [PubMed] [Google Scholar]
- 24.Standeven AM, Wetterhahn KE. Carcinogenesis. 1992;13:1319–1324. doi: 10.1093/carcin/13.8.1319. [DOI] [PubMed] [Google Scholar]
- 25.Mikalsen A, Alexander J, Ryberg D. Chem.-Biol. Interact. 1989;69:175–192. doi: 10.1016/0009-2797(89)90076-8. [DOI] [PubMed] [Google Scholar]
- 26.Jennette KW. J. Am. Chem. Soc. 1982;104:874–875. [Google Scholar]
- 27.Shi X, Dong Z, Dalal NS, Gannett PM. Biochim. Biophys. Acta. 1994;1226:65–72. doi: 10.1016/0925-4439(94)90060-4. [DOI] [PubMed] [Google Scholar]
- 28.Shi XL, Dalal NS. J. Inorg. Biochem. 1990;40:1–12. doi: 10.1016/0162-0134(90)80034-u. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki Y, Fukuda K. Arch. Toxicol. 1990;64:169–176. doi: 10.1007/BF02010721. [DOI] [PubMed] [Google Scholar]
- 30.Myers CR, Myers JM, Carstens BP, Antholine WE. Toxic Subst. Mech. 2000;19:25–51. [Google Scholar]
- 31.Jannetto PJ, Antholine WE, Myers CR. Toxicology. 2001;159:119–133. doi: 10.1016/s0300-483x(00)00378-4. [DOI] [PubMed] [Google Scholar]
- 32.Borthiry GR, Antholine WE, Kalyanaraman B, Myers JM, Myers CR. Free Radic. Biol. Med. 2007;42:738–755. doi: 10.1016/j.freeradbiomed.2006.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Myers CR, Myers JM. Carcinogenesis. 1998;19:1029–1038. doi: 10.1093/carcin/19.6.1029. [DOI] [PubMed] [Google Scholar]
- 34.Pratt PF, Myers CR. Carcinogenesis. 1993;14:2051–2057. doi: 10.1093/carcin/14.10.2051. [DOI] [PubMed] [Google Scholar]
- 35.Liu KJ, Shi X. Mol. Cell. Biochem. 2001;222:41–47. [PubMed] [Google Scholar]
- 36.Becker N, Chang-Claude J, Frentzel-Beyme R. Br. J. Ind. Med. 1991;48:675–683. doi: 10.1136/oem.48.10.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakagawa K, Matsubara T, Kinoshita I, Tsuchiya E, Sugano H, Hirano T. Lung Cancer. 1984;24:301–310. [Google Scholar]
- 38.Franchini I, Magnani F, Mutti A. Scand. J. Work Environ. Health. 1983;9:247–252. doi: 10.5271/sjweh.2413. [DOI] [PubMed] [Google Scholar]
- 39.Stern RM. Arch. Environ. Health. 1983;38:148–155. doi: 10.1080/00039896.1983.10543996. [DOI] [PubMed] [Google Scholar]
- 40.Blankenship LJ, Carlisle DL, Wise JP, Orenstein JM, Dye LE, 3rd, Patierno SR. Toxicol. Appl. Pharmacol. 1997;146:270–280. doi: 10.1006/taap.1997.8237. [DOI] [PubMed] [Google Scholar]
- 41.Wise JP, Leonard JC, Patierno SR. Mutat. Res. 1992;278:69–79. doi: 10.1016/0165-1218(92)90287-a. [DOI] [PubMed] [Google Scholar]
- 42.Pias EK, Aw TY. Cell Death Differ. 2002;9:1007–1016. doi: 10.1038/sj.cdd.4401064. [DOI] [PubMed] [Google Scholar]
- 43.Myers CR, Myers JM. J. Bacteriol. 1997;179:1143–1152. doi: 10.1128/jb.179.4.1143-1152.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stearns DM, Wetterhahn KE. Chem. Res. Toxicol. 1994;7:219–230. doi: 10.1021/tx00038a016. [DOI] [PubMed] [Google Scholar]
- 45.Molyneux MJ, Davies MJ. Carcinogenesis. 1995;16:875–882. doi: 10.1093/carcin/16.4.875. [DOI] [PubMed] [Google Scholar]
- 46.Borthiry GR, Antholine WE, Kalyanaraman B, Myers JM, Myers CR. Free Radic. Biol. Med. 2007;42:738–755. doi: 10.1016/j.freeradbiomed.2006.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pratt PF, Myers CR. Carcinogenesis. 1993;14:2051–2057. doi: 10.1093/carcin/14.10.2051. [DOI] [PubMed] [Google Scholar]
- 48.Soderdahl T, Enoksson M, Lundberg M, Holmgren A, Ottersen OP, Orrenius S, Bolcsfoldi G, Cotgreave IA. FASEB J. 2003;17:124–126. doi: 10.1096/fj.02-0259fje. [DOI] [PubMed] [Google Scholar]
- 49.Mueller CF, Widder JD, McNally JS, McCann L, Jones DP, Harrison DG. Circ. Res. 2005;97:637–644. doi: 10.1161/01.RES.0000183734.21112.b7. [DOI] [PubMed] [Google Scholar]
- 50.Shi X, Dalal NS. Arch. Biochem. Biophys. 1990;277:342–350. doi: 10.1016/0003-9861(90)90589-q. [DOI] [PubMed] [Google Scholar]
- 51.Hagen WR. Adv. Inorg. Chem. 1992;38:165–222. [Google Scholar]
- 52.Werth MT, Tang S-F, Formicka G, Zeppezauer M, Johnson MK. Inorg. Chem. 1995;34:218–228. [Google Scholar]
- 53.Levina A, Zhang L, Lay PA. Inorg. Chem. 2003;42:767–784. doi: 10.1021/ic020621o. [DOI] [PubMed] [Google Scholar]
- 54.Beinert H, Albracht SP. Biochim. Biophys. Acta. 1982;683:245–277. doi: 10.1016/0304-4173(82)90003-9. [DOI] [PubMed] [Google Scholar]
- 55.Wise SS, Holmes AL, Ketterer ME, Hartsock WJ, Fomchenko E, Katsifis S, Thompson WD, Wise JP., Sr Mutat. Res. 2004;560:79–89. doi: 10.1016/j.mrgentox.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 56.Mace K, Gonzalez FJ, McConnell IR, Garner RC, Avanti O, Harris CC, Pfeifer AM. Mol. Carcinogen. 1994;11:65–73. doi: 10.1002/mc.2940110203. [DOI] [PubMed] [Google Scholar]
- 57.Baruthio F. Biol. Trace Element Res. 1992;32:145–153. doi: 10.1007/BF02784599. [DOI] [PubMed] [Google Scholar]
- 58.Reddel RR, Ke Y, Gerwin BI, McMenamin MG, Lechner JF, Su RT, Brash DE, Park JB, Rhim JS, Harris CC. Cancer Res. 1988;48:1904–1909. [PubMed] [Google Scholar]
- 59.Van Vleet T, Macé K, Coulombe RJ. Cancer Res. 2002;62:105–112. [PubMed] [Google Scholar]
- 60.Frampton MW, Ghio AJ, Samet JM, Carson JL, Carter JD, Devlin RB. Am. J. Physiol. 1999;277:L960–L967. doi: 10.1152/ajplung.1999.277.5.L960. (Lung Cell. Mol. Physiol. 21) [DOI] [PubMed] [Google Scholar]
- 61.Gurr JR, Wang AS, Chen CH, Jan KY. Toxicology. 2005;213:66–73. doi: 10.1016/j.tox.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 62.Ghio AJ, Carter JD, Dailey LA, Devlin RB, Samet JM. Am. J. Physiol. 1999;276:L933–L940. doi: 10.1152/ajplung.1999.276.6.L933. (Lung Cell. Mol. Physiol. 20) [DOI] [PubMed] [Google Scholar]
- 63.Van Vleet TR, Klein PJ, Coulombe RA., Jr J. Toxicol. Environ. Health A. 2002;65:853–867. doi: 10.1080/00984100290071216. [DOI] [PubMed] [Google Scholar]
- 64.Hall PdM, Stupans I, Burgess W, Birkett DJ, McManus ME. Carcinogenesis. 1989;10:521–530. doi: 10.1093/carcin/10.3.521. [DOI] [PubMed] [Google Scholar]
