Abstract
Using a new mAb raised against the mouse neuroepithelium, we have identified and cDNA-cloned prominin, an 858-amino acid-containing, 115-kDa glycoprotein. Prominin is a novel plasma membrane protein with an N-terminal extracellular domain, five transmembrane segments flanking two short cytoplasmic loops and two large glycosylated extracellular domains, and a cytoplasmic C-terminal domain. DNA sequences from Caenorhabditis elegans predict the existence of a protein with the same features, suggesting that prominin is conserved between vertebrates and invertebrates. Prominin is found not only in the neuroepithelium but also in various other epithelia of the mouse embryo. In the adult mouse, prominin has been detected in the brain ependymal layer, and in kidney tubules. In these epithelia, prominin is specific to the apical surface, where it is selectively associated with microvilli and microvilli-related structures. Remarkably, upon expression in CHO cells, prominin is preferentially localized to plasma membrane protrusions such as filopodia, lamellipodia, and microspikes. These observations imply that prominin contains information to be targeted to, and/or retained in, plasma membrane protrusions rather than the planar cell surface. Moreover, our results show that the mechanisms underlying targeting of membrane proteins to microvilli of epithelial cells and to plasma membrane protrusions of non-epithelial cells are highly related.
The generation of specific domains in membranes is one of the fundamental principles in the organization of eukaryotic cells. Specific membrane domains not only provide the basis for most of the functions common to all cells, they also contribute in an essential way to the origin of cellular diversity. A paradigmatic example is the plasma membrane of epithelial cells. This membrane is characterized by two major domains, the apical and basolateral domain, which show a distinct protein and lipid composition. Insight into the generation and maintenance of apical and basolateral plasma membrane domains has come from a large number of investigations, most of which have been carried out with “classical” epithelia of kidney, liver, and gut, and with cell lines derived therefrom (1–5).
Comparatively little is known about the generation and maintenance of apical and basolateral plasma membrane domains in neuroepithelial cells, which constitute the inner layer of the neural tube of the vertebrate embryo and give rise to all neurons and macroglial cells of the central nervous system. Neuroepithelial cells show features of an apical–basal polarity (6, 7), and thus their apical and basolateral plasma membranes are likely to be distinct in molecular composition. Indeed, two brush border enzymes and megalin, a low density lipoprotein-receptor family member, have been detected on the apical, but not basolateral, surface of neuroepithelial cells of the neural tube (8, 9).
At the neural tube stage, however, the neuroepithelium no longer contains tight junctions (6), which separate apical and basolateral plasma membrane domains of epithelial cells and prevent the lateral diffusion of transmembrane proteins and lipids of the extracellular membrane leaflet between these two membrane domains (10). This raises the question as to the mechanism that underlies the maintenance of apical localization of membrane components upon down-regulation of tight junctions. An answer to this question may lie in the existence of plasma membrane subdomains (11). In epithelial cells with a brush border, a characteristic subdomain of the apical plasma membrane are the microvilli (12, 13), which enlarge the apical surface and thereby increase the resorptive capacity of these cells.
Microvilli-like protrusions of the plasma membrane are also found in non-epithelial cells. In leukocytes, for example, such protrusions have been implicated in cell-to-cell adhesion, mediating the initial contact to endothelial cells (14). The functional diversity between microvilli of epithelial and non-epithelial cells is reflected in the differences in membrane protein composition (brush border enzymes in the case of resorptive epithelial cells, cell adhesion molecules in the case of leukocytes). Moreover, key components of the cytoskeleton, a major organizer of these plasma membrane subdomains (15, 16), differ between microvilli of epithelial and non-epithelial cells, such as the cytosolic actin-binding protein villin that occurs only in epithelial cells (17). Interestingly, villin expressed in non-epithelial cells is targeted to, and induces the formation of, plasma membrane protrusions (18). It is important to determine whether a transmembrane protein specific to microvilli of epithelial cells is also targeted to plasma membrane protrusions after expression in non-epithelial cells. This would be indicative of a highly conserved mechanism underlying the formation of this type of membrane subdomain and could provide a general explanation for plasma membrane polarity in the absence of tight junctions.
In the present study, we have investigated this issue. We generated a panel of mAbs against the mouse telencephalic neuroepithelium, one of which specifically stains microvilli on the apical surface of this and other epithelia. The cell biological and molecular characterization of the corresponding antigen, prominin, reveals a high degree of conservation between epithelial and non-epithelial cells of membrane protein targeting to plasmalemmal subdomains.
METHODS
mAbs.
