Abstract
We report here that wild-type Escherichia coli can grow on the chitin disaccharide, N,N′-diacetylchitobiose (GlcNAc)2, as the sole source of carbon. Transposon mutants were isolated that were unable to ferment (GlcNAc)2 but grew normally on the monosaccharide GlcNAc. One such mutant was used to screen a wild-type E. coli genomic cosmid library for restoration of (GlcNAc)2 fermentation. A partial sequence analysis of the isolated fragment mapped the clone to the (previously sequenced) E. coli genome between 39.0 and 39.2 min. The nucleotide ORFs at this region had been previously assigned to code for a “cryptic” cellobiose utilization (cel) operon. We report here, however, that functional analysis of the operon, including growth and chemotaxis, reveal that it encodes a set of proteins that are not cryptic, but are induced by (GlcNAc)2 and catabolize the disaccharide. We therefore propose to rename the cel operon as the chb (N,N′-diacetylchitobiose) operon, with the letter designation of the genes of the operon to be reassigned consistent with the nomenclature based on functional characterization of the gene products as follows: celA to chbB, celB to chbC, celC to chbA, celD to chbR, and celF to chbF. Furthermore, sequencing evidence indicates that the operon contains an additional gene of unknown function to be designated as chbG. Thus, the overall gene sequence is to be named chbBCARFG.
Cellulose and chitin are the two most abundant organic compounds found in nature, composed of β-1,4-linked glucose and β-1,4-linked N-acetylglucosamine residues, respectively. Microorganisms have developed sophisticated mechanisms for the catabolism of these polymers (1–4). Each is degraded by specific hydrolases, and bacteria have evolved systems for the uptake of the resultant mono- and disaccharides. Functional genes for cellobiose utilization have been described in certain strains of the Gram-negative bacteria Escherichia coli (5) and Klebsiella (6), as well as in Gram-positives such as Cellulomonas uda (1) and Bacillus stearothermophilus (7), and the structures of several cellulases have been determined (8). One of the best characterized systems for cellobiose uptake is encoded by the E. coli cryptic cel operon (9–12). Cryptic operons have been defined as silent genes, not expressed in wild-type organisms, but requiring a mutation(s) for activation (13).
Chitin catabolism has been described in a wide variety of organisms (14), including many marine bacteria (15–17). The major product of virtually all known bacterial chitinases is the disaccharide N,N′-diacetylchitobiose (GlcNAc)2. We have reported a specific (GlcNAc)2 permease in the Gram-negative chitinolytic marine bacterium Vibrio furnissii (18).
In this paper, we report that E. coli catabolizes (GlcNAc)2 and that the genes responsible for its uptake are the previously isolated cryptic cel operon (11, 12), which we propose to rename as the chb (N,N′-diacetylchitobiose) operon.
MATERIALS AND METHODS
Reagents for bacterial media were obtained from Difco, J.T. Baker, and BBL Microbiology Systems. Molecular biology reagents were obtained from New England Biolabs, United States Biochemical, and Stratagene. Radioisotopes were purchased from DuPont/NEN. Pure crystalline (GlcNAc)2 was prepared and will be described elsewhere. Other buffers and reagents were of the highest purity available commercially.
E. coli strain XL1-Blue MR was purchased from Stratagene, and E. coli strain LR-175 (19), Salmonella typhimurium strain LT2 (20), and other strains were obtained from the American Type Culture Collection. Bacterial strains were grown at 37°C in Luria broth (LB) or on Luria agar plates supplemented with ampicillin (50 μg/ml) and tetracycline (15 μg/ml) where appropriate for selection of recombinant E. coli. Fermentation was assayed by streaking cells on plates containing one of the following: (i) Difco MacConkey agar base, (ii) Difco Levine EMB (eosin Y/methylene blue) agar (without lactose), or (iii) minimal/tetrazolium (M9 salts with 1.5% agar, 0.5 mM thiamine, 0.01% Casamino acids, and 10 mM 2,3,5-triphenyltetrazolium chloride), each of which was supplemented with the indicated carbon source at 10 mM. Plates were incubated at 37°C for 15–20 hr before being scored. Growth curves were obtained by growing strains overnight in LB and inoculating 1:100 into the indicated media.
