Abstract
Initiation and control of replication of the broad-host-range plasmid RK2 requires two plasmid-encoded elements, the replication origin (oriV) and the initiation protein TrfA. Purified TrfA is largely in the form of a dimer; however, only the monomeric form of the protein can bind specifically to the direct repeats (iterons) at the RK2 origin. The largely dimeric form of wild-type TrfA is inactive in the initiation of replication of RK2 in an in vitro replication system reconstituted from purified components. However, preincubation of the TrfA protein with the ClpX molecular chaperone isolated from Escherichia coli activates the initiator protein for replication in the purified system. We further observed that ClpX, in an ATP-dependent reaction, greatly increases the proportion of TrfA monomers and, therefore, the ability of this protein to bind to iterons localized within RK2 origin. Finally, a copy-up mutant of the TrfA protein which is largely in the monomer form is active in the reconstituted in vitro replication system, and its activity is not affected by ClpX.
Molecular chaperones have been shown to be involved in both the assembly and the disassembly of DNA replication complexes. Originally, it was demonstrated that the Escherichia coli chaperone proteins DnaK, DnaJ, and GrpE are essential for the displacement of the λP protein of the prepriming complex during the initiation of replication of bacteriophage λ (1). These proteins were also shown to activate the replication initiation protein (Rep) of plasmids P1 (2, 3) and F (4) for binding to nucleotide sequence repeats (iterons) in the origin (ori) of DNA replication. RepA, a replication initiation protein of plasmid P1, can also be activated by the ClpA chaperone, a member of the Clp ATP-ase family of proteins (5). Another member of the Clp protein family, the 46-kDa ClpX protein (6), is involved in activation of the bacteriophage λ initiation protein λO (7) and disassembly of the tetramer form of bacteriophage Mu transposase (8, 9). The ClpX protein can act both as a molecular chaperone and as a protease together with proteolytic subunits (ClpP) (for review, see ref. 10). It has also been shown that both the λO (6) and the MuA (8) initiation proteins and the addiction protein Phd of plasmid P1 (11) are substrates for proteolytic degradation by the ClpXP complex.
Studies on chaperone involvement in plasmid Rep protein activation have been limited to plasmid replicons naturally occurring in E. coli (for review, see ref. 12). Purified plasmid Rep proteins are largely in the dimer form; however, several lines of evidence have shown that it is the monomer form of the protein that specifically binds to the origin of replication (13–16). RK2 is a broad-host-range plasmid that can be transferred to and maintained in a wide range of Gram-negative bacteria (17). Controlled replication of RK2 in these bacteria requires only two plasmid-encoded elements: the origin of replication (oriV) and the Rep protein (TrfA) (18–20). All other elements required for RK2 DNA replication are host-encoded factors (21–24). The purified form of the TrfA protein has been shown to exist largely as a dimer in solution (25–27); however, only the monomer form can bind to the 17-bp iteron sequences localized within the RK2 replication origin (16). TrfA can be activated for iteron binding in vitro by treatment with 4 M guanidine-HCl, and the activated form of the protein has been identified as a monomer (16). An RK2 plasmid carrying a double mutation in the trfA gene, designated trfA 254D/267L, exhibits an elevated copy number in an E. coli host (28). The mutant TrfA 254D/267L protein has been purified and found to be predominantly in the monomeric form (A. E. Toukdarian and D. R. Helinski, unpublished results). In comparison with the mainly dimeric form of wild-type TrfA, the mutant protein exhibits a substantially greater specific activity in binding to the iterons in vitro (28).
Results presented in this paper show that the wild-type TrfA protein is activated by ClpX for DNA replication initiation in vitro. This activation is the result of monomerization of the TrfA dimers by ClpX and, consequently, a greatly increased activity of the protein for binding to the iterons of the RK2 origin of replication.
MATERIALS AND METHODS
Bacterial Strains, Proteins, DNA, and Reagents.
The E. coli strains used in this study were: C600 (thr, leu, thi, laY, supE44, tonA) (29), SG20250, the parent strain of SG20080 (clpX∷kan), SG22098 (clpP∷cm), SG22097 (clpX∷kan, clpP∷cm), SG22093 (clpA∷kan), and SG22100 (clpB∷kan) (30). Highly purified proteins were used for the various assays. The ClpX chaperone protein was a kind gift from Dr. Maciej Zylicz (Gdansk University). E. coli DNA polymerase III holoenzyme was kindly provided by Dr. Michael O’Donnell (Cornell University Medical College). Purified preparations of His6-TrfA, His6-TrfA 254D/267L, and E. coli DnaA protein were kindly provided by Dr. Aresa Toukdarian (University of California, San Diego) and Dr. Alessandra Blasina (Scripps Research Institute). Radiolabeled His6-TrfA and His6-TrfA 254D/267L were purified essentially as described previously (16, 28) from E. coli C600 cells grown in M9 medium with the ICN Tran35S-LABEL metabolic reagent. E. coli DnaB and DnaC proteins were purified as previously described (31, 32). pTJS42 and pSP6 are minireplicons of plasmid RK2 and contain the five-iteron oriV region (25, 33). Commercially obtained proteins and reagents were: DNA gyrase, DnaG primase, HU protein, and ssDNA-binding protein purchased from Enzyco, Inc. (Boulder, CO); BSA (fraction V), ovalbumin, carbonic anhydrase, creatine phosphate, creatine kinase, and rNTPs from Sigma; Klenow fragment of E. coli DNA polymerase from New England Biolabs; Bio-Gel P-100 from Bio-Rad; dNTPs from Pharmacia; and [methyl-3H]dTTP from ICN.
RK2 Replication Reaction Reconstituted with Purified Proteins.
The reaction mixture (25 μl) for RK2 oriV plasmid replication was assembled as previously described (24) and contains: 40 mM Hepes/KOH, pH 8.0; 25 mM, Tris-HCl, pH 7.4; 80 μg/ml BSA; 4% sucrose; 4 mM DTT; 11 mM magnesium acetate; 2 mM ATP; 50 μM concentrations of each dNTP; [methyl-3H]TTP (150 cpm/pmol); 500 μM (each) CTP, GTP, and UTP; 20 μM creatine phosphate; 20 μg/ml creatine kinase; 230 ng of ssDNA-binding protein; 120 ng of DNA gyrase; 1600 ng of DnaB; 100 ng of DnaG primase; 55 ng of DNA pol III core subunit; 55 ng of τ subunit; 15 ng of β subunit; 10 ng of ψ complex; 600 ng of DnaA; 800 ng of DnaC; 300 ng of supercoiled DNA carrying oriV (pTJS42); and 400 ng of His6-TrfA 254D/267L. Reactions were assembled on ice and then incubated at 32°C for 30 min. Reactions were stopped by placing on ice and adding 0.1 M sodium pyrophosphate and 10% TCA. Total nucleotide incorporation (picomoles) was measured by liquid scintillation counting after filtration onto Whatman GF/C glass fiber filters.
RK2 in Vitro Replication with the Use of Crude Extracts.
Reactions were performed as described (23). The five-iteron RK2 origin plasmid pTJS42 and crude cell extracts (FII) from E. coli C600, and the various mutant strains were prepared as previously described (23). Both His6-TrfA and the copy-up mutant His6-TrfA 254D/267L proteins were used in vitro for the initiation of replication.
Gel Mobility Shift Assay.
Incubations were carried out under the in vitro replication conditions described above. The preincubation mixture (12 μl) containing His6-TrfA (1 μg) or mutant His6-TrfA 254D/267L (0.5 μg) in 40 mM Hepes/KOH, pH 8.0; 25 mM Tris-HCl, pH 7.4; 80 μg/ml BSA; 4% sucrose; 4 mM DTT; 11 mM magnesium acetate; 2 mM ATP, 20 μM creatine phosphate; and 20 μg/ml creatine kinase was incubated for 10 min at 32°C with or without the ClpX protein (1.5 μg). After preincubation, the reactions were diluted in the same buffer to obtain various concentrations of the wild-type or mutant His6-TrfA proteins, and a 32P-labeled 393-bp DNA fragment containing the RK2 five-iteron origin was added. The reactions were incubated for 15 min at 32°C followed by the addition of Ficoll 4000 (Pharmacia) to 2.5% (vol/vol). The reactions were loaded on a 5% polyacrylamide gel. Gels were prepared in Tris-borate/EDTA buffer and after electrophoresis were dried and exposed for autoradiography as described previously (25). The DNA probe was labeled with the Klenow fragment of E. coli DNA polymerase.
Size Exclusion Chromatography.
To analyze monomeric and dimeric forms of TrfA protein, a column gel filtration method with Bio-Gel P-100 resin was used. The reaction mixture was the same as used for the preincubation step during the gel mobility shift assay except that it contained radiolabeled His6-TrfA (0.1 μg) or mutant His6-TrfA 254D/267L (0.1 μg) and unlabeled ClpX (1.5 μg) protein. Incubations were carried out for 20 min at 32°C. After incubation, the reactions were diluted in the incubation buffer to a volume of 20 μl and then run through a Bio-Gel P-100 column (0.5 × 19 cm) equilibrated at room temperature in a column buffer (20 mM Tris, pH 7.4; 50 mM KCl; 5 mM MgCl2; 1 mM DTT; 0.1% Nonidet P-40; and 0.5 M NaCl). Four-drop fractions (50 μl) were collected and analyzed by liquid scintillation counting.
RESULTS
Replication Initiation Activity in Vitro of Wild-Type and Mutant 254D/267L TrfA Proteins.
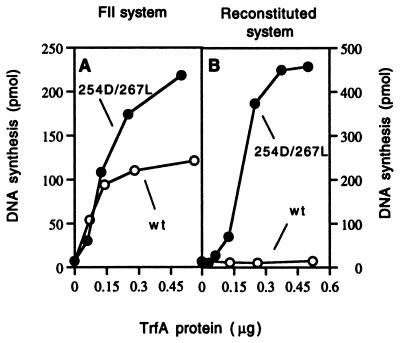
We determined the activity of the purified wild-type and mutant RK2 replication initiator proteins in two in vitro replication systems: a crude cell extract system (FII) and a reconstituted system assembled from purified components. In the FII system, all replication components except for the TrfA protein and the supercoiled DNA template containing a five-iteron RK2 minimal origin were provided by a crude bacterial extract prepared from E. coli C600 cells. Both the wild-type His6-TrfA protein and the copy-up His6-TrfA 254D/267L mutant protein were active in this replication system, however, DNA synthesis with the mutant protein was ∼2-fold higher in comparison with the wild-type protein at the highest TrfA concentrations (Fig. 1A). In contrast, wild-type His6-TrfA protein was not active in the replication system reconstituted from purified components, whereas in this system the His6-TrfA 254D/267L copy-up mutant protein was highly active (Fig. 1B). These results suggested that a specific component(s) present in the crude extract was absent in the reconstituted system and that the largely dimeric wild-type His6-TrfA protein requires this component(s) for initiation activity.
Figure 1.
In vitro replication activity of the TrfA protein. Reactions were carried out under standard replication conditions as described in Materials and Methods with the use of the five-iteron plasmid pTJS42 and increasing amounts of wild-type His6-TrfA or the His6-TrfA 254D/267L protein as indicated. Total nucleotide incorporation was determined after 30 min. incubation at 32°C. (A) Replication with a crude extract (FII) prepared from E. coli C600. (B) Replication with an in vitro system reconstituted with purified components.
ClpX Chaperone Protein Activates Wild-Type TrfA for Initiation of Replication in Vitro.
ClpX is a member of the Clp family of proteins which is highly conserved and has been identified in a wide range of organisms (10). We investigated the influence of the molecular chaperone ClpX on the replication of the broad-host-range plasmid RK2 and specifically its effect on the initiation activity of the TrfA protein. The wild-type His6-TrfA protein which was not active in the reconstituted replication system was found, after preincubation with the ClpX chaperone, to exhibit in vitro replication activity (Fig. 2). The preincubation step resulted in a level of wild-type His6-TrfA protein replication activity approximating that obtained with the crude extract system, i.e., ∼50% of maximum nucleotide incorporation obtained for the copy-up mutant His6-TrfA 254D/267L (Fig. 2). In contrast, the replication activity of the predominantly monomeric His6-TrfA 254D/267L mutant protein was not increased by preincubation with the ClpX chaperone.
Figure 2.
ClpX activates TrfA for in vitro replication. The wild-type His6-TrfA (0.84 μg) or His6-TrfA 254D/267L (0.34 μg) were preincubated for 10 min at 32°C in a preincubation mixture (12 μl) containing 40 mM Hepes/KOH, pH 8.0; 25 mM Tris/HCl, pH 7.4; 80 μg/ml BSA; 4% sucrose; 4 mM DTT; 11 mM magnesium acetate; 2 mM ATP; 500 μM (each) CTP, GTP, and UTP; 8 μM creatine phosphate; 20 μg/ml creatine kinase; 300 ng oriV (pTJS42); and various amounts of ClpX as indicated. After preincubation, the other essential purified components of the standard replication system were added, and the replication mixtures were incubated at 32°C for an additional 30 min. Reactions were stopped, and total nucleotide incorporation was measured.
ClpX Activates TrfA for Binding to Iterons.
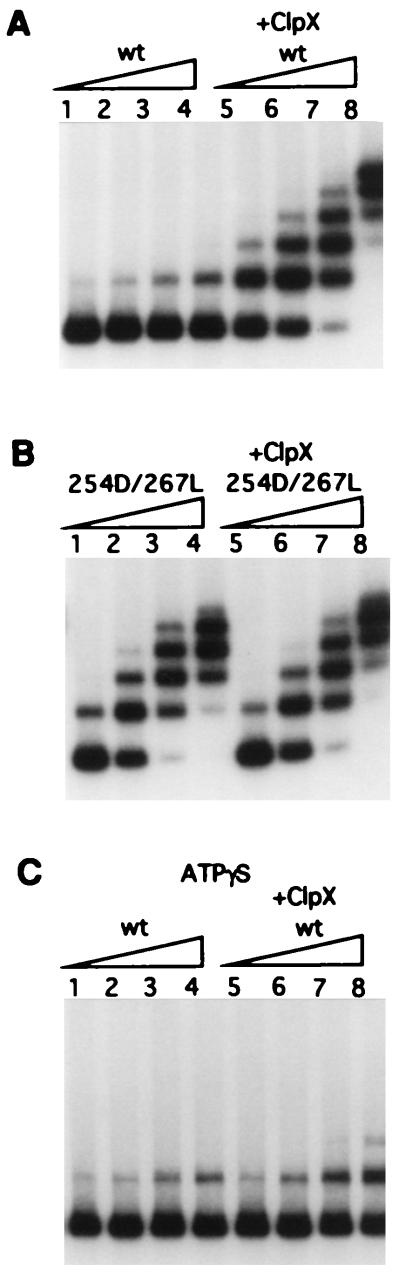
Using the gel mobility shift assay, we determined the affect of the ClpX chaperone on the ability of the RK2 initiation protein to bind to the oriV iteron sequences. The probe used in these experiments was a 32P-labeled linear DNA fragment containing the RK2 minimal origin region which includes five-iteron sequences specific for TrfA protein binding. It was shown earlier that both the wild-type TrfA and the copy-up mutant protein can bind to this linear fragment and produce five retarded bands in the gel mobility shift assay (28). However, a considerably higher concentration of wild-type TrfA, in comparison with the mutant TrfA 254D/267L, was necessary to achieve the same degree of retardation (28). Similarly, in our experiments a much higher specific activity for iteron binding for the copy-up mutant His6-TrfA254D/267L was observed in comparison with the activity for iteron binding shown by the wild-type protein His6-TrfA (Fig. 3 A and B). However, after preincubation of the wild-type His6-TrfA with the ClpX chaperone, the specific activity for iteron binding was increased manyfold and approached the level of specific activity obtained for the mutant protein (Fig. 3A). In contrast, using this assay, there was little or no effect of preincubation of His6-TrfA 254D/267L mutant protein with the ClpX chaperone on binding activity (Fig. 3 A and B). If ATPγS was used in the activation reaction in place of ATP, increased iteron binding by His6-TrfA after preincubation with ClpX was not observed (Fig. 3C).
Figure 3.
ClpX activates TrfA for binding to the RK2 replication origin wild-type His6-TrfA (1 μg) (A and C) or His6-TrfA 254D/267L (0.5 μg) (B) were preincubated as described in Materials and Methods. The preincubation mixtures were then diluted, and different amounts of either the wild-type or the 254D/267L TrfA protein were incubated further for 15 min at 32°C with a 32P-labeled minimal origin DNA fragment of RK2. Increasing amounts: 1.25 ng, 2.5 ng, 5 ng, 10 ng of wild-type His6-TrfA (lanes 1 to 4 and 5 to 8, A and C) and 0.3 ng, 0.6 ng, 1.25 ng, 2.5 ng of His6-TrfA 254D/267L (lanes 1 to 4 and 5 to 8, B), respectively, were used for the incubations with the labeled fragment. After incubation, samples were loaded on a 5% polyacrylamide gel and after electrophoreses were dried and exposed for autoradiography as described in Materials and Methods. (C) Results of the experiment carried out in the presence of ATPγS (2 mM) instead of ATP and the ATP regeneration system .
ClpX Monomerize TrfA in an ATP-Dependent Reaction.
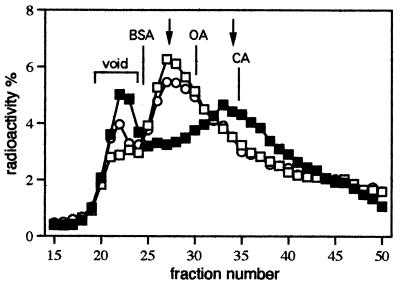
To determine whether ClpX activation of TrfA binding is caused by ClpX monomerization of TrfA dimers, gel exclusion chromatography analysis was carried out. After incubation of 35S-radiolabeled His6-TrfA wild-type protein followed by gel filtration on a Bio-Gel P-100 column, the peak fractions of protein were found at a position corresponding to a globular protein with an approximate mass of 54 kDa (Fig. 4). This position is roughly what is expected for the dimer form of the TrfA molecule (74 kDa). NaCl (0.5 M) and 0.1% Nonidet P-40 were present during the column separation to reduce the re-formation of dimers from monomers. Under the same experimental conditions, the addition of ClpX chaperone to the incubation mixture resulted in peak fractions of TrfA at a position corresponding to a 29-kDa protein, approximately the size of the monomer TrfA form (37 kDa) (Fig. 4). Protein observed in the void fractions may correspond to TrfA associated with the ClpX chaperone or TrfA aggregates. If ATPγS was used instead of ATP during ClpX incubation with TrfA, the location of the peak fractions of the TrfA protein corresponded once again to the dimeric form of the protein. In a control experiment, incubation of the mutant His6-TrfA 254D/267L protein with or without ClpX resulted in peak fractions after chromatography corresponding to the position expected for the monomer form (results not show).
Figure 4.
ClpX chaperone monomerizes TrfA in an ATP-dependent reaction. Column gel exclusion chromatography using Bio-Gel P-100 was performed as described in Materials and Methods after incubation of 35S-radiolabeled wild-type His6-TrfA with or without ClpX chaperone (1.5 μg). After column separation, the amount of radioactivity was determined for each fraction. TrfA protein profiles after incubation in the presence of ClpX and ATP (2 mM) (▪); ClpX and ATPγS (2 mM) (□); and ATP (2 mM) alone (○). The positions of peak fractions of the molecular weight markers are indicated: BSA (66 kDa), ovalbumin (OA) (44 kDa), and carbonic anhydrase (CA) (29 kDa). Arrows, positions of the TrfA dimer and monomer forms.
RK2 Can Be Established in E. coli clp Mutants.
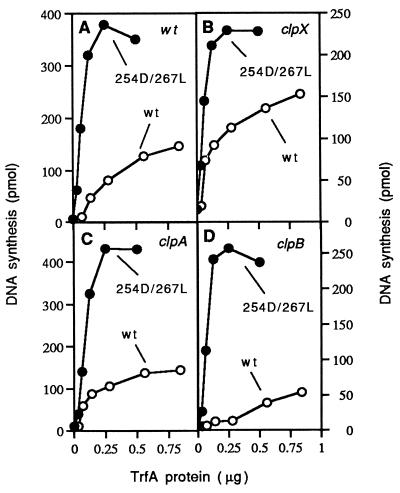
Mutations in genes encoding chaperones in E. coli have been found to affect the replication of plasmid replicons (12). To determine whether or not mutations that inactivate specific clp genes affect RK2 replication, we tested the efficiency of transformation and the copy number of mini-RK2 plasmids in different E. coli clp mutants. In addition, the replication activity of mini-RK2 replicons was examined using crude extracts prepared from these strains. No significant difference in the efficiency of transformation of the RK2 minimal five-iteron origin replicon, pTJS42, or the eight-iteron minireplicon, pTJS26, in a clpX, clpP, clpA, clpB, or a double clpX, clpP mutant strain was detected (Table 1). Also no significant difference in copy number of RK2 minireplicons in the E. coli clpX mutant was observed when compared with the wild-type strains by clonal analysis (results not shown). Finally, the wild-type His6-TrfA protein exhibited similar specific activity in the in vitro replication system using crude extracts (FII) prepared from either the E. coli parent strain or mutant clpX or clpA strains relative to the specific replication activity observed with the mutant His6-TrfA 254D-267L protein using the same extracts (Fig. 5). Interestingly, the replication activity of the wild-type His6-TrfA protein using crude extracts prepared from an E. coli clpB mutant was considerably lower (Fig. 5D). The mutant His6-TrfA 254D/267L protein exhibited a higher replication activity in the crude extracts for all the strains tested when compare with replication activity of the wild-type His6-TrfA protein (Fig. 5).
Table 1.
Establishment of RK2 minireplicons in various E. coli clp mutants
| Strain | Relevant phenotype | Transformation efficiency
|
||
|---|---|---|---|---|
| pTJS26 | pTJS42 | pBR322 | ||
| SG 20250 | wt | 5.4 × 104 | 1.5 × 103 | 9.7 × 104 |
| SG 20080 | clpX::kan | 5.2 × 104 | 1.4 × 103 | 9.1 × 104 |
| SG 22098 | clpP::cm | 4.6 × 104 | 1.2 × 103 | 8.6 × 104 |
| SG 22097 | clpX::kan, clpP::cm | 1.2 × 105 | 2.2 × 103 | 2.1 × 105 |
| SG 22093 | clpA::kan | 1.4 × 105 | 3.3 × 103 | 2.4 × 105 |
| SG 22100 | clpB::kan | 5.3 × 104 | 2.3 × 103 | 1.2 × 105 |
The transformation efficiencies were estimated for 1 μg of supercoiled plasmid DNA purified by using alkaline lysis and double CsCl/ethidium bromide equilibrium centrifugation. pTJS42 and pTJS26 are RK2 minireplicons containing five and eight iterons, respectively. Plasmid pBR322 DNA was used as a control.
Figure 5.
TrfA is active in the replication of the RK2 origin in crude extracts prepared from various E. coli clp mutants. The in vitro replication reactions with various amounts of wild-type His6-TrfA or His6-TrfA 254D/267L were performed under standard replication conditions as described in Materials and Methods with the use of extracts prepared from E. coli isogenic bacterial strains: (A) SG20250, (B) SG22080 (clpX∷kan), (C) SG22093 (clpA∷kan), and (D) SG 22100 (clpB∷kan). Replication activity was measured after incubation for 60 min at 32°C.
DISCUSSION
Binding of the monomer form of the TrfA protein to the iteron sequences localized within the RK2 replication origin results in a formation of a specific nucleoprotein structure and an open complex (34). It has been shown that the largely dimeric wild-type TrfA protein can be activated for iteron binding by guanidine-HCl treatment (16). In addition, the copy-up mutant protein, TrfA 254D/267L, was found to be mainly in the monomeric form (A. E. Toukdarian and D.R.H., unpublished results) and to exhibit a substantially elevated iteron-binding activity (28). The copy-up mutant protein is not further activated by guanidine-HCl treatment (A. E. Toukdarian and D.R.H., unpublished results). The results presented here show that the ClpX chaperone can activate wild-type TrfA protein but cannot further activate the mutant 254D/267L protein for in vitro replication using a reconstituted system (Fig. 2). In addition, we demonstrate that the ClpX protein in an ATP-dependent reaction increases the specific affinity of the TrfA protein for iteron binding (Fig. 3). Furthermore, gel exclusion chromatography analysis showed that incubation of the wild-type protein with the ClpX protein in the presence of ATP greatly increases the proportion of wild-type TrfA monomers (Fig. 4). The ATP dependency for the activation is similar to the ClpX-dependent activation of the nucleoprotein replicative intermediate during transposition of phage Mu (8, 9), the ClpX-dependent desegregation of the bacteriophage λ initiation protein λO (7), and the activation of the plasmid P1 initiator RepA protein by the ClpA chaperone (5, 35). Studies with the mini-P and mini-F replicons suggest that chaperones can affect specific DNA binding by Rep proteins by (i) dissociating of Rep dimers to the monomer form (2, 36) and (ii) increasing the specific binding activity of Rep monomers (37, 38). The dimer form of the wild-type TrfA protein is relatively stable. A 100-fold dilution of the largely dimeric wild-type TrfA protein does not affect iteron binding; however, guanidine-HCl treatment significantly increased specific iteron-binding activity (16). Our results show that the ClpX chaperone is able to monomerize a substrate protein. In the case of the TrfA protein, this monomerization by ClpX results in activation of the protein for the initiation of replication. The possibility that TrfA monomers can be further activated by ClpX chaperone requires additional study. ELISA experiments using TrfA and ClpX proteins and a ClpX antibody confirm a physical interaction between these two proteins (I.K. and M. Zylicz, unpublished results). Deletion analysis has shown that the sequence at the carboxyl-terminal end is important for targeting the MuA protein for ClpX disassemble and degradation (8). It will be of particular interest to test various TrfA mutants for ClpX-dependent activation, especially because the carboxyl terminal ends of TrfA, MuA, and other ClpX substrates show some homology (T. Baker, personal communication).
It is clear that the TrfA protein can be found in at least two forms: a monomer form that serves as a replication initiator; and a dimer form that may be involved in the formation of a coupling complex (16, 23, 28). Evidence has been obtained for a handcuffing or iteron inhibition model for the control of RK2 plasmid copy number (23, 28). According to this model, plasmid copy number is controlled by the formation of a nucleoprotein complex in which two plasmid molecules are coupled at the origins regions by the TrfA protein, and this coupled complex is unable to initiate replication. It is possible that chaperones influences RK2 replication by changing the TrfA monomer/dimer ratio which in turn could affect both replication initiation and iteron coupling.
It is of interest that we did not find a ClpX requirement for RK2 replication in vivo or in crude extracts (Table 1, Fig. 5). In addition, the RK2 plasmid can be maintained in E. coli dnaJ and dnaK null mutant strains (A. E. Toukdarian and D.R.H., unpublished results). It was demonstrated previously that two groups of molecular chaperones can perform similar roles during the replication initiation of plasmid P1. DnaK, DnaJ, and GrpE or ClpA, a member of the Clp protein family, can activate the RepA protein for iteron binding (5). It is likely, therefore, that in vivo the ClpX protein can be substituted by homologous or other chaperones in the monomerization of TrfA. A ClpX homologue, ClpY, has been identified recently in E. coli (39). It is also possible that different chaperone homologues carry out TrfA activation in distantly related bacterial species. The TrfA protein may contain certain structural properties as a universal substrate for chaperone activation; otherwise this required preinitiation step would be bacterial host-limited. The chaperone-dependent reactions during replication initiation of bacteriophage λ (1, 40), phage Mu (8, 9), plasmid P1 (2, 5, 37, 41, 42), or F (4) naturally occur in a narrow range of bacterial hosts. It has been proposed that RK2 replication initiation in different bacterial hosts may depend on a host-nonspecific interaction of the host DnaA protein with one or more of the DnaA boxes at the plasmid replication region, a specific interaction between the DnaA protein and other host replication proteins, and a host-nonspecific activation of the DnaB helicase as a result of TrfA iteron binding and the induction of specific nucleoprotein changes at the RK2 origin (24). The results presented in this work indicate that an additional preinitiation step, i.e., the monomerization of TrfA by a chaperone protein, is a requirement for RK2 replication. It remains to be determined whether or not this step is carried out by homologous or different chaperones in the wide range of Gram-negative bacteria that stably maintain plasmid RK2.
Acknowledgments
We thank Dr. M. Zylicz, Dr. M. O’Donnell, Dr. A. Toukdarian, and Dr. A. Blasina for generous gifts of purified proteins. We are grateful to Dr. A. Toukdarian for helpful discussions during the course of this work and for critically reading the manuscript. We thank R. Neves for help in the preparation of the manuscript. This work was supported by National Institutes of Health Research Grant AI-07194 and by Polish State Committee for Scientific Research Grant 6P04A01712.
References
- 1.Liberek K, Georgopoulos C, Zylicz M. Proc Natl Acad Sci USA. 1988;85:6632–6636. doi: 10.1073/pnas.85.18.6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wickner S, Skowyra D, Hoskins J, McKenney K. Proc Natl Acad Sci USA. 1992;89:10345–10349. doi: 10.1073/pnas.89.21.10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sozhamannan S, Chattoraj D K. J Bacteriol. 1993;175:3546–3555. doi: 10.1128/jb.175.11.3546-3555.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kawasaki Y, Wada C, Yura T. Mol Gen Genet. 1990;220:277–282. doi: 10.1007/BF00260494. [DOI] [PubMed] [Google Scholar]
- 5.Wickner S, Gottesman S, Skowyra D, Hoskins J, McKenney K, Maurizi M R. Proc Natl Acad Sci USA. 1994;91:12218–12222. doi: 10.1073/pnas.91.25.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wojtkowiak D, Georgopoulos C, Zylicz M. J Biol Chem. 1993;268:22609–22617. [PubMed] [Google Scholar]
- 7.Wawrzynow A, Wojtkowiak D, Marszalek J, Banecki B, Jonsen M, Graves B, Georgopoulos C, Zylicz M. EMBO J. 1995;14:1867–1877. doi: 10.1002/j.1460-2075.1995.tb07179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levchenko I, Luo L, Baker T A. Genes Dev. 1995;9:2399–2408. doi: 10.1101/gad.9.19.2399. [DOI] [PubMed] [Google Scholar]
- 9.Kruklitis R, Welty D J, Nakai H. EMBO J. 1996;15:935–944. [PMC free article] [PubMed] [Google Scholar]
- 10.Wawrzynow A, Banecki B, Zylicz M. Mol Microbiol. 1996;21:895–899. doi: 10.1046/j.1365-2958.1996.421404.x. [DOI] [PubMed] [Google Scholar]
- 11.Lehnherr H, Yarmolinsky M. Proc Natl Acad Sci USA. 1995;92:3274–3277. doi: 10.1073/pnas.92.8.3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chattoraj D K. Genet Eng NY. 1995;17:81–98. [PubMed] [Google Scholar]
- 13.Manen D, Upegui-Gonzalez L C, Caro L. Proc Natl Acad Sci USA. 1992;89:8923–8927. doi: 10.1073/pnas.89.19.8923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishiai M, Wada C, Kawasaki Y, Yura T. Proc Natl Acad Sci USA. 1994;91:3839–3843. doi: 10.1073/pnas.91.9.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wickner S, Hoskins J, McKenney K. Proc Natl Acad Sci USA. 1991;88:7903–7907. doi: 10.1073/pnas.88.18.7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toukdarian A E, Helinski D R, Perri S. J Biol Chem. 1996;271:7072–7078. doi: 10.1074/jbc.271.12.7072. [DOI] [PubMed] [Google Scholar]
- 17.Thomas C M, Helinski D R. In: Promiscuous Plasmids of Gram-Negative Bacteria. Thomas C M, editor. San Diego, CA: Academic Press; 1989. pp. 1–25. [Google Scholar]
- 18.Figurski D H, Helinski D R. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas C M, Meyer R, Helinski D R. J Bacteriol. 1980;141:213–222. doi: 10.1128/jb.141.1.213-222.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidhauser T J, Helinski D R. J Bacteriol. 1985;164:446–455. doi: 10.1128/jb.164.1.446-455.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaylo P, Turjman N, Bastia D. J Bacteriol. 1987;169:4703–4709. doi: 10.1128/jb.169.10.4703-4709.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pinkney M, Diaz R, Lanka E, Thomas C M. J Mol Biol. 1988;203:927–938. doi: 10.1016/0022-2836(88)90118-0. [DOI] [PubMed] [Google Scholar]
- 23.Kaittell B L, Helinski D R. Proc Natl Acad Sci USA. 1991;88:1389–1393. doi: 10.1073/pnas.88.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konieczny, I. & Helinski, D. R. (1997) J. Biol. Chem., in press. [DOI] [PubMed]
- 25.Perri S, Helinski D R, Toukdarian A. J Biol Chem. 1991;266:12536–12543. [PubMed] [Google Scholar]
- 26.Lin J, Helinski D R. J Bacteriol. 1992;174:4110–4119. doi: 10.1128/jb.174.12.4110-4119.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cereghino J L, Helinski D R. J Biol Chem. 1993;268:24926–24932. [PubMed] [Google Scholar]
- 28.Blasina A, Kittell B L, Toukdarian A E, Helinski D R. Proc Natl Acad Sci USA. 1996;93:3559–3564. doi: 10.1073/pnas.93.8.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Appleyard R K. Genetics. 1954;39:440–452. doi: 10.1093/genetics/39.4.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gottesman S, Clark W P, de Crecy-Lagard V, Maurizi M R. J Biol Chem. 1993;268:22618–22626. [PubMed] [Google Scholar]
- 31.Kobori J A, Kornberg A. J Biol Chem. 1982;257:13763–13769. [PubMed] [Google Scholar]
- 32.Arai K, Yasuda S, Kornberg A. J Biol Chem. 1981;256:5247–5252. [PubMed] [Google Scholar]
- 33.Schmidhauser T J, Filutowicz M, Helinski D R. Plasmid. 1983;9:325–330. doi: 10.1016/0147-619x(83)90010-0. [DOI] [PubMed] [Google Scholar]
- 34.Konieczny, I., Doran, K. S., Helinski, D. R. & Blasina, A. (1997) J. Biol. Chem. 20173–20178. [DOI] [PubMed]
- 35.Pak M, Wickner S. Proc Natl Acad Sci USA. 1997;94:4901–4906. doi: 10.1073/pnas.94.10.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skowyra D, Wickner S. J Biol Chem. 1993;268:25296–25301. [PubMed] [Google Scholar]
- 37.DasGupta S, Mukhopadhyay G, Papp P P, Lewis M S, Chattoraj D K. J Mol Biol. 1993;232:23–34. doi: 10.1006/jmbi.1993.1367. [DOI] [PubMed] [Google Scholar]
- 38.Chattoraj D K, Ghirlando R, Park K, Dibbens J A, Levis M, S. Genes to Cells. 1996;1:189–199. doi: 10.1046/j.1365-2443.1996.d01-235.x. [DOI] [PubMed] [Google Scholar]
- 39.Missiakas D, Schwanger F, Betton J M, Georgopoulos C, Raina S. EMBO J. 1996;15:6899–6909. [PMC free article] [PubMed] [Google Scholar]
- 40.Zylicz M. Philos Trans R Soc London B. 1993;339:271–277. doi: 10.1098/rstb.1993.0025. ; discussion 277–278. [DOI] [PubMed] [Google Scholar]
- 41.Tilly K, Yarmolinsky M. J Bacteriol. 1989;171:6025–6029. doi: 10.1128/jb.171.11.6025-6029.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wickner S, Hoskins J, McKenney K. Nature (London) 1991;350:165–167. doi: 10.1038/350165a0. [DOI] [PubMed] [Google Scholar]