Abstract
The ribozyme RNase P absolutely requires divalent metal ions for catalytic function. Multiple Mg2+ ions contribute to the optimal catalytic efficiency of RNase P, and it is likely that the tertiary structure of the ribozyme forms a specific metal-binding pocket for these ions within the active-site. To identify base moieties that contribute to catalytic metal-binding sites, we have used in vitro selection to isolate variants of the Escherichia coli RNase P RNA with altered specificities for divalent metal. RNase P RNA variants with increased activity in Ca2+ were enriched over 18 generations of selection for catalysis in the presence of Ca2+, which is normally disfavored relative to Mg2+. Although a wide spectrum of mutations was found in the generation-18 clones, only a single point mutation was common to all clones: a cytosine-to-uracil transition at position 70 (E. coli numbering) of RNase P. Analysis of the C70U point mutant in a wild-type background confirmed that the identity of the base at position 70 is the sole determinant of Ca2+ selectivity. It is noteworthy that C70 lies within the phylogenetically well conserved J3/4-P4-J2/4 region, previously implicated in Mg2+ binding. Our finding that a single base change is sufficient to alter the metal preference of RNase P is further evidence that the J3/4-P4-J2/4 domain forms a portion of the ribozyme’s active site.
Despite the occurrence of a wide variety of structures and mechanisms among catalytic RNAs (ribozymes), most are metalloenzymes that require divalent metal cations for catalytic function (1). Divalent metals are thought to play two critical roles in ribozyme function. First, they promote the proper folding of RNA tertiary structures. Second, metals can participate directly in catalysis by activating nucleophiles, stabilizing transition states, and stabilizing leaving groups (1–3). Small catalytic RNAs, such as the hammerhead ribozyme, can function with a wide variety of metal species. In general, the differences in catalytic efficiency that the hammerhead RNA exhibit in response to different metal species can be attributed to the nature of the metal cations (i.e., differences in the pKa values of coordinated water molecules) rather than to the RNA itself (4, 5). In contrast, larger ribozymes, such as the group I self-splicing intron and RNase P are much more stringent in their metal requirements. RNase P, for instance, requires Mg2+ for optimal activity, although Mn2+ can substitute with a marginal loss of activity (6–9). The only other divalent cation reported to stimulate RNase P activity is Ca2+, which does so with a 104-fold reduction in the rate of the catalytic step relative to Mg2+ (10, 11). Similarly, group I introns function only in the presence of Mg2+ or Mn2+ (12). The preferences shown by large ribozymes for particular species of divalent metal cannot be explained solely by the chemical properties of the metals. For instance, based on the pKa values of Ca2+- and Mg2+-bound water molecules, the catalytic activity of RNase P should vary only 25-fold between the two metals (3). Rather, stringent metal specificities suggest that these ribozymes form unique structures that can discriminate between various species of metal, as has been observed in the crystal structures of several RNA–metal complexes (13–15).
Although providing a wealth of structural information, x-ray crystallographic data cannot a priori determine whether a given metal-binding site is functionally important and, if so, whether it is required for RNA folding, catalysis, or both. The functions of metal-binding sites instead must be explored by other means, such as deoxy- or phosphorothioate-modification-interference techniques, which can identify 2′-OH or phosphate groups (respectively) that coordinate functionally important metals (16–23). For example, a cluster of phosphate oxygens within a phylogenetically well conserved domain of RNase P (helix P4 and the flanking J2/4 element) has been identified by phosphorothioate interference as being critical for catalysis (21). In structural models of the ribozyme–substrate complex, helix P4 is positioned immediately adjacent to the site of pre-tRNA cleavage (24, 25). Harris and Pace (21) thus have proposed that helix P4 binds Mg2+ ions that comprise a portion of the enzyme’s active site.
In addition to phosphate oxygens, it is likely that some base moieties of an RNA are involved in the formation of specific metal-binding pockets. These bases may coordinate Mg2+ (either directly or through metal-bound water) or form structures with 2′ hydroxyl or phosphate groups properly positioned to bind metal ions specifically. One means of identifying base residues that interact with metals is through in vitro selection, which allows the screening of large and complex populations of RNA sequence variants (26–28). Using this approach, Pan and Uhlenbeck (29, 30) and Williams et al. (31) have isolated artificial ribozymes that self-cleave in the presence of Pb2+ or Mg2+, respectively. Similarly, Ciesiolka and Yarus (32, 33) have selected small RNA structures capable of specifically binding Zn2+. The identification of conserved nucleotides and/or structural motifs in such artificially created phylogenies can suggest testable models for metal–RNA interactions, as well as explore the varieties of metal-binding pockets.