Transposon Mutagenesis.
Transposon mutagenesis was performed using a lysate from E. coli strain MC4100 P1Clr100Cm containing the mini-mμ lac–tetracycline-resistant transposable element (kind gift of B. Bassler, Princeton University, Princeton, NJ). Phage P1 transduction into E. coli strain XL1-Blue MR was performed according to standard procedures (21). Transductants were screened for loss of (GlcNAc)2 fermentation on MacConkey medium supplemented with 10 mM (GlcNAc)2 and 15 μg/ml tetracycline.
Construction of XL1-Blue MR Genomic Library.
A cosmid library was constructed by using bacterial genomic DNA from E. coli strain XL1-Blue MR as described (22). Library construction, including conditions for partial genomic DNA restriction (using Sau3A) and ligation into the cosmid vector SuperCos1 were performed according to the supplier’s recommendations (Stratagene). The ligation mixture was packaged into λ phage by using GigaPack Gold III packaging extract (Stratagene) and tranfected into various E. coli strains according to the supplier’s recommendations.
DNA Manipulations.
DNA preparation and analysis, restriction enzyme digestions, ligation, and transformations were performed using standard techniques (22). Double-stranded DNA prepared from the recombinant clone pES1 was partially sequenced by the dideoxynucleotide method using the United States Biochemical Sequenase V2.0 sequencing kit (23, 24). Sequencing primers were designed on the basis of the nucleotide sequence of the vector, Supercos1, flanking the insert, and were 5′-CCATTATTATCATGACATTAA-3′, sequencing clockwise (CW), and 5′-GTCCGTGGAATGAACAATGG-3′, sequencing counterclockwise (CCW). The analyses of DNA sequences were conducted with the GCG sequence analysis package (Version 7, Genetics Computer Group, Madison WI). The database used for nucleotide sequence similarity searches (using fasta) was GenBank, release 79.
Swarm Plate Assay for E. coli Chemotaxis.
Chemotaxis was assayed by the ability of cells to form swarm rings on “swarm plates” containing 0.25% agar, M9 salts, 0.5 mM thiamine, and the indicated carbon source, usually at a concentration of 5–10 mM (25, 26). E. coli were inoculated at the center of the swarm plate with a colony taken from a rich broth (LB) plate. The plates were incubated at 30°C for 20–36 hr and then photographed. Fast swarmers of each strain were selected by several passages on rich broth (LB) swarm plates, using cells successively taken from the edge of swarm rings.
RESULTS
(GlcNAc)2 Fermentation by Gram-Negative Bacteria.
Various strains of Gram-negative bacteria were tested for their ability to ferment the disaccharide (GlcNAc)2, and the results with MacConkey base agar supplemented with 10 mM (GlcNAc)2 are summarized in Table 1. In this medium positive colonies turn red. Pink colonies were scored as +/−, and positives ranging from deep red centers (of colonies) to completely red colonies were scored as + and ++, respectively. It should be noted that the fermentation response was weaker than that observed with monosaccharides such as GlcNAc or glucose, which is presumably a reflection of the rate of fermentation. With the exception of HB101, all E. coli strains tested were positive, including LR-175, which is defective for the GlcNAc permease (19).
Table 1.
Fermentation of sugars by bacterial strains
| Bacterial strain | (GlcNAc)2 | GlcNAc | Glucose |
|---|---|---|---|
| Escherichia coli | |||
| XL1-Blue MR | + | +++ | +++ |
| LR-175 | + | − | +++ |
| Xm1.4 | − | +++ | +++ |
| Xm1.4:pCBU7.3 | ++ | +++ | +++ |
| DH5A | + | +++ | +++ |
| K-12 | + | +++ | +++ |
| EMG-29 | + | +++ | +++ |
| EMG-30 | + | +++ | +++ |
| HB101 | − | +++ | +++ |
| Salmonella typhimurium | |||
| LT2 | +/− | +++ | +++ |
| 3507 | + | +++ | +++ |
| Vibrio furnissii* | |||
| SR1514 | ++ | ++ | ++ |
| SR1519 | ++ | ++ | ++ |
| Enterobacter cloacae | |||
| A1.3 | ++ | +++ | +++ |
The ability of bacterial strains to ferment glucose, GlcNAc, and (GlcNAc)2 was assayed by streaking cells on MacConkey agar base indicator media supplemented with 10 mM carbohydrate, and individual colonies were scored as follows: −, negative; +/−, dark pink colonies; +, deep red center of colonies; ++, completely red colonies; and +++, completely red colonies with surrounding red zones.
V. furnissii strains were tested by using Levine EMB agar (without lactose) supplemented with 10 mM carbohydrate.
The response range was tested with MacConkey agar base containing 1–20 mM (GlcNAc)2, and maximal response was observed at 10 mM. Fermentation was assayed with other indicator media, including Levine EMB agar (without lactose) and minimal/tetrazolium medium, each supplemented with (GlcNAc)2. Neither test medium gave as clear results as seen with the MacConkey base agar.
E. coli strain XL1-Blue MR was chosen for further study because (i) it displayed a consistent and strong response in (GlcNAc)2 fermentation, (ii) the strain is easily amenable to genetic manipulation, and (iii) it is one of the host strains recommended for use in phage-cosmid library screening (Stratagene).
Transposon Mutagenesis and Screening of E. coli Strain XL1-Blue MR.
Transposon mutagenesis of XL1-Blue MR was conducted as described in Materials and Methods. Five thousand transposon mutants were screened on MacConkey agar plates supplemented with 10 mM (GlcNAc)2, and four nonfermenting (white) colonies were isolated. All four fermented both GlcNAc and glucose (data not shown). One mutant strain, designated Xm1.4, was used for further study.
Complementation of Xm1.4 by Using Wild-Type E. coli XL1-Blue MR Genomic Cosmid Library.
A λ genomic cosmid library was constructed from E. coli strain XL1-Blue MR and screened in Xm1.4 as described in Materials and Methods. A total of 4,000 colonies were screened on MacConkey base agar supplemented with 10 mM (GlcNAc)2 and 50 μg/ml ampicillin, and 3 positive clones were selected. Restriction analysis of the 3 isolates showed inserts ranging from 25 to 30 kb, with the isolates sharing many similar-sized bands when tested with several different restriction enzymes (data not shown). The isolate with the smallest size insert (25 kb), designated pRQ1.1, was chosen for further study. Because the cosmid vector, SuperCos1, contained two EcoRI sites flanking the XbaI insertion site, pRQ1.1, was subcloned by digestion of the full-length clone with EcoRI. Many fragments were obtained, including SuperCos1 containing EcoRI ends. The mixture was ligated under conditions favoring reinsertion of one fragment into SuperCos1. The ligation mixture was screened for complementation of the mutation in strain Xm1.4, and the smallest insert thus isolated was 7.3 kb (pES1).
Sequencing of the Ends of pES1 and Identification of the (GlcNAc)2 Catabolic Operon as the “Cryptic” Cellobiose Operon.
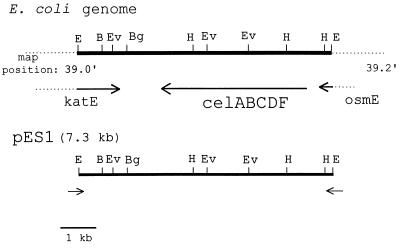
Portions of the nucleotide sequences of the 7.3-kb E. coli insert in SuperCos1 were determined as described in Materials and Methods. Sequencing primers were designed for both ends of the vector, SuperCos1, and used to sequence in the direction of the insert. The primers yielded 225 bp of clockwise (CW) and 250 bp of counterclockwise (CCW) sequence relative to the SuperCos1 sequence (Fig. 1). A blast search of the GenBank database resulted in almost identical matches of the two sequences to the region around the E. coli chromosome at 39.1–39.2 min, with sequence CW having 100% identity to a region in the katE gene and sequence CCW having 99% identity to a region in the osmE gene. Furthermore, a restriction analysis of pES1 showed a complete match to the results predicted by the sequence at the E. coli region shown in Fig. 1.
Figure 1.
Comparison of pES1 to sequenced E. coli genomic map position at 39 min. The isolated cosmid clone capable of restoring (GlcNAc)2 utilization in the E. coli transposon mutant strain, Xm1.4, was subcloned to 7.3 kb in the vector Supercos1. Using primers designed to the ends of Supercos1 and reading in the direction of the insert from both directions, we gathered the following sequence data: (i) clockwise (CW), 250 bp; and (ii) counterclockwise (CCW), 225 bp. A blast search of the nucleotide sequence database revealed greater than 99% identity to a region in the osmE gene (CW) and the katE gene (CCW), respectively. Restriction analysis of the clone was in agreement with that reported for the E. coli genome at the region illustrated. Restriction sites shown include B = BamHI, Bg = BglII, E = EcoRI, Ev = EcoRV, and H = HindIII. All other restriction enzymes tested gave results in agreement with the predicted sequence and included the following: AatII, AccI, AvaI, Bsp106, HpaI, KpnI, NdeI, NheI, NotI, PstI, PvuI, SalI, SphI, and XmnI.
An analysis of the ORFs of this region showed that it corresponded to the previously described “cryptic” cellobiose operon (Fig. 1).
Interpretations of these and the following results are presented in Discussion.
Growth of Wild-Type, Mutant, and Complemented Mutant Strains on Lactate, (GlcNAc)2, or Cellobiose.
The ability of wild-type E. coli XL1-Blue MR, the transposon mutant Xm1.4, and the complemented transposon mutant (Xm1.4:pES1) to ferment a variety of sugars was assayed using MacConkey base agar. The results were as follows: (i) all strains could ferment glucose and GlcNAc; (ii) wild type and the complemented mutant were capable of fermenting (GlcNAc)2, whereas the mutant could not; and (iii) the β-glucosides cellobiose, salicin, and arbutin were not fermented under any conditions tested.
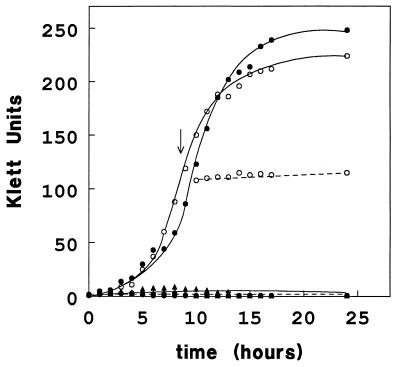
The capability of the three strains to grow on (GlcNAc)2 or cellobiose as the sole carbon source was also tested (Fig. 2). These results show that: (i) The growth rates of all three strains were comparable on lactate and GlcNAc (data not shown). (ii) Wild-type E. coli and the transformant Xm1.4:pES1 can grow on (GlcNAc)2, whereas the mutant, Xm1.4, cannot (Fig. 2). (iii) None of the strains, even the mutant harboring the “cel” operon-containing plasmid (Xm1.4:pES1), can grow on cellobiose. (iv) Preinduction of Xm1.4:pES1 (Fig. 2) or wild type (data not shown) by growth on (GlcNAc)2 does not result in detectable growth on cellobiose. In the latter experiment cells were grown to mid-logarithmic phase on (GlcNAc)2 and then washed into minimal medium containing cellobiose.
Figure 2.
Growth of E. coli strains on (GlcNAc)2 and cellobiose. E. coli strains XL1-Blue MR (wild type, •), Xm1.4 (transposon mutant, ▴), and Xm1.4:pES1 (transposon mutant harboring the 7.3-kb clone, ○) were tested for their ability to grow on (GlcNAc)2 (solid lines) and cellobiose (dashed lines) as sole carbon sources. Minimal media (M9 salts) supplemented with 0.5 mM thiamine and 0.05% Casamino acids containing either 10 mM (GlcNAc)2 or 10 mM cellobiose were inoculated using a 1:100 dilution of overnight cultures grown in LB. Growth was monitored over the indicated time course by using a Klett photoelectric colorimeter with a no. 54 green filter (550 nm). At the arrow, an aliquot of Xm1.4:pES1 cells [preinduced by growth to mid-logarithmic phase on (GlcNAc)2] was harvested and washed three times with an equal volume of minimal medium and then resuspended in minimal medium containing 10 mM cellobiose.
Swarm Plates.
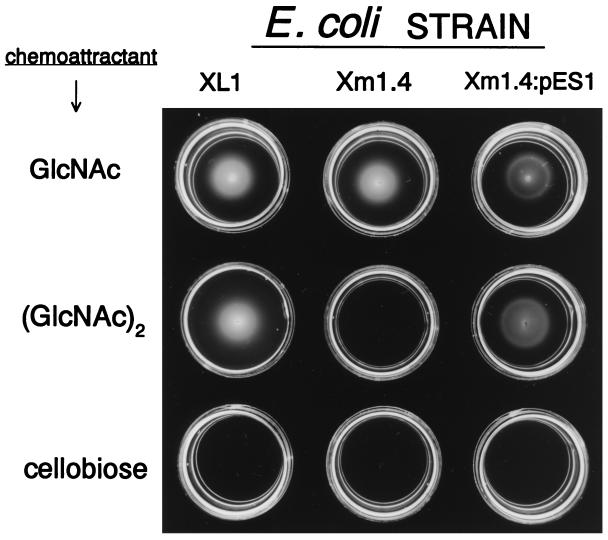
Bacterial chemotaxis was assayed by the use of swarm plates as described in the Materials and Methods. In this assay, bacteria are inoculated in the center of soft agar plates containing the putative chemoattractant. As the cells utilize the test substrate a gradient is formed, and if the compound is a chemoattractant, the cells migrate from the point of inoculation outwards, forming a characteristic ring. If the test compound can be metabolized but is not an attractant, such as lactate, bacterial growth will be visible at the point of inoculation without any swarm rings. Chemotaxis of the wild-type stain XL1-Blue, the transposon mutant Xm1.4, and the transformant Xm1.4:pES1 toward GlcNAc, (GlcNAc)2, and cellobiose is presented in Fig. 3. These data show the following: (i) All three strains formed swarm rings when tested with GlcNAc. (ii) The mutant could neither grow on nor form swarm rings with (GlcNAc)2. (iii) The wild type and transformant harboring the “cel” operon could grow and form swarm rings on (GlcNAc)2. (iv) No rings (or growth) were visible with cellobiose, even after prolonged incubation.
Figure 3.
E. coli chemotaxis toward GlcNAc, (GlcNAc)2, and cellobiose in swarm plates. Low percentage agar plates were prepared containing 10 mM GlcNAc, (GlcNAc)2, or cellobiose as the sole carbon source. E. coli strains are indicated above the respective columns and include XL1-Blue MR (wild type), Xm1.4 (transposon mutant), and Xm1.4:pES1 (transposon mutant harboring the 7.3-kb clone). The chemoattractants tested are indicated by the designated rows. Cells were inoculated at the center of each plate and allowed to swarm for 36 hr at 30°C.
Swarm rings were also visible with (GlcNAc)3, but not with any higher chitin oligosaccharides tested, such as (GlcNAc)4 and (GlcNAc)5.
DISCUSSION
We report here that a variety of wild-type E. coli strains express an inducible system for the transport and catabolism of (GlcNAc)2. The (GlcNAc)2 catabolic operon was cloned by complementation of a (GlcNAc)2-nonfermenting transposon mutant and was identified as the previously designated cryptic cel operon. However, the isolated wild-type allele results only in (GlcNAc)2 and not cellobiose catabolism, as confirmed by fermentation and growth studies. Even under conditions when the wild-type operon was fully induced, little to no growth on cellobiose was detected. Furthermore, E. coli was shown to be chemotactic toward (GlcNAc)2 and (GlcNAc)3 but not to cellobiose, and the swarming response was contingent upon the presence of the intact “cel” operon. A kinetic and molecular description of the (GlcNAc)2 permease, and characterization of phospho-(GlcNAc)2 as the product of transport, will be reported elsewhere.