Telencephali of ten 12-day-old NMRI mouse embryos (E12, ≈1 mg protein), which consist mostly of neuroepithelium, were homogenized in 1 ml PBS, mixed with crushed nitrocellulose filter (1 cm2 in 0.5 ml PBS) as adjuvant, and injected i.p. into a LouXSD rat. The rat was re-injected after 5 weeks and 11 weeks with the same material prepared from 14 embryos each. Rat spleen cells were fused with Ag 8.653 mouse myeloma cells (19). Ten days after the fusion, hybridoma supernatants were screened by light microscopic immunocytochemistry on cryosections of E12 mouse brains. mAb 13A4 was enriched from hybridoma supernatant by ammonium sulfate precipitation. The mAb 13A4 recognizes its antigen in mouse but not rat, chicken, and Drosophila.
Isolation of cDNA Clones.
An oligo(dT)-primed λ ZAP-II cDNA expression library from adult mouse kidney, kindly provided by M. Zerial (European Molecular Biology Laboratory), was screened following standard procedures (20) with mAb 13A4. One positive clone (clone 3Ab1) was isolated from 9 × 105 recombinant λ phages, and its 2,845-bp EcoRI–XhoI insert was recovered in pBluescript SK phagemid (Stratagene) by in vivo excision. The 5′ region of the cDNA was obtained by rapid amplification of cDNA ends (RACE) using the mouse kidney Marathon-Ready cDNA kit (CLONTECH). The cDNA-specific 3′ primers used were 5′-CTGCTTAGGCTTGGTCTGATGC-3′ and 5′-CTGCTGAGCGAATTCTCG-3′ for the first and second PCRs, respectively. The 5′-RACE product was then digested with NotI and PstI and the resulting 976-bp fragment was subcloned into the corresponding sites of pBluescript KS. To identify potential PCR errors, four RACE clones (clones 4, 4B, 18, and 19) were isolated from two independent PCRs. Both strands of clone 3Ab1 and the four RACE clones were completely sequenced (Sequenase version 2.0, United States Biochemical).
Expression Vector and Transfection.
The eukaryotic expression plasmid pRc/CMV-prominin was generated by ligating the PstI–XbaI fragment from clone 3Ab1 and the HindIII–PstI fragment derived from clone 4B by PCR (see below) into the pRc/CMV vector (Invitrogen) opened with HindIII and XbaI. The HindIII restriction site in the clone 4B-derived fragment had been generated during PCR amplification by using the oligonucleotide 5′-CCAGAAGGATCCTCAAGCTTGAGAGATCAGGC-3′ as a primer 5′ to the translation initiation codon of the prominin cDNA, T3 as 3′ primer and clone 4B as template. All PCR products were verified by sequencing. CHO cells were transfected with the pRc/CMV-prominin plasmid, or the pRc/CMV vector as control, using the calcium phosphate precipitation procedure. Pools of stable transfectants obtained by selection using G418 were incubated for 17 h with 5 mM sodium butyrate to induce the expression of the transgene in the cells prior to use.
Miscellaneous.
All other methods followed standard protocols as indicated in the figure legends.
RESULTS
Prominin, an Antigen Specific to the Apical Surface of Epithelial Cells and Enriched in Microvilli.
A rat was immunized with telencephalic neuroepithelium of 12-day-old mouse embryos (E12), and several mAbs were isolated based on the expression pattern of the corresponding antigens revealed by immunohistochemistry of E12 mouse brain. One of these antibodies, mAb 13A4, stained specifically the apical (ventricular), but not the basolateral, side of neuroepithelial cells of the E12 mouse embryo (Fig. 1A). mAb 13A4 immunoreactivity was also observed on the apical side of other embryonic epithelia (lung buds, gut, urether buds; data not shown). In the adult mouse, 13A4 immunoreactivity was detected at the apical side of the ependymal layer of the brain (data not shown) and at the apical (lumenal) side of kidney tubules (Fig. 1B), but not in the other tissues examined (gut, lung, liver, pituitary, adrenal, heart, and spleen; data not shown).
Figure 1.