In vitro selection has also been employed to change the spectrum of divalent metals with which the group I ribozyme is catalytically active (34, 35). Lehman and Joyce (34) identified a complex suite of seven mutations that greatly improved the activity of group I intron in Ca2+. Several of these mutations are located in proximity to the GTP-binding site and so probably are near the active site of the ribozyme. It is unlikely that site-directed mutagenesis would have uncovered the requisite combination of nucleotide changes obtained by this selection.
In this paper we report the in vitro selection of RNase P ribozymes with altered metal specificity. Because RNase P normally acts in trans, we used a mutagenized population of self-cleaving pre-tRNA-RNase P RNA conjugates (36) to select for enhanced catalytic activity in the presence of Ca2+, which is normally disfavored relative to Mg2+. In contrast to the results reported for the group I intron, we find that a single-nucleotide change within the core of RNase P is sufficient to alter substantially the metal selectivity of this ribozyme.
MATERIALS AND METHODS
The procedures used to prepare guanosine monophosphorothioate (GMPS)-initiated RNA transcripts, couple GMPS-RNA to iodoacetyl-derivatized agarose (Sulfolink, Pierce), select active TP292 variants, reamplify cDNA, and clone cDNA libraries were performed as described (Fig. 1; ref. 37). However, in the selection experiments described, 100 mM CaCl2 (pH 8) was used to elute active ribozymes from the agarose beads. The selection was made progressively more stringent by decreasing the time of incubation in the presence of Ca2+: round 1, 25 min; rounds 2–9, 5 min; rounds 10–13, 1 min; rounds 14–18, 30 sec. Error-prone PCR followed the protocol of Cadwell and Joyce (38).
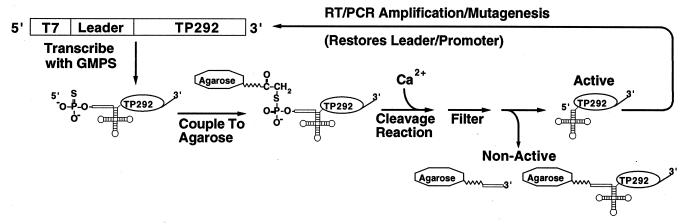
Figure 1.
RNase P in vitro selection scheme. GMPS, guanosine monophosphorothioate. Agarose, Sulfolink coupling gel (Pierce). See text for details.
Activity assays were performed, as described (36, 37), in 3 M NH4OAc/44.5 mM Tris/16 mM Pipes. Reactions in CaCl2 (100 mM, unless specified) were performed at pH 8, which is optimal for TP292 self-cleavage. To accurately measure rates in MgCl2 (25 mM, unless specified), it was necessary to slow the reactions (≈100-fold) by assaying cleavage at pH 6 (11). First-order reaction rate constants were determined by measuring the slopes of plots of ln[(S/So) − (S∞/So)] vs. time (S = [RNA precursor], So = initial [RNA], and S∞= [RNA] at >5 half-lives). Average S∞/So values for the data in Table 1 are as follows: TP292 Ca2+ = 0.3 ± .04; TP292 Mg2+ = 0.2 ± .02; TP292-C70U Ca2+ = 0.2 ± .02; TP292-C70U Mg2+ = 0.4 ± 0.1. The diversity index used in Fig. 3 is calculated as (H/Hmax), where H equals the Shannon entropy at a given position within the sequence alignment −Σplog2(p); p = frequency of occurrence of a given base or gap at a given position; ref. 39). Hmax = log2(5) for five character states (G, A, U, C, −). Natural RNase P RNA sequences were obtained from the RNase P RNA database (40).
Table 1.
Self-cleavage rate constants in Ca2+ and Mg2+*
| RNA | kCa | kMg | kCa/kMg |
|---|---|---|---|
| TP292 | .062 ± .02 min−1 | 10.2 ± 2 | 1† |
| TP292-C70U | .26 ± .03 | .48 ± 1 | 91 |
First-order rate constants measured at pH 8 for Ca2+ and pH 6 for Mg2+.
Normalized value.