- 65.Shi X, Dalal NS. FEBS Lett. 1990;276:189–191. doi: 10.1016/0014-5793(90)80539-u. [DOI] [PubMed] [Google Scholar]
- 66.Liu KJ, Jiang J, Swartz HM, Shi X. Arch. Biochem. Biophys. 1994;313:248–252. doi: 10.1006/abbi.1994.1384. [DOI] [PubMed] [Google Scholar]
- 67.Sugiyama M, Tsuzuki K, Haramaki N. Mutat. Res. 1993;299:95–102. doi: 10.1016/0165-1218(93)90086-s. [DOI] [PubMed] [Google Scholar]
- 68.Liebross RH, Wetterhahn KE. Carcinogenesis. 1992;13:2113–2120. doi: 10.1093/carcin/13.11.2113. [DOI] [PubMed] [Google Scholar]
- 69.Sugiyama M, Tsuzuki K. FEBS Lett. 1994;341:273–276. doi: 10.1016/0014-5793(94)80471-0. [DOI] [PubMed] [Google Scholar]
- 70.Yuann J-MP, Liu KJ, Hamilton JW, Wetterhahn KE. Carcinogenesis. 1999;20:1267–1275. doi: 10.1093/carcin/20.7.1267. [DOI] [PubMed] [Google Scholar]
- 71.Aiyar J, Borges KM, Floyd RA, Wetterhahn KE. Toxicol. Environ. Chem. 1989;22:135–148. [Google Scholar]
- 72.Shi XL, Dalal NS. Free Radic. Res. Commun. 1990;10:17–26. doi: 10.3109/10715769009145929. [DOI] [PubMed] [Google Scholar]
- 73.Martin BD, Schoenhard JA, Sugden KD. Chem. Res. Toxicol. 1998;11:1402–1410. doi: 10.1021/tx9801559. [DOI] [PubMed] [Google Scholar]
- 74.Giard DJ, Aaronson SA, Todaro GJ, Arnstein P, Kersey JH, Dosik H, Parks WP. J. Natl. Cancer Inst. 1973;51:1417–1423. doi: 10.1093/jnci/51.5.1417. [DOI] [PubMed] [Google Scholar]
- 75.Croce MV, Colussi AG, Price MR, Segal-Eiras A. Pathol. Oncol. Res. 1999;5:197–204. doi: 10.1053/paor.1999.0212. [DOI] [PubMed] [Google Scholar]
- 76.Nanavaty UB, Pawliczak R, Doniger J, Gladwin MT, Cowan MJ, Logun C, Shelhamer JH. Exp. Lung Res. 2002;28:591–607. doi: 10.1080/01902140260426715. [DOI] [PubMed] [Google Scholar]
- 77.Codd R, Lay PA. J. Am. Chem. Soc. 2001;123:11799–11800. doi: 10.1021/ja0160501. [DOI] [PubMed] [Google Scholar]
- 78.Codd R, Lay PA. J. Am. Chem. Soc. 1999;121:7864–7876. [Google Scholar]
- 79.Branca M, Dessi' A, Kozlowski H, Micera G, Swiatek J. J. Inorg. Biochem. 1990;39:217–226. [Google Scholar]
- 80.Leonard SS, Roberts JR, Antonini JM, Castranova V, Shi X. Mol. Cell. Biochem. 2004;255:171–179. doi: 10.1023/b:mcbi.0000007273.23747.67. [DOI] [PubMed] [Google Scholar]
- 81.Elias Z, Poirot O, Baruthio F, Daniere MC. Carcinogenesis. 1991;12:1811–1816. doi: 10.1093/carcin/12.10.1811. [DOI] [PubMed] [Google Scholar]
- 82.Xie H, Holmes AL, Wise SS, Gordon N, Wise JP., Sr Chem. Res. Toxicol. 2004;17:1362–1367. doi: 10.1021/tx0498509. [DOI] [PubMed] [Google Scholar]
- 83.Wise JP, Sr, Wise SS, Little JE. Mutat. Res. 2002;517:221–229. doi: 10.1016/s1383-5718(02)00071-2. [DOI] [PubMed] [Google Scholar]
- 84.Wise JPS, Stearns DM, Wetterhahn KE, Patierno SR. Carcinogenesis. 1994;15:2249–2254. doi: 10.1093/carcin/15.10.2249. [DOI] [PubMed] [Google Scholar]
- 85.Wise SS, Schuler JH, Holmes AL, Katsifis SP, Ketterer ME, Hartsock WJ, Zheng T, Wise JP., Sr Environ. Mol. Mutagen. 2004;44:156–162. doi: 10.1002/em.20044. [DOI] [PubMed] [Google Scholar]