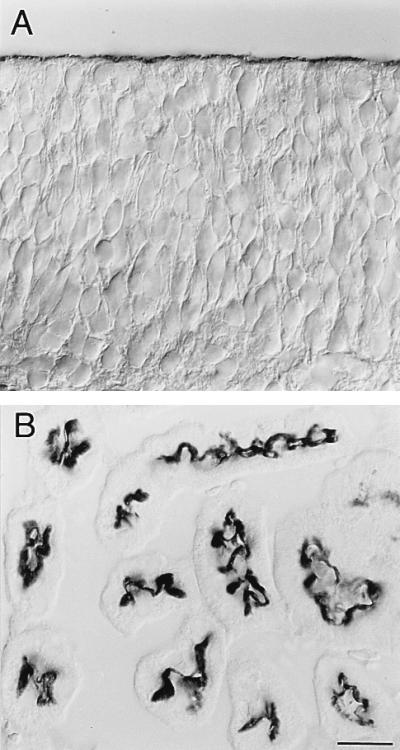
Immunoperoxidase localization of the 13A4 antigen (prominin) in the neuroepithelium and kidney tubules. Triton X-100-permeabilized cryosections (8 μm) of paraformaldehyde-fixed telencephalic neuroepithelium of a 12-day-old mouse embryo (A) and adult mouse kidney cortex (B) were stained with mAb 13A4 (1 μg/100 μl) followed by peroxidase-coupled goat anti-rat IgG/IgM and observed using Nomarski optics. In A, the apical and the basal side of the neuroepithelium are at the top and bottom, respectively. Immunoreactivity is confined to the apical, ventricular side. In B, most tubules are cross-sectioned. Immunoreactivity is restricted to their apical, lumenal side. (Bars = 20 μm.)
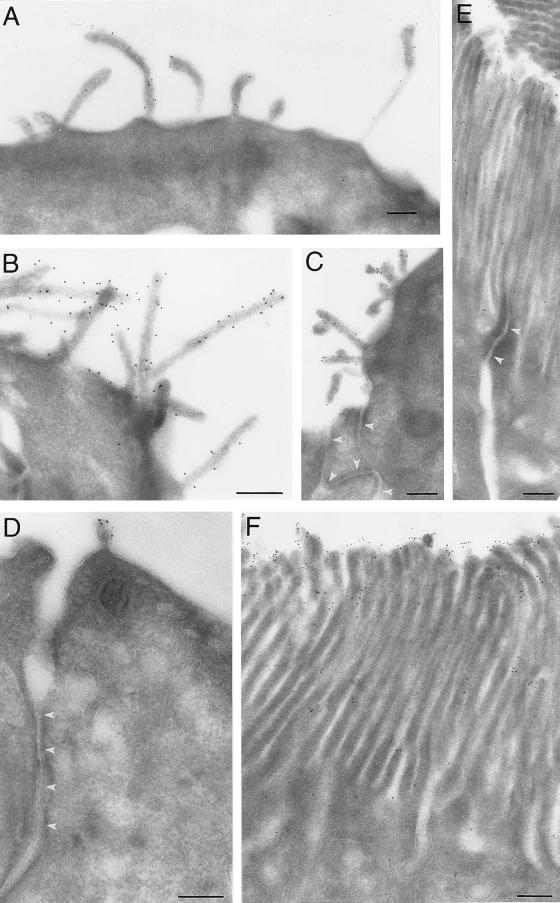
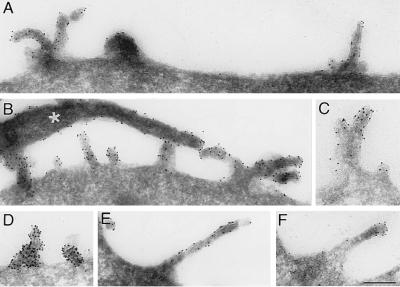
The subcellular localization of the 13A4 antigen in mouse E9–10 neuroepithelial cells and adult kidney proximal tubule cells was investigated by immunogold electron microscopy (Fig. 2). Strong labeling was observed over the kidney brush border membrane, where 13A4 immunoreactivity appeared to be concentrated toward the tips of the microvilli (Fig. 2 E and F). Remarkably, in neuroepithelial cells, whose apical plasma membrane contains fewer microvilli than the kidney brush border, 13A4 immunoreactivity was associated mostly, if not exclusively, with microvilli and plasma membrane protrusions, and was not detected in the planar areas of the apical plasma membrane (Fig. 2 A–D). Because of this preferential localization, we propose to call the 13A4 antigen “prominin” (from the Latin word “prominere,” to stand out, to be prominent).
Figure 2.
Immunoelectron microscopy (26) of the 13A4 antigen (prominin) on the apical surface of neuroepithelial cells and the brush border of kidney tubules. Ultrathin cryosections (26) of paraformaldehyde-fixed tissue were stained with mAb 13A4 (0.5–1 mg/ml) followed by rabbit anti-rat IgG/IgM and 9-nm protein A-gold. Neuroepithelial cells of 9-day-old (A and C) and 10-day-old (B and D) mouse embryos and proximal tubule cells of adult mouse kidney (E and F) are shown. In neuroepithelial cells (A–D), immunoreactivity is confined to protrusions of the apical plasma membrane including microvilli-like structures. Junctional complexes are indicated by white arrowheads in C and D. In the kidney proximal tubule (E and F), immunoreactivity is restricted to the brush border where it appears to be concentrated at the tips of the microvilli. A tight junction is indicated by white arrowheads in E. (Bars = 0.5 μm.)
Prominin Is a Membrane-Associated 115-kDa Glycoprotein.
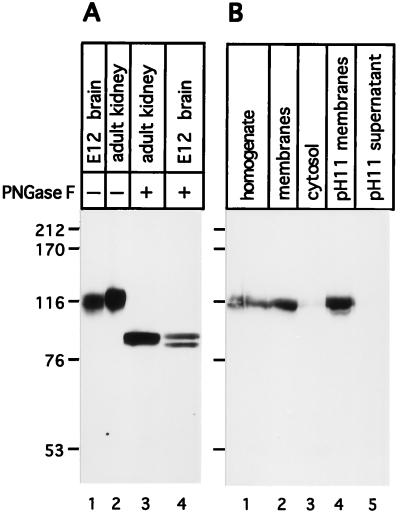
Immunoblotting of E12 mouse brain and adult mouse kidney using mAb 13A4 showed that prominin has a mean apparent molecular weight of 115 kDa (Fig. 3A, lanes 1 and 2). Deglycosylation of brain and kidney prominin with peptide-N-glycosidase F (PNGase F) yielded a 94-kDa band (Fig. 3A, lanes 3 and 4), indicating that prominin is N-glycosylated. (The additional 88-kDa band seen upon deglycosylation of brain prominin represents a C-terminally truncated form; data not shown.)
Figure 3.
Glycosylation and membrane association of prominin. (A) Membranes (50 μg protein) prepared from either the brains of 12-day-old mouse embryos (E12) or adult mouse kidneys (40,000 × g, 30 min) were dissolved in SDS, incubated in the absence (−) or presence (+) of 0.8 units of PNGase F (Boehringer Mannheim) and analyzed by immunoblotting using mAb 13A4 (1 μg/ml) followed by the ECL system (Amersham). (B) A homogenate of adult mouse kidney was fractionated into a total membrane and cytosol fraction and the total membranes were treated with carbonate at pH 11 (27), yielding pH11 membranes and the pH11 supernatant. Corresponding aliquots of these fractions were analyzed by immunoblotting using mAb 13A4.
Upon high-speed centrifugation of mouse kidney homogenate (Fig. 3B, lane 1), prominin was quantitatively recovered in the membrane pellet (Fig. 3B, lanes 2 and 3) from which it was not extracted by carbonate at pH 11 (Fig. 3B, lanes 4 and 5). This result shows that prominin is tightly membrane-associated. The same result was obtained with E12 mouse brain (data not shown).
cDNA Cloning and Sequence Analysis of Prominin.
Immunological screening of a mouse kidney λ ZAP cDNA library with mAb 13A4 led to the isolation of a cDNA clone with a ≈2.8-kb insert (clone 3Ab1, nt 819–3,663). Its cDNA sequence showed an ORF that, however, did not appear to be full-length. The 5′ end of the cDNA was therefore obtained by RACE (clone 4B, nt 1–976). Consistent with the length of the overlapping clones 3Ab1 and 4B [3,663 nt without poly(A) tail], Northern blot analysis of adult mouse kidney poly(A)+ RNA using cDNA fragments from either clone 3Ab1 (nt 1,061–2,019) or clone 4B (nt 1–405) as probes revealed a single band of ≈4.0 kb (data not shown). To validate that the cDNA sequence encodes the 13A4 antigen, we raised rabbit antisera against prominin fragments (derived from glutathione S-transferase fusion proteins) that corresponded to either amino acids 183–426 or amino acids 807–858 (C-terminal domain) of the predicted ORF (Fig. 4). In immunoblots of mouse kidney membranes, both antisera recognized a 115-kDa protein whose electrophoretic mobility was indistinguishable from that recognized by mAb 13A4 (data not shown).
Figure 4.
Amino acid sequence of mouse prominin deduced from the corresponding cDNA. Doubly underlined italic letters, putative signal peptide; doubly underlined plain letters, predicted membrane-spanning segments; bold letters, cysteine cluster and N-glycosylation sites.
The mouse prominin cDNA contains an ORF from nt 189 to 2,762, which encodes a 858-amino acid protein with a predicted molecular weight of 96,222 (Fig. 4). The sequence surrounding the putative translational start codon meets the consensus criteria for initiation of translation. An in-frame stop codon is present 5′ at nt 168–170. The 3′ untranslated region contains a polyadenylation consensus sequence at nt 3,644–3,649.
Hydropathy plot analysis of mouse prominin reveals six hydrophobic regions (underlined in Fig. 4). The first, N-terminal hydrophobic segment most likely constitutes a signal peptide. A potential signal peptidase cleavage site is found between Ser-19 and Glu-20 (Fig. 4). Cleavage at this site would generate a 839-amino acid protein with a predicted molecular weight of 94,303, which would be in agreement with the molecular mass (94 kDa) of the N-deglycosylated prominin molecule (Fig. 3A). The other five hydrophobic regions (Fig. 4) constitute presumptive transmembrane segments (M1–M5).
Prominin contains eight potential N-glycosylation sites, of which five are located between the M2 and M3 segments and three between the M4 and M5 segments (Fig. 4). The reduction in the molecular mass of prominin by ≈20 kDa after N-deglycosylation suggests that at least some of these sites are actually glycosylated. This in turn implies that the two large hydrophilic domains containing the N-glycosylation sites are located extracellularly.
The prominin cDNA shows no striking homology to other nucleotide sequences in the EMBL and GenBank databases, except for several expressed sequence tag sequences, one (89% identity) from adult mouse brain (accession no. W49223) and seven (64–70% identity) from either adult human retina (accession nos. W22150, W22511, W25808, W26483, W27006) or other human sources (accession nos. R32598, R36499).
Protein database searches using the blast network service (21) at either National Center for Biotechnology Information or Ecole Polytechnique Federal de Lausanne servers with the prominin protein sequence as a probe revealed a 20% identity and a 44% similarity to F08B12.1 (accession no. Z68104), an 866-residue protein predicted from sequences compiled in the context of the Caenorhabditis elegans genome project (22). Interestingly, whereas the homology between prominin and F08B12.1 appears to be uniformly distributed along the sequence, each of the six cysteine residues in the extracellular loops of prominin (likely to form disulfide bridges) are conserved in position, and the hydropathy plots of the two proteins are strikingly similar (data not shown), suggesting that F08B12.1 is the C. elegans homologue of mouse prominin.
Expression of Prominin in CHO Cells.
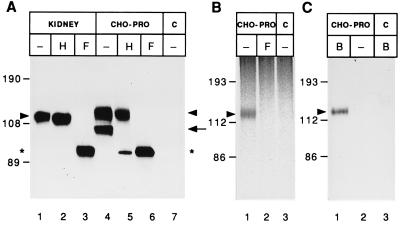
We generated a full-length cDNA clone from clones 3Ab1 and 4B and expressed it under the control of the cytomegalovirus (CMV) promoter by stable transfection of CHO cells. Immunoblotting with mAb 13A4 revealed two bands of 123 kDa (Fig. 5A, lane 4, arrowhead) and 104 kDa (Fig. 5A, lane 4, arrow). No immunoreactivity was detected when CHO cells were transfected with the expression vector alone (Fig. 5A, lane 7). Deglycosylation indicated that the difference between the 123- and 104-kDa form of prominin expressed in CHO cells and the 115-kDa form of prominin present in kidney (Fig. 5A, lane 1) was due to differential N-glycosylation. The 123-kDa form found in transfected CHO cells, like the 115-kDa form in kidney, was largely resistant to endo H treatment (Fig. 5A, lanes 2 and 5). This indicated that it had passed through the medial Golgi, and was consistent with this form of prominin being present at the plasma membrane. In contrast, the 104-kDa form found in transfected CHO cells was converted to a 94-kDa form upon endo H treatment (Fig. 5A, lane 5, asterisk), consistent with its localization in the endoplasmic reticulum or an early Golgi compartment. N-deglycosylation by PNGase F of the 123-kDa and 104-kDa forms of prominin found in transfected CHO cells yielded the same 94-kDa product (Fig. 5A, lane 6) as that obtained in the case of endogenous prominin of kidney (Fig. 5A, lane 3). These results show that the composite cDNA clone expressed in CHO cells encoded the full-length form of prominin.
Figure 5.
Expression of prominin in CHO cells. CHO cells were stably transfected with either the expression vector containing the mouse prominin cDNA (CHO-PRO) or, as a control, vector DNA alone (lanes C). (A) Lysates from the CHO cells and, for comparison, from adult mouse kidney membranes were incubated in the absence (−) or presence of 10 milliunits endo H (H) or 1 unit PNGase F (F) (Boehringer Mannheim), and analyzed by immunoblotting with mAb 13A4. Arrowheads, endo H-resistant, PNGase F-sensitive form; arrow, endo H- and PNGase F-sensitive form; asterisks, product after N-deglycosylation. (B) The CHO cells were labeled for 2 h with [35S]sulfate, followed by immunoprecipitation of prominin with the antiserum against the C-terminal domain and incubation of the solubilized immunoprecipitate without (−) or with (F) 1 unit PNGase F. (C) The CHO cells were incubated for 1 h at 4°C without (−) or with (lanes B) sulfo-NHS-LC-biotin (27), solubilized, and biotinylated proteins adsorbed to streptavidin-Agarose followed by immunoblot analysis of the adsorbed material using the antiserum against the C-terminal domain (1:5000).
Consistent with its resistance to endo H digestion, the 123-kDa form produced by prominin-transfected CHO cells underwent sulfation (Fig. 5B, lane 1, arrowhead), a posttranslational modification occurring in the trans-Golgi network. Most, if not all, of the sulfate appeared to be bound to N-linked carbohydrate residues, as shown by its removal upon PNGase F digestion (Fig. 5B, lane 2). Surface biotinylation of transfected CHO cells provided direct evidence that the 123-kDa form, but not the 104-kDa form, of prominin reached the cell surface (Fig. 5C, lane 1, arrowhead).
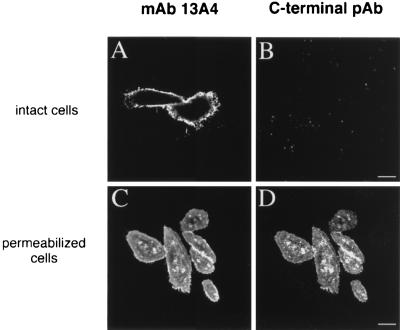
The rabbit antiserum against the C-terminal 52 amino acids was used as a topological probe in immunofluorescence experiments on either intact or permeabilized, prominin-transfected CHO cells. Upon exposure of intact cells to this antiserum, no cell surface staining was detected (Fig. 6B), in contrast to the strong immunofluorescence seen with mAb 13A4 (Fig. 6A). The latter implies that the 13A4 epitope is located on the extracellular side of the plasma membrane. When fixed cells were permeabilized with Triton X-100, cells that stained with mAb 13A4 (Fig. 6C) were also stained with the antiserum against the C-terminal domain (Fig. 6D). These data show that the C-terminal domain of prominin is located on the cytoplasmic side of the plasma membrane.
Figure 6.
Intracellular localization of the C-terminal domain of prominin. CHO cells were stably transfected with the expression vector containing the mouse prominin cDNA. Intact cells at 4°C (27) (A and B) or paraformaldehyde-fixed, Triton X-100-permeabilized cells (C and D) were incubated with the mAb 13A4 (10 μg/ml) and the antiserum against the C-terminal domain (1:3000), followed by appropriate fluorescein- and rhodamine-conjugated secondary antibodies and double immunofluorescence analysis using confocal microscopy. Single optical sections at the level of the coverslip are shown. (Bars = 10 μm.)
Prominin Is Targeted to Plasma Membrane Protrusions When Expressed in Nonepithelial Cells.
Given the localization of prominin in microvilli and related plasma membrane protrusions of the apical surface of epithelial cells (Fig. 2), it was of interest to investigate whether in the transfected fibroblasts prominin would be distributed randomly over the entire plasma membrane or be enriched in plasma membrane protrusions. Immunogold EM of CHO cells expressing prominin revealed the preferential localization of prominin in protrusions of the plasma membrane (Fig. 7). In numerous cases, gold particles were concentrated at microspike-like membrane protrusions, whereas the neighboring planar areas of the plasma membrane were labeled to a much lesser extent or not at all (Fig. 7 A and B). This preferential localization was more obvious in cells with moderate levels of prominin expression than in highly expressing cells, although it could be detected even in the latter case (Fig. 7D). Whereas in most cases the labeling for prominin was distributed along the entire length of the plasma membrane protrusion (Fig. 7E), in a few instances prominin immunoreactivity appeared to be concentrated toward its tip (Fig. 7 C and F). Immunogold labeling for prominin was also observed along the plasma membrane of filopodia (Fig. 7B, asterisk), where it appeared to be stronger than over the planar areas of the cell body plasma membrane. Comparison of the plasma membrane protrusions of mock-transfected and prominin-transfected CHO cells by phalloidin staining and cell surface fluorescence using wheat germ agglutinin did not reveal an obvious quantitative or qualitative difference (data not shown). However, because of the morphological heterogeneity of the CHO cells, the question of whether or not prominin can induce or stabilize the formation of plasma membrane protrusions could not be resolved and requires further investigation.
Figure 7.
Immunoelectron microscopy of prominin on the plasma membrane of transfected, paraformaldehyde-fixed CHO cells. Ultrathin cryosections were stained with mAb 13A4 followed by rabbit anti-rat IgG/IgM and 9-nm protein A-gold. The white asterisk in B indicates a filopodium extending from the cell shown in this panel. All panels are the same magnification. (Bar in F = 0.2 μm.)
DISCUSSION
We have identified and characterized prominin, a novel polytopic membrane protein specifically localized in microvilli of the apical surface of various epithelial cells. From its sequence and the N-glycosylation and antibody accessibility analyses, prominin (after cleavage of the putative signal sequence) is predicted to consist of an extracellular N-terminal domain, five transmembrane domains separating two small cytoplasmic and two large extracellular loops, and a cytoplasmic C-terminal domain. The occurrence of an ORF in C. elegans that is highly related to mouse prominin not only suggests that prominin is conserved between vertebrates and invertebrates, but also broadens the spectrum of approaches that can be taken to investigate the as yet unknown function of this protein.
A key feature regarding the subcellular localization of prominin in epithelial cells is its selective occurrence, within the apical plasma membrane, in specific subdomains. In the adult kidney, prominin is specifically associated with the brush border membrane, apparently concentrated toward the tips of the microvilli. In neuroepithelial cells, it is selectively associated with microvilli-like protrusions, rather than the planar areas, of the apical plasma membrane. The latter association presumably explains why prominin is selectively found at the apical surface of neuroepithelial cells not only at the neural plate stage (E8 mouse embryo; data not shown), when the neuroepithelium contains functional tight junctions (6) which are known to prevent lateral diffusion of transmembrane proteins between the apical and basolateral domains, but also after neural tube closure (E9 mouse embryo; Fig. 2), when functional tight junctions have disappeared (6).
Remarkably, when prominin is expressed in non-epithelial (CHO) cells, it also becomes enriched in plasma membrane subdomains—i.e., filopodia, lamellipodia, and microspikes. The targeting of this apical membrane protein is therefore different from that of another, influenza virus hemagglutinin, which was distributed uniformly over the surface of non-epithelial (BHK) cells (23). Our observations imply that (i) prominin contains dual targeting information, for the apical plasma membrane domain as such (as shown after expression in MDCK cells; D.C., K. Röper, and W.B.H., unpublished data) and for the microvillar subdomain within the apical plasma membrane of epithelial cells, and (ii) the microvillar targeting information in prominin can be “decoded” in non-epithelial cells, resulting in its accumulation in plasma membrane protrusions. As to the mechanism underlying its accumulation of prominin in microvilli and related plasma membrane protrusions, prominin may interact with the actin cytoskeleton, which is known to function as an organizer of microvilli, filopodia, lamellipodia, and microspikes (15–17). Certain actin binding proteins such as fimbrin (24) and ezrin (25) are found specifically in microvilli of epithelial cells and related plasma membrane protrusions of non-epithelial cells. It is also conceivable that these plasma membrane protrusions exhibit a specific lipid composition/organization, which differs from that of the planar region of the plasma membrane, and that prominin (perhaps via its transmembrane segments) has a preference for these lipids.
On a more general note, our observations reveal an unexpected degree of conservation of membrane protein targeting to microvilli of epithelial cells and to related plasma membrane protrusions of non-epithelial cells. Membrane protein targeting to microvilli appears to reflect a cell-type-specific adaptation of a process common to all cells.
Acknowledgments
We thank Dr. M. Zerial (European Molecular Biology Laboratory) for the kidney λ ZAP cDNA library, Dr. Patrick Argos (European Molecular Biology Laboratory) for advice with biocomputing, Dr. Antje Niehaus and Manfred Griesheimer for advice and help with the production of mAb, Dr. Matthew Hannah for advice with confocal microscopy, Alan Summerfield for artwork, and Drs. Kai Simons, Eeva Aaku-Saraste, Matthew Hannah, and Christoph Thiele for helpful comments on the manuscript. A.W. was the recipient of a fellowship from the Graduiertenkolleg Neurobiologie of the Deutsche Forschungsgemeinschaft and acknowledges the advice by Drs. Andreas Faissner and Jacqueline Trotter during her Ph.D. thesis. D.C. was the recipient of a fellowship from the Medical Research Council of Canada. W.B.H. was supported by grants from the Deutsche Forschungsgemeinschaft (SFB 317 and SFB 352).
Footnotes
Abbreviation: RACE, rapid amplification of cDNA ends.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF026269).
References
- 1.Nelson W J. Science. 1992;258:948–955. doi: 10.1126/science.1439806. [DOI] [PubMed] [Google Scholar]
- 2.Rodriguez-Boulan E, Powell S K. Annu Rev Cell Biol. 1992;8:395–427. doi: 10.1146/annurev.cb.08.110192.002143. [DOI] [PubMed] [Google Scholar]
- 3.Simons K, Dupree P, Fiedler K, Huber L A, Kobayashi T, Kurzchalia T, Olkkonen V, Pimplikar S, Parton R, Dotti C. Cold Spring Harbor Symp Quant Biol. 1992;57:611–619. doi: 10.1101/sqb.1992.057.01.067. [DOI] [PubMed] [Google Scholar]
- 4.Eaton S, Simons K. Cell. 1995;82:5–8. doi: 10.1016/0092-8674(95)90045-4. [DOI] [PubMed] [Google Scholar]
- 5.Mays R W, Nelson W J, Marrs J A. Cold Spring Harbor Symp Quant Biol. 1995;60:763–773. doi: 10.1101/sqb.1995.060.01.082. [DOI] [PubMed] [Google Scholar]
- 6.Aaku-Saraste E, Hellwig A, Huttner W B. Dev Biol. 1996;180:664–679. doi: 10.1006/dbio.1996.0336. [DOI] [PubMed] [Google Scholar]
- 7.Huttner W B, Brand M. Curr Opin Neurobiol. 1997;7:29–39. doi: 10.1016/s0959-4388(97)80117-1. [DOI] [PubMed] [Google Scholar]
- 8.Spencer-Dene B, Thorogood P, Nair S, Kenny J A, Harris M, Henderson B. Development (Cambridge, UK) 1994;120:3213–3226. doi: 10.1242/dev.120.11.3213. [DOI] [PubMed] [Google Scholar]
- 9.Willnow T E, Hilpert J, Armstrong S A, Rohlmann A, Hammer R E, Burns D K, Herz J. Proc Natl Acad Sci USA. 1996;93:8460–8464. doi: 10.1073/pnas.93.16.8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schneeberger E E, Lynch R D. Am J Physiol. 1992;262:L647–L661. doi: 10.1152/ajplung.1992.262.6.L647. [DOI] [PubMed] [Google Scholar]
- 11.Vega-Salas D E, Salas P J I, Gunderson D, Rodriguez-Boulan E. J Cell Biol. 1987;104:905–916. doi: 10.1083/jcb.104.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kerjaschki D, Noronha-Blob L, Sacktor B, Farquhar M G. J Cell Biol. 1984;98:1505–1513. doi: 10.1083/jcb.98.4.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Danielsen M E, van Deurs B. J Cell Biol. 1995;131:939–950. doi: 10.1083/jcb.131.4.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.von Andrian U H, Hasslen S R, Nelson R D, Erlandsen S L, Butcher E C. Cell. 1995;82:989–999. doi: 10.1016/0092-8674(95)90278-3. [DOI] [PubMed] [Google Scholar]
- 15.Louvard D. Curr Opin Cell Biol. 1989;1:51–57. doi: 10.1016/s0955-0674(89)80036-5. [DOI] [PubMed] [Google Scholar]
- 16.Condeelis J. Annu Rev Cell Biol. 1993;9:411–444. doi: 10.1146/annurev.cb.09.110193.002211. [DOI] [PubMed] [Google Scholar]
- 17.Louvard D, Kedinger M, Hauri H P. Annu Rev Cell Biol. 1992;8:157–195. doi: 10.1146/annurev.cb.08.110192.001105. [DOI] [PubMed] [Google Scholar]
- 18.Friederich E, Huet C, Arpin M, Louvard D. Cell. 1989;59:461–475. doi: 10.1016/0092-8674(89)90030-5. [DOI] [PubMed] [Google Scholar]
- 19.Faissner A, Kruse J. Neuron. 1990;5:627–637. doi: 10.1016/0896-6273(90)90217-4. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook M, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 21.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 22.Wilson R, Ainscough R, Anderson K, Baynes C, Berks M, et al. Nature (London) 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]
- 23.Peränen J, Auvinen P, Virta H, Wepf R, Simons K. J Cell Biol. 1996;135:153–167. doi: 10.1083/jcb.135.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bretscher A, Weber K. J Cell Biol. 1980;86:335–340. doi: 10.1083/jcb.86.1.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berryman M, Gary R, Bretscher A. J Cell Biol. 1995;131:1231–1242. doi: 10.1083/jcb.131.5.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Griffiths G. Fine Structure Immunocytochemistry. Heidelberg: Springer; 1993. [Google Scholar]
- 27.Pimplikar S W, Huttner W B. J Biol Chem. 1992;267:4110–4118. [PubMed] [Google Scholar]