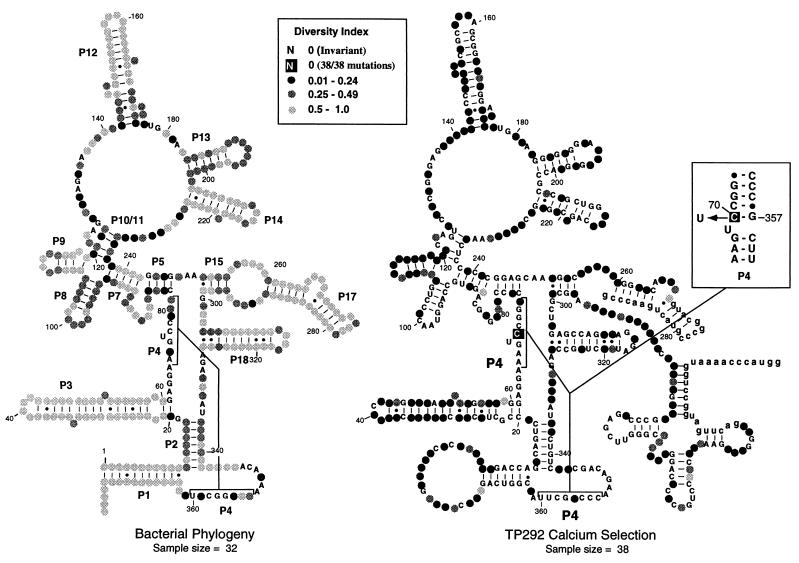
Figure 3.
Distribution of mutations identified in round-18 clones. The genetic variability of each nucleotide position within RNase P/TP292 is summarized for both natural bacterial phylogeny (Left; sequences obtained from the RNase P RNA database; ref. 40) and the Ca2+ selection (Right). The diversity of sequence at each position was calculated as H/Hmax, where H is the Shannon entropy [−Σplog(p)] and Hmax is log(5), for five character states (G, A, U, C, −). Higher sequence variability (i.e., a higher diversity index) is denoted by more lightly shaded circles, and invariant positions are denoted by capital letters. The boxed base at position 70 of TP292 signifies that all round-18 clones contained a mutation at this position. The boxed Inset (Right) shows the position of the C-to-U transition at position 70, within the context of helix P4.
RESULTS
Selection for Enhanced Activity in Ca2+.
In vitro selection experiments require the ability to separate functional from nonfunctional RNAs in a randomized population. Because the native RNase P ribozyme is a true enzyme, in that its structure is not altered by the catalytic cycle, such sorting is precluded. As a substrate for selection, therefore, we used a self-cleaving conjugate between E. coli RNase P RNA and a pre-tRNA substrate (Fig. 3); characterization of the RNA used in this study, TP292, has been described (36). Self-cleavage by TP292 produces a free 5′ leader element along with a concomitantly shortened mature-tRNA-RNase P RNA conjugate molecule. Functional and nonfunctional molecules can be distinguished, therefore, on the basis of the catalytic release of the 5′ leader. TP292 undergoes rapid and efficient self-cleavage in vitro with a first-order reaction rate similar to that of the native reaction (36); it is thus an appropriate construct for selections designed to modify catalytic activity.
In the selection system used in this study (Fig. 1), randomly mutagenized TP292 RNA molecules are covalently linked, via their 5′ leader elements, to a solid support (Sulfolink agarose, Pierce). Catalytically active molecules are released from the matrix upon self-cleavage, whereas nonactive molecules remain bound. Cleavage is initiated by addition of CaCl2 to the covalently bound population of RNA, and after elution of active molecules, full-length TP292 cDNA (i.e., with T7 promoter and 5′ leader element restored) is regenerated by reverse transcription and PCR.
TP292 variants with enhanced catalytic activity in Ca2+ were enriched over 18 generations of selection and reamplification. Random mutations were introduced into the initial RNA pool as well as the rounds 1–12 pools by error-prone PCR (38). In a control experiment, we determined that an average of 1.4 mutations were incorporated into each PCR product per generation. At this level of mutagenesis, the initial population of TP292 sequence variants (≈4 × 1013 molecules) was sufficiently complex to encompass all possible quadruple mutations.
In each round of selection, the activity of the population was estimated by measuring the amount of radiolabeled RNA eluted from the column, by either scintillation counting or phosphorimaging of denaturing polyacrylamide gels. Whole-population self-cleavage activity assays were also performed in parallel for the RNA libraries obtained in every third generation (the RNAs in these assays were not bound to agarose). Fig. 2 shows self-cleavage assays in which the relative activities of the RNA populations are reflected by the ratios of product and precursor RNAs, as resolved by gel electrophoresis. At the population level, the rates of cleavage in Ca2+ decreased 3-fold during the first six rounds of selection. However, in the subsequent 12 rounds of selection, Ca2+ activity steadily increased and surpassed the activities of both native TP292 and the initial, mutagenized RNA population.
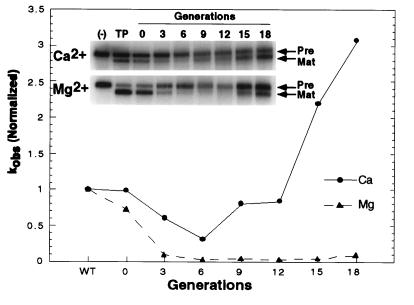
Figure 2.
Evolution of catalytic activity in Ca2+ or Mg2+. Apparent rate constants (kobs) were determined by single-time-point assays of self-cleavage for every third generation of RNA. Measurements were within the linear ranges of the reactions. Precursor and product RNAs were quantitated by phosphorimaging. Values were normalized to the rate constants determined for native TP292 in either Ca2+ or Mg2+. (n.b. the absolute value of kMg/kCa for native TP292 is ≈1.6 × 104). (Inset) Autoradiographs of single-time-point, self-cleavage assays of selected RNA populations performed in either Ca2+ (100 mM CaCl2, pH 8.0) or Mg2+ (25 mM MgCl2, pH 6.0). (−) TP292 RNA incubated in the absence of divalent metal. TP, native TP292 RNA; Pre, full-length TP292 RNA; Mat, TP292 RNA minus the 5′ leader element.
Surprisingly, although no explicit counterselection against Mg2+ activity was performed, the activity of the selected RNA pools in Mg2+ decreased through the first 12 rounds of selection. Although activity in Mg2+ rose slightly between rounds 12 and 18, the RNAs in these pools remained less active in Mg2+ than native TP292. The overall loss of activity in Mg2+ is significant for two reasons. First, it demonstrates that the self-cleavage activity in Ca2+ was not because of trace contaminants of Mg2+ or Mn2+. Second, loss of Mg2+ activity means that the increase in Ca2+ activity is not the result of mutations that simply improve catalytic activity, regardless of the metal (i.e., “up” mutations). Rather, 18 rounds of selection for Ca2+ activity produced a population of TP292 variant RNAs with altered specificity for divalent metal ions.
Sequence Analysis of Round 18 Clones.
Between rounds 16 and 18, little increase in Ca2+ activity of the bulk population was observed, and so following round 18, a portion of the cDNA library was cloned to examine individual RNA species. Full-length sequences were determined for 38 round-18 clones. The spectrum of mutations identified in these clones is summarized in Fig. 3, in which nucleotides have been grouped according to the degree of sequence diversity found at each position. Also shown is a similar depiction of the naturally occurring phylogenetic variability at each position of bacterial RNase P. On average, each TP292 gene was found to contain 16 point mutations. Although mutations were identified throughout the length of TP292, their distribution was not random. In general, few mutations were found in phylogenetically well conserved regions of the RNA (e.g., helix P4). Thirty-three of the 39 phylogenetically invariant positions within the bacterial RNase P RNA remained invariant in the artificially selected TP292 molecules. Presumably, mutations within these elements diminished catalytic activity and were eliminated; thus, the core structure of the ribozyme was maintained throughout the selection. In contrast, phylogenetically variable structures (e.g., helices P3, P9, and P12) as well as structural elements unique to TP292 (e.g., the nucleotides linking the ribozyme to its substrate and those in the loop that links the native 5′ and 3′ ends in P1) accumulated far more mutations than the more well conserved domains.
Only one residue was changed in every round-18 clone: C at position 70 to U (C70U; Fig. 3). Recovery of C70U in every round-18 clone strongly implicates this mutation as critical for the observed alteration in metal specificity of the selected population of RNAs. Intriguingly, C70 is a phylogenetically invariant nucleotide in helix P4, at the core of RNase P (24, 25). Because helix P4 is implicated in binding catalytically important Mg2+ ions (21), it was important to assess the contribution of the single C70U mutation to the altered metal preference exhibited by all the round-18 RNAs.
A Single Mutation Is Sufficient to Activate Ca2+ Activity.
To directly assess the involvement of the C70U mutation in enhanced Ca2+ activity, C70U was introduced into an otherwise native TP292 construct, creating TP292-C70U. RNAs were prepared from native TP292 and TP292-C70U constructs, and their rates of self-cleavage were determined in the presence of 100 mM Ca2+ or 25 mM Mg2+ (optimal for TP292 cleavage). As summarized in Fig. 4A and Table 1, the single C70U mutation is sufficient to increase the activity of TP292 in Ca2+ approximately 4-fold, relative to native TP292. Indeed, the rate of TP292-C70U self-cleavage was comparable to, or exceeded, the rates of cleavage measured for individual clones from the round-18 library. In contrast, the mutations C70A, C70G, and C71U (which converts an adjacent, invariant C residue to U) individually reduce catalytic activity in Ca2+ (Fig. 4A; data not shown). In addition to activating Ca2+ activity, the C70U mutation also resulted in diminished self-cleavage activity in the presence of Mg2+ and Mn2+ (Table 1 and data not shown). In Mg2+, the rate of TP292-C70U self-cleavage was reduced greater than 20-fold. Thus, the single TP292-C70U mutation is sufficient to alter the metal specificity (kCa/kMg) of TP292 by approximately 90-fold. No other metals tested (Ba2+, Cd2+, Co2+, Cu2+, Ni2+, Pb2+, Sr2+, and Zn2+) increased the activity of TP292-C70U relative to native TP292 (data not shown).
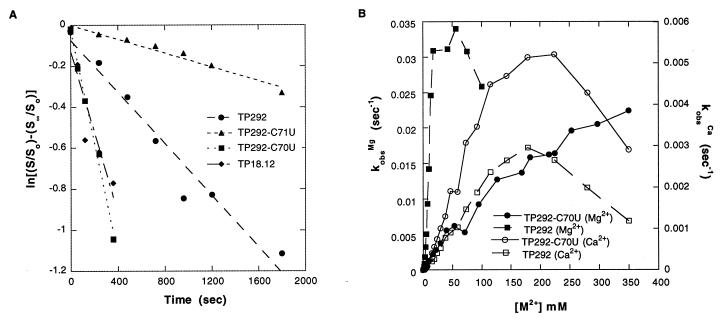
Figure 4.
Kinetic analysis of TP292 variants. All reactions were performed under standard conditions (Materials and Methods; ref. 36). (A) Reaction time courses of TP292 variants performed in CaCl2. TP292-C70U, site-directed C-to-U mutation at position 70, as found in all round-18 clones. TP292-C71U, site-directed C-to-U mutation at position 71. TP18.12, TP292 clone from round 18 of selection. (B) Titration of TP292-C70U and native TP292 with MgCl2 or CaCl2.
To more fully characterize the TP292-C70U mutation, self-cleavage rates were measured over a range of Ca2+ and Mg2+ concentration (Fig. 4B). The activities of TP292 and TP292-C70U RNAs exhibited similar responses to titration with Ca2+, both maximal at approximately 200 mM Ca2+ (normalized, the two curves are superimposable). Despite these similarities, the TP292-C70U RNA was uniformly more active than TP292 at each point of the titration. This indicates that the effect of the C70U mutation was not simply to increase the binding of Ca2+ by the ribozyme. In contrast, titration with Mg2+ revealed that TP292-C70U requires greatly elevated Mg2+ concentrations relative to TP292 for optimal activity (>350 mM vs. 25–50 mM). Thus, the diminished activity of TP292-C70U that was determined at 25 mM Mg2+ (Table 1) can be largely restored by increased Mg2+, suggesting that the C70U mutation disrupts the affinity of TP292 for Mg2+.
DISCUSSION
RNase P functions optimally only in the presence of the divalent metal ions Mg2+ and Mn2+. Although Ca2+ also can promote catalysis, it results in activity that is several orders of magnitude less efficient than Mg2+ and Mn2+ (≈104; ref. 11). In this study we have used in vitro selection to isolate RNase P RNA variants with increased catalytic activity in Ca2+. Our goal was to identify RNase P nucleotides involved in binding the divalent metals that participate in catalysis. The selection experiment used a self-cleaving pre-tRNA-RNase P RNA conjugate, TP292, the activity of which is comparable to native RNase P (36). TP292 activity also favors Mg2+ and Mn2+ over Ca2+ (kMg/kCa ≈ 1.6 × 104), demonstrating that the metal specificity of the conjugate RNA is similar to that of the native RNase P reaction.
TP292 variants with increased catalytic activity in Ca2+ were isolated over 18 rounds of selection. Over the course of the first six rounds, the RNA pools became progressively less active in both Ca2+ and Mg2+. We attribute this diminution of activity to a relatively low stringency of selection coupled with the continuous introduction of deleterious mutations into the population via PCR mutagenesis; the stringency of selection initially was not sufficient to remove all unfavorable mutations from the population. The concomitant loss of activity in both cations, despite the absence of a counterselection against Mg2+-induced self-cleavage, further supports this idea. Following round 6, however, the Ca2+ activity of the RNA pools steadily increased and by round 18 had plateaued at a level approximately 3-fold higher than the Ca2+ activity of native TP292. In contrast, Mg2+ activity of the selected RNAs remained well below native TP292 activity throughout the course of the selection. This selection experiment thus produced a population of TP292 variants with altered specificity for divalent ions.
Although the round-18 clones that were sequenced (n = 38) contained an average of 16 mutations per clone, only one mutation was common to all clones: a C-to-U transition at position 70. Analysis of C70U in an otherwise native TP292 context demonstrates that this mutation is sufficient to enhance Ca2+ activity to an extent equal to, or greater than, individual clones from the round-18 library. Furthermore, TP292-C70U results in diminished catalytic activity in Mg2+ and Mn2+. Interestingly, several of the round-18 clones were significantly less active in Mg2+ than was TP292-C70U, but exhibited activity in Ca2+ that was comparable to TP292-C70U. In these clones, mutations other than C70U must impact the level of Mg2+-dependent catalysis without affecting the basal level of catalysis in Ca2+. We are unable to discern any patterns in the mutations found in these clones that would explain this phenomenon.
The overall effect of the TP292-C70U mutation is to increase Ca2+-dependent activity by 4-fold and decrease activity in Mg2+ by more than 20-fold, relative to native TP292. Thus, the 1.6 × 104-fold preference of TP292 for Mg2+ over Ca2+ is reduced to 1.8 × 102-fold by a single point mutation, TP292-C70U. Based on the titration experiments shown in Fig. 4B, the change in specificity induced by C70U resulted from two distinct effects: (i) a decrease in the Mg2+-binding affinity of the ribozyme that is most straightforwardly interpreted as the loss of specific liganding elements and (ii) an increase in the ability of TP292-C70U to use Ca2+ in catalysis, rather than to bind Ca2+. The diminished specificity that results from the TP292-C70U mutation is specific for Ca2+, not caused simply by a “loosening” of the interactions through which RNase P normally selects functional metals, as evidenced by the fact that no other divalent metal tested was activated by the C70U mutation (data not shown).
Comparison to Selections with Group I Intron.
The RNase P and group I intron RNAs, the only large RNAs that have been subjected to metal-dependent selection experiments, respond differently to similar selection schemes (34, 35). In general, the structure and function of the group I intron seems to be much more adaptable than is RNase P RNA. Selection for Ca2+ activity resulted in group I RNAs that are several orders of magnitude more active in Ca2+ than the native RNA, but produced only moderate gains in the Ca2+ activity of RNase P. The winning solution found in the group I intron Ca2+ selection was a composite of seven mutations, several of which independently resulted in increased catalytic activity (35). In addition to mutations that altered metal specificity (kCa/kMg), a number of mutations in the group I intron simply increased the overall catalytic activity of the ribozyme, regardless of metal species (35). Fixation of these gain-of-activity mutations resulted in a population of RNAs with increased fitness in Mg2+ (35). In contrast, only a single mutation, C70U, was found to increase Ca2+-dependent activity in RNase P; no similar “up” mutations have been identified in RNase P (this work; ref. 37). The frequency of mutations in the winning populations of group I and RNase P RNAs are similar (4.7% vs. 3.8% mutagenesis), indicating that roughly equivalent volumes of sequence space were explored in the two selections. The fact that RNase P could not evolve to accommodate Ca2+ as well as the group I intron suggests that the structure of RNase P, particularly its active site, is much more constrained than is that of the group I intron.
Structural Implications.
The cytosine at position 70 of bacterial RNase P RNA is normally paired with a guanosine at position 357. The TP292-C70U mutation should, therefore, introduce a U⋅G wobble pair into the center of helix P4. The guanosine at position 357 is also critical for improved activity in Ca2+; no other base at this position will enhance Ca2+ activity in combination with C70U (or in the native context; data not shown). Changing the adjacent C71-G356 base pair to a U⋅G mismatch diminished the activity of TP292 in Ca2+ (Fig. 4A), indicating that enhanced Ca2+ activity is specific to the 70/357 pair within helix P4.
The finding that a base within helix P4 influences the ribozyme’s specificity for catalytic metal ions is significant for several reasons. First, the strict phylogenetic conservation of P4 (as well as the flanking J3/4 and J2/4 elements; ref. 41) suggests that this structure must serve a critical function in RNase P activity. The results of another TP292 in vitro selection experiment indicate that optimal catalytic activity stringently requires the native structure in the J3/4-P4-J2/4 region (37). Second, phosphorothioate modification-interference studies have identified four phosphate oxygens at both ends of helix P4 that are somehow involved in catalysis; the rescue of activity by Mn2+ implies that at least one of these phosphate oxygens coordinates a Mg2+ ion that participates in catalysis (21). Finally, molecular modeling of RNase P RNA tertiary structure, based on a large library of crosslinking and phylogenetic constraints, positions helix P4 within the core of the ribozyme and in close proximity to the scissile bond of its pre-tRNA substrate (24, 25). Thus, several lines of evidence indicate that J3/4-P4-J2/4 is intimately involved in forming the active site of RNase P; it is likely that the C70U mutation uncovered in the Ca2+ selection experiment mediates a direct effect on active-site structure.
How, then, might the single base change at position 70 alter the structure of the RNase P active site in a way that better accommodates Ca2+? Introduction of a U⋅G wobble pair into an A-form helix would be expected to alter helical structure in three specific ways (42, 43): (i) the major groove of the helix would be widened in the immediate vicinity of the U⋅G pair; (ii) the 4-amino group of cytosine would be replaced by the 4-keto group of uracil, which extends into the major groove; and (iii) the 2-amino group of guanosine would protrude further into the minor groove. One effect of widening the major groove of P4 would be to increase the interstrand phosphate distances across the groove. Because hydrated Ca2+ ions are larger than hydrated Mg2+ ions (44), coordination of Ca2+ to interstrand phosphates might be improved by the wider major groove. However, we do not favor this explanation because the U70⋅G357 pair cannot be replaced by the isosteric C70⋅A357 pair, which should similarly distort helical structure (data not shown). Therefore, it is likely that the pattern of hydrogen bonds presented by P4 is altered by the U⋅G pairing in such a way that Ca2+ more effectively participates in catalysis. Because the affinity of TP292-C70U for Ca2+ is similar to that of native TP292 (Fig. 4B), the U⋅G pair does not simply enhance the ability of TP292 to bind Ca2+. Rather, the U⋅G pair seems to improve the ability of TP292 to use Ca2+ during the catalytic step of the self-cleavage reaction, for example, by inducing local conformational changes in the RNA that better position Ca2+-bound water molecules for nucleophilic attack, or by altering the electronic state of Ca2+, thereby changing the pKa of the nucleophile. In any case, more rigorous biochemical analysis is needed to test these various hypotheses.
In summary, previous phosphorothioate modification interference studies have implicated a subset of the J3/4-P4-J2/4 phosphates in binding divalent metal ions essential for catalysis (21). Through in vitro RNA selection, we have found that a single nucleotide change within helix P4 is sufficient to alter the preference of RNase P for catalytic metals. Together, these findings imply that the phylogenetically well conserved J3/4-P4-J2/4 domain forms a portion of the active site of RNase P.
Acknowledgments
We thank the members of the Pace and Ellington labs for much helpful advice. We thank Dr. Drew Smith and Dr. Brian Thomas for thoughtful comments. This work was supported by American Cancer Society Postdoctoral Fellowship PF-3933 (D.N.F.) and National Institutes of Health Grant GM 34527 (N.R.P.).
References
- 1.Yarus M. FASEB J. 1993;7:31–39. doi: 10.1096/fasebj.7.1.8422972. [DOI] [PubMed] [Google Scholar]
- 2.Pan T, Long D M, Uhlenbeck O C. In: The RNA World. Gesteland R F, Atkins J F, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1993. pp. 271–302. [Google Scholar]
- 3.Smith D. In: The Biological Chemistry of Magnesium. Cowan J A, editor. New York: VCH; 1995. pp. 111–136. [Google Scholar]
- 4.Dahm S C, Uhlenbeck O C. Biochemistry. 1991;30:9464–9469. doi: 10.1021/bi00103a011. [DOI] [PubMed] [Google Scholar]
- 5.Dahm S C, Derrick W B, Uhlenbeck O C. Biochemistry. 1993;32:13040–13045. doi: 10.1021/bi00211a013. [DOI] [PubMed] [Google Scholar]
- 6.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 7.Gardiner K J, Marsh T L, Pace N R. J Biol Chem. 1985;260:5415–5419. [PubMed] [Google Scholar]
- 8.Guerrier-Takada C, Haydock K, Allen L, Altman S. Biochemistry. 1986;25:1509–1515. doi: 10.1021/bi00355a006. [DOI] [PubMed] [Google Scholar]
- 9.Surratt C K, Carter B J, Payne R C, Hecht S M. J Biol Chem. 1990;265:22513–22519. [PubMed] [Google Scholar]
- 10.Smith D, Burgin A B, Haas E S, Pace N R. J Biol Chem. 1992;267:2429–2436. [PubMed] [Google Scholar]
- 11.Smith D, Pace N R. Biochemistry. 1993;32:5273–5281. doi: 10.1021/bi00071a001. [DOI] [PubMed] [Google Scholar]
- 12.Grosshans C, Cech T R. Biochemistry. 1989;28:6888–6894. doi: 10.1021/bi00443a017. [DOI] [PubMed] [Google Scholar]
- 13.Jack A, Ladner J E, Rhodes D, Brown R S, Klug A. J Mol Biol. 1977;111:315–328. doi: 10.1016/s0022-2836(77)80054-5. [DOI] [PubMed] [Google Scholar]
- 14.Quigley G J, Teeter M M, Rich A. Proc Natl Acad Sci USA. 1978;75:64–68. doi: 10.1073/pnas.75.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cate J H, Gooding A R, Podell E, Zhou K, Golden B L, Kundrot C E, Cech T R, Doudna J A. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 16.Waring R B. Nucleic Acids Res. 1989;17:10281–10293. doi: 10.1093/nar/17.24.10281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruffner D E, Uhlenbeck O C. Nucleic Acids Res. 1990;18:6025–6029. doi: 10.1093/nar/18.20.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Christian E C, Yarus M. J Mol Biol. 1992;228:743–758. doi: 10.1016/0022-2836(92)90861-d. [DOI] [PubMed] [Google Scholar]
- 19.Hardt W D, Warnecke J M, Erdmann V A, Hartmann R K. EMBO J. 1995;14:2935–2944. doi: 10.1002/j.1460-2075.1995.tb07293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaur R K, Krupp G. Nucleic Acids Res. 1993;21:21–26. doi: 10.1093/nar/21.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harris M E, Pace N R. RNA. 1995;1:210–218. [PMC free article] [PubMed] [Google Scholar]
- 22.Piccirilli J A, Vyle J S, Caruthers M H, Cech T R. Nature (London) 1993;361:85–88. doi: 10.1038/361085a0. [DOI] [PubMed] [Google Scholar]
- 23.Warnecke J M, Furste J P, Hardt W, Erdmann V A, Hartmann R K. Proc Natl Acad Sci USA. 1996;93:8924–8928. doi: 10.1073/pnas.93.17.8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harris M E, Nolan J M, Malhotra A, Brown J W, Harvey S C, Pace N R. EMBO J. 1994;13:3953–3963. doi: 10.1002/j.1460-2075.1994.tb06711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harris M E, Kazantsev A V, Chen J-L, Pace N R. RNA. 1997;3:561–576. [PMC free article] [PubMed] [Google Scholar]
- 26.Szostak J W, Ellington A D. In: The RNA World. Gesteland R F, Atkins J F, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1993. pp. 511–534. [Google Scholar]
- 27.Breaker R R, Joyce G F. TIBTECH. 1994;12:268–274. doi: 10.1016/0167-7799(94)90138-4. [DOI] [PubMed] [Google Scholar]
- 28.Kumar P K R, Ellington A E. FASEB J. 1995;9:1183–1195. doi: 10.1096/fasebj.9.12.7672511. [DOI] [PubMed] [Google Scholar]
- 29.Pan T, Uhlenbeck O C. Biochemistry. 1992;31:3887–3895. doi: 10.1021/bi00131a001. [DOI] [PubMed] [Google Scholar]
- 30.Pan T, Uhlenbeck O C. Nature (London) 1992;358:560–563. doi: 10.1038/358560a0. [DOI] [PubMed] [Google Scholar]
- 31.Williams K P, Ciafre S, Tocchini-Valentini G P. EMBO J. 1995;14:4551–4557. doi: 10.1002/j.1460-2075.1995.tb00134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ciesiolka J, Gorski J, Yarus M. RNA. 1995;1:538–550. [PMC free article] [PubMed] [Google Scholar]
- 33.Ciesiolka J, Yarus M. RNA. 1996;2:785–793. [PMC free article] [PubMed] [Google Scholar]
- 34.Lehman N, Joyce G F. Nature (London) 1993;361:182–185. doi: 10.1038/361182a0. [DOI] [PubMed] [Google Scholar]
- 35.Lehman N, Joyce G F. Curr Biol. 1993;3:723–734. doi: 10.1016/0960-9822(93)90019-k. [DOI] [PubMed] [Google Scholar]
- 36.Frank D N, Harris M E, Pace N R. Biochemistry. 1994;33:10800–10808. doi: 10.1021/bi00201a030. [DOI] [PubMed] [Google Scholar]
- 37.Frank D N, Ellington A E, Pace N R. RNA. 1996;2:1179–1188. [PMC free article] [PubMed] [Google Scholar]
- 38.Cadwell R C, Joyce G F. PCR Methods Appl. 1992;2:28–33. doi: 10.1101/gr.2.1.28. [DOI] [PubMed] [Google Scholar]
- 39.Yockey H P. Information Theory and Molecular Biology. Cambridge, MA: Cambridge Univ. Press; 1992. [Google Scholar]
- 40.Brown J W, Haas E S, Gilbert D G, Pace N R. Nucleic Acids Res. 1994;22:3660–3662. doi: 10.1093/nar/22.17.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haas E S, Brown J W, Pitulle C, Pace N R. Proc Natl Acad Sci USA. 1994;91:2527–2531. doi: 10.1073/pnas.91.7.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Puglisi J D, Wyatt J R, Tinoco I. Biochemistry. 1990;1990:4215–4226. doi: 10.1021/bi00469a026. [DOI] [PubMed] [Google Scholar]
- 43.Holbrook S R, Cheong C, Tinoco I, Kim S-H. Nature (London) 1991;353:579–581. doi: 10.1038/353579a0. [DOI] [PubMed] [Google Scholar]
- 44.Cowan J A. Inorganic Biochemistry: An Introduction. New York: VCH; 1993. [Google Scholar]