The “functional” cellobiose catabolic operon (designated cryptic), was originally isolated by subjecting wild-type E. coli to extreme conditions such prolonged incubation (months) of cells on plates containing cellobiose (10–12). Mutations in the sugar-specific proteins of the wild-type operon would be required to alter the sugar specificity of the operon. It is likely that a series of mutations ultimately resulted in cellobiose utilization.
Indeed, recent sequencing data from the E. coli genome database (27) have covered the region spanning the cel operon (39.1–39.2 min), and a comparison of the two sequences reveals several differences between the genomic database sequence and that originally reported for the cel operon (11). These differences are clustered in the permease and in the putative phospho-β-glucosidase. Furthermore, in almost all cases the ability to catabolize cellobiose required “activation” of the operon, either by disruption of the repressor coding region through integration of an insertion element or by a point mutation of the repressor that results in cellobiose recognition (12).
Our results, therefore, show that wild-type enteric bacteria, E. coli, S. typhimurium, and E. cloacae, can ferment and grow on (GlcNAc)2 as the sole carbon source, and that in E. coli the disaccharide is a chemoattractant (the other species were not tested). The cel operon was identified as encoding the necessary proteins required for (GlcNAc)2 catabolism, and we suggest that the apparent crypticity and change in the sugar specificity of the operon resulted from mutations in the relevant genes. We therefore propose that the term “cel” be changed to chb (N,N′-diacetylchitobiose) operon.
The renaming of the cel operon also involves changes of the letter designations of each gene as listed in Table 2. Detailed functional characterization of some of the chb gene products will be described elsewhere. The reasons for the reassignments follow from an attempt to keep the gene names consistent with the nomenclature of the gene products. For example, the chbB, C, and A protein products have been characterized as being homologous to the IIB, IIC, and IIA “domains” of phosphotransferase (PTS)-mediated permeases, respectively (for review, see ref. 28). Sequence information (27) also has revealed that the operon contains no termination loop after chbF, but instead appears to encode an additional protein product of unknown function, in a gene designated chbG, after which a termination loop can be found.
Table 2.
Proposed renaming of the cel operon
| Previous designation | New designation | Putative function |
|---|---|---|
| celA | chbB | IIB domain of PTS permease* |
| celB | chbC | IIC domain of PTS permease* |
| celC | chbA | IIA domain of PTS permease* |
| celD | chbR | Repressor |
| celF | chbF | Phospho-chitobiase |
| — | chbG | Unknown |
These proteins are homologous to the indicated “domain” nomenclature assigned to the phosphotransferase (PTS)-mediated permeases (28).
Acknowledgments
We thank Dr. Sankar Adhya for helpful suggestions. This work was supported by Grants GM51215 and GM38759 from the National Institute of General Medical Sciences and by Grant NA46RG0091 from the National Oceanic and Atmospheric Administration to Maryland Sea Grant. This paper is publication 1515 from the McCollum–Pratt Institute.
ABBREVIATIONS
- GlcNAc
N-acetylglucosamine
- (GlcNAc)n
β-1,4-linked oligomers of GlcNAc
- PTS
phosphoenolpyruvate:glycose phosphotransferase system
References
- 1.Coughlan M P, Mayer F. In: The Prokaryotes. 2nd Ed. Balows A, Truper H G, Dworkin M, Harder W, Schleifer K H, editors. New York: Springer; 1991. pp. 460–516. [Google Scholar]
- 2.Tracy M V. Rev Pure Appl Chem. 1957;7:1–14. [Google Scholar]
- 3.Flach J, Pilet P E, Jolles P. Experientia. 1992;48:701–716. doi: 10.1007/BF02124285. [DOI] [PubMed] [Google Scholar]
- 4.Warren R A J. Annu Rev Microbiol. 1996;50:183–212. doi: 10.1146/annurev.micro.50.1.183. [DOI] [PubMed] [Google Scholar]
- 5.Faunce W, Hall B G. J Bacteriol. 1987;169:2713–2717. doi: 10.1128/jb.169.6.2713-2717.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Al-Zaag A. J Biotechnol. 1989;12:79–86. [Google Scholar]
- 7.Lai X, Ingram L O. J Bacteriol. 1993;175:6441–6450. doi: 10.1128/jb.175.20.6441-6450.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohmiya K, Sakka K, Karita S, Kimura T. Biotech Genet Eng Rev. 1997;14:365–414. doi: 10.1080/02648725.1997.10647949. [DOI] [PubMed] [Google Scholar]
- 9.Hall B G, Betts P W, Kricker M J. Mol Biol Evol. 1986;3:389–402. doi: 10.1093/oxfordjournals.molbev.a040406. [DOI] [PubMed] [Google Scholar]
- 10.Kricker M, Hall B G. Genetics. 1987;115:419–429. doi: 10.1093/genetics/115.3.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parker L L, Hall B G. Genetics. 1990;124:455–471. doi: 10.1093/genetics/124.3.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parker L L, Hall B G. Genetics. 1990;124:473–482. doi: 10.1093/genetics/124.3.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hall B G, Yokoyama S, Calhoun D H. Mol Biol Evol. 1983;1:109–124. doi: 10.1093/oxfordjournals.molbev.a040300. [DOI] [PubMed] [Google Scholar]
- 14.Muzzarelli R A A, editor. Chitin Enzymology: Second International Symposium on Chitin Enzymology. Italy: Atec Edizioni; 1996. [Google Scholar]
- 15.Sommerville C C, Colwell R R. Proc Natl Acad Sci USA. 1993;90:6751–6755. doi: 10.1073/pnas.90.14.6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keyhani N O, Roseman S. J Biol Chem. 1996;271:33414–33424. doi: 10.1074/jbc.271.52.33414. [DOI] [PubMed] [Google Scholar]
- 17.Keyhani N O, Roseman S. J Biol Chem. 1996;271:33425–33432. doi: 10.1074/jbc.271.52.33425. [DOI] [PubMed] [Google Scholar]
- 18.Keyhani N O, Wang L X, Lee Y C, Roseman S. J Biol Chem. 1996;271:33409–33413. doi: 10.1074/jbc.271.52.33409. [DOI] [PubMed] [Google Scholar]
- 19.Sprenger G A, Lengeler J W. J Bacteriol. 1984;157:39–45. doi: 10.1128/jb.157.1.39-45.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cordaro J C, Roseman S. J Bacteriol. 1972;112:17–29. doi: 10.1128/jb.112.1.17-29.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silhavy T J, Berman M L, Enquist L W. Experiments with Gene Fusions. Plainview, NY: Cold Spring Harbor Lab. Press; 1984. [Google Scholar]
- 22.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1996. , Vols. I and II and Supplements. [Google Scholar]
- 23.Sanger F, Niklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.United States Biochemical. Protocols for DNA Sequencing with Sequenase (V 2.0) T7 DNA Polymerase. 7th Ed. Cleveland, OH: United States Biochemical; 1993. [Google Scholar]
- 25.Adler J. Sci Am. 1976;234(4):40–47. doi: 10.1038/scientificamerican0476-40. [DOI] [PubMed] [Google Scholar]
- 26.Bassler B, Gibbons P, Roseman S. Biochem Biophys Res Commun. 1989;161:1172–1176. doi: 10.1016/0006-291x(89)91365-x. [DOI] [PubMed] [Google Scholar]
- 27.Blattner F R, Plunkett G, Bloch C A, Perna N T, Burluand V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 28.Postma P W, Lengeler J W, Jacobson G R. Microbiol Rev. 1993;57:543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]