Abstract
Many examples of extreme virus resistance and posttranscriptional gene silencing of endogenous or reporter genes have been described in transgenic plants containing sense or antisense transgenes. In these cases of either cosuppression or antisense suppression, there appears to be induction of a surveillance system within the plant that specifically degrades both the transgene and target RNAs. We show that transforming plants with virus or reporter gene constructs that produce RNAs capable of duplex formation confer virus immunity or gene silencing on the plants. This was accomplished by using transcripts from one sense gene and one antisense gene colocated in the plant genome, a single transcript that has self-complementarity, or sense and antisense transcripts from genes brought together by crossing. A model is presented that is consistent with our data and those of other workers, describing the processes of induction and execution of posttranscriptional gene silencing.
The coat protein genes of many plant viruses have been transformed into a wide range of plant species to obtain viral protection. In some studies, the expression of the protein has been responsible for the resistance (1), but in a number of cases, the resistance has been demonstrated to occur at the RNA level (2–4). The expression of virus-derived sense or antisense RNA in transgenic plants conferring RNA-mediated virus resistance appears to induce a form of posttranscriptional gene silencing (PTGS) (4, 5). The PTGS mechanism is typified by the highly specific degradation of both the transgene mRNA and the target RNA, which contains either the same or complementary nucleotide sequences. If the transgene contains viral sequences, then virus genomic RNA containing these sequences cannot accumulate in the plant (2–5). To explain the effectiveness of transgene mRNA silencing of same-sense endogenous gene transcripts or viral genomic RNA, it has been suggested that a plant-encoded RNA-dependent RNA polymerase (6, 7) makes a complementary strand from the transgene mRNA and that the small cRNAs potentiate the degradation of the target RNA. Antisense RNA and the hypothetical cRNAs have been proposed to act by hybridizing with the target RNA to either make the hybrid a substrate for double-stranded (ds) RNases or arrest the translation of the target RNA (4).
We sought to test whether the introduction of gene constructs that produced mRNA transcripts capable of forming a duplex would be more or less effective at generating PTGS than constructs producing either sense or antisense mRNA alone. If the critical event in silencing requires the reaction of a free RNA that is complementary to, or the same polarity as, the target RNA, then simultaneously introducing RNA molecules of both polarities should result in less effective silencing. However, if dsRNA itself is the trigger that induces the silencing mechanism, then simultaneous expression of both polarities should silence gene expression more effectively than expression of either polarity alone.
MATERIALS AND METHODS
Plasmid Construction.
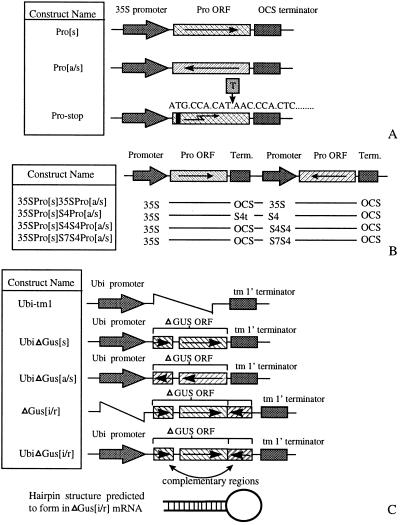
Standard gene cloning methods (8) were used to make the gene constructs, a summary of which is shown in Fig. 1. The plasmids for tobacco transformation were derived from pART 7 and pART 27 (9), and those for rice transformation were derived from pVec4 (10). The protease (Pro) gene was obtained by reverse transcription–PCR from an Australian O type isolate of potato virus Y (PVY) by using the forward primer (5′-TTGCACAAGCTTTGTTTTG-3′) and the reverse primer (5′-GTGATAAAGCTTTGAAGATTGATTTAATG-3′). Pro[s]-stop ([s], sense) was generated by using the same forward primer and the stop reverse primer: 5′-CCCAAGCTTATAATGCCACATTAACCCACTCAAAGTTTG-3′. The enhanced (S4S4 and S7S4) and unenhanced (S4) subterranean clover stunt virus promoters (11), subterranean clover stunt virus 4 terminator (11), and Agrobacterium tumor morphology large gene terminator (tm1′) were derived from pComponent4 (12), pComponent7 (12), and pTRA151 (13).
Figure 1.
Gene constructs. (A) Gene constructs Pro[s], Pro[a/s], and Pro[s]-stop. Pro[s] contains the Pro ORF in a sense orientation and is capable of expressing Pro protein, Pro[a/s] contains the Pro ORF in an antisense orientation, and Pro[s]-stop is the same as Pro[s] except that a thymidine residue has been inserted, making a stop codon (at the fourth codon) and a frameshift. (B) Constructs designed to express both Pro[s] and Pro[a/s] mRNAs. The Pro[s] is controlled in all constructs by the cauliflower mosaic virus 35S promoter; the terminators of Pro[s] and the promoters controlling the Pro[a/s] gene are shown. OCS, octopine synthase; term., terminator. (C) Constructs used to express ΔGUS mRNA in rice. The ΔGUS ORF has a 231-base deletion to prevent production of active GUS protein. With the exception of Gus[i/r] (i/r, inverted repeat), which is promoterless, the constructs are controlled by the ubiquitin promoter. UbiΔGus[s] and UbiΔGus[a/s] contain the ΔGUS ORFs in a sense and an antisense orientation, respectively. The 3′ region of the transcription unit of UbiΔGus[i/r] and ΔGus[i/r] is complementary to the 5′ region of the ΔGUS transcript. The predicted hairpin structure of such an mRNA is shown at the bottom.
The Δβ-glucuronidase (GUS) gene was constructed by digestion of a complete GUS-encoding sequence with EcoRV and religation. This generated a 231-bp deletion.
Plant Transformation.
Nicotiana tabaccum cv. W38 tissue was transformed and regenerated into whole plants essentially as described by Ellis et al. (14). Rice (Oryza sativa cv. Taipei 309) was transformed essentially as described by Wang et al. (15).
For rice supertransformation, calli derived from mature seeds of hygromycin-resistant rice plants constitutively expressing GUS were incubated for 2 days with Agrobacteria, which contained various binary vector constructs; next the calli were placed on callusing media containing bialaphos (10 mg/liter), hygromycin (50 mg/liter), and Timentin (SmithKline Beecham; 150 mg/liter). During the next 4 weeks, bialaphos-resistant calli developed. These calli were maintained on hygromycin- and bialaphos-containing media for another 4 weeks before being assayed for GUS activity.
Analysis of Transgenic Plants.
DNA extraction and Southern blot hybridization analysis were carried out essentially as described by McAlister et al. (16). Total DNA (15 μg per sample) was used for electrophoresis. RNA extractions and Northern blot hybridization analysis were carried out following the Qiagen RNeasy and Amersham LIFE SCIENCE Megaprime instruction manuals. For PVY Pro strand-specific probes, the random nonamer primers were replaced with PVY sense complimentary primers (5′-CTTTGGGAGTGCTGTC-3′ and 5′-GATGAGGATGATGTCC-3′) or PVY antisense complimentary primers (5′-GCCACATAACCCACTCAAA-3′ and 5′-GGACATCATCCTCATC-3′). The total RNA was extracted from leaves frozen in liquid nitrogen immediately after harvesting. The age of the plants used varied from 6 weeks to 4 months. Total RNA (15 μg per sample) was used for electrophoresis. To monitor equal loading of mRNA in Northern blots, the filters were stripped by boiling twice in 0.1% (wt/vol) SDS and then probed for actin mRNA with a Megaprime probe derived from pACT. This plasmid contains an exon sequence of the tobacco actin gene cloned by PCR from tobacco by using the primers 5′-ACAACAGAATTCGAGGGATATGCTTTGCC-3′ and 5′-ACAACAGAATTCGATATCCACGTCGCACTTC-3′.
Rice calli were tested for GUS activity by using the histochemical stain X-glucuronide essentially as described by Jefferson et al. (17).
Virus Resistance Assays.
Tobacco plants, at the 5- to 10-leaf stage, were inoculated with sap extracts from PVY-infected leaf material (5 ml of 0.1 M sodium phosphate buffer, pH 7.0, per g of tissue) and scored for symptoms from 5 to 21 days after inoculation. PVY-specific ELISA was carried out on leaf material 21 days after inoculation according to the Agdia (Elkhart, IN). instruction sheet.
RESULTS
Tobacco Plants Containing Sense or Antisense Constructs.
About 50 independent transgenic tobacco lines were produced in two independent experiments for each of three constructs, Pro[s], Pro[a/s], and Pro[s]-stop (Fig. 1). All three constructs were controlled by the cauliflower mosaic virus 35S promoter and octopine synthase terminator; Pro[s] and Pro[a/s] contained the PVY nuclear inclusion Pro ORF in the sense and antisense orientations, respectively. The Pro[s]-stop construct contained the PVY Pro ORF in the sense orientation but with a stop codon three codons downstream from the initiation codon. This mutation should prevent production of the Pro protein. When the transgenic plants were inoculated with PVY, 5 of 57 Pro[s] plant lines, 1 of 54 Pro[a/s] plant lines, and 10 of 49 Pro[s]-stop plant lines were immune to the virus. Because many of the Pro[s]-stop lines, encoding untranslatable transgene mRNA, showed immunity, this cannot have been conferred by the PVY Pro protein.
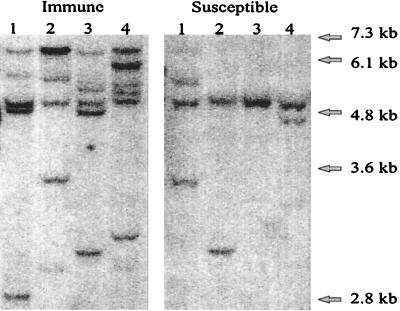
Southern blot analysis (Fig. 2) showed that each of the 16 immune plants contained three to nine copies of the transgene, whereas each of 15 susceptible plants (a random sample) contained one to three copies.
Figure 2.
Southern blot analysis of HindIII-digested DNA (15 μg per lane) extracted from four PVY-susceptible and four PVY-immune tobacco plants containing Pro[s]-stop transgenes. The filters were hybridized with a 32P-labeled probe produced from a gel-purified DNA fragment of the 35S promoter. Each band represents one gene copy and locus.
PVY Immune Plants Containing Sense and Antisense Pro Genes at the Same Locus.
To investigate whether expression of both a sense and an antisense Pro mRNA in the same plant is more or less effective at inducing PVY immunity than expression of either a sense or antisense Pro mRNA alone, tobacco plants were generated that contained gene constructs that encoded the Pro sequence in the sense or antisense orientation or in both orientations. A range of promoters was used to control the antisense gene (Fig. 1B). The plants were challenged with PVY, scored for symptoms, and tested for PVY replication by ELISA. Each plant showed one of three different phenotypes: obvious mottling of leaves 5–10 days after inoculation and the presence of high levels of virus (susceptible); small patches of mottling or chlorotic lesions that appeared more than 14 days after inoculation (resistant); the virus was only detectable in tissues showing symptoms; and no symptoms and no virus accumulation (immune). Less than 15% of the Pro[s] or Pro[a/s] lines were resistant or immune to PVY, but 44–54% of the lines containing both sense and antisense genes showed resistance or immunity (Table 1). This result suggests that the sense and antisense mRNA in the same cell enhance, rather than interfere with, each other in eliciting PVY resistance or immunity.
Table 1.
Resistance to PVY infection of transgenic tobacco plants containing sense, antisense, or both sense and antisense PVY Pro genes
| Plant genotype | No. of immune lines | Copy no. of immune | No. of resistant lines | Copy no. of resistant | No. of susceptible* lines | Copy no. of susceptible* | Total of plants |
|---|---|---|---|---|---|---|---|
| 35SPro[s] | 2 | 1/6 | 2 | 1/5 | 23 | 11/12/13/14/15 | 27 |
| 35SPro[a/s] | 1 | 8 | 0 | − | 24 | 0/11/15/24/8 | 25 |
| 35SPro[s]35SPro[a/s] | 10 | 1/2/2/2/3/3/3/3/4/6 | 2 | 1/1 | 15 | 1/1/2/2/6 | 27 |
| 35SPro[s] S4Pro[a/s] | 11 | 1/1/1/1/2/2/3/3/4/4/6 | 2 | 2/6 | 11 | 2/3/5/8/– | 24 |
| 35SPro[s]S4S4Pro[a/s] | 7 | 1/2/2/2/2/4/5 | 7 | 1/1/1/1/1/1/2 | 12 | 1/1/1/1/7 | 26 |
| 35SPro[s]S7S4Pro[a/s] | 7 | 2/2/2/4/5/7/12 | 4 | 1/2/2/3 | 13 | 1/1/1/1/3 | 25 |
Copy number of five different susceptible lines, picked at random, were determined. Boldface type indicates plants used in crossing experiments; superscript denotes the identification number assigned to these plants, e.g., 24 is Pro[a/s]4.
The effectiveness of RNA-mediated virus resistance has been elegantly shown (2, 3) to correlate with high transgene copy number (three to eight), and this correlation was also apparent in our first experiment described above. Therefore, the number of transgene copies in the plants showing immunity and in five susceptible plants per construct was determined (Table 1). With the exception of the Pro[s] and Pro[a/s] constructs, there was no convincing correlation between high copy number and immunity. Indeed, 4 of the 11 immune 35SPro[s]S4Pro[a/s] plants each contained a single sense plus antisense copy. This finding suggests that the increased effectiveness of sense plus antisense constructs is not simply because of increased copy number.
The inheritance of the immunity phenotype was examined in progeny from selfed 35SPro[s]S4Pro[a/s], 35SPro[s]35SPro[a/s], 35SPro[s], and 35SPro[a/s] plants (Table 2). The progeny from susceptible T0 plants showed complete susceptibility to PVY regardless of the gene construct they contained. The progeny from the one immune 35SPro[a/s] plant and the one immune 35SPro[s] plant showed inheritance ratios of 2:18 (immune/susceptible). Southern analysis of the progeny from the 35SPro[a/s] showed that some susceptible progeny had apparently inherited all eight genes, indicating that plants with identical genotypes had different phenotypes; this is not uncommon among progeny of plants showing PTGS. However, the progeny of 35SPro[s]S4Pro[a/s] and 35SPro[s]35SPro[a/s] plants all had ratios close to the expected 3:1 or 15:1 (immune/susceptible) segregation ratios. This result indicates that, unlike some of the simple sense or antisense constructs, the sense plus antisense constructs gave stable expression of PVY immunity, which is inherited in a Mendelian way.
Table 2.
Inheritance of resistance to PVY infection of transgenic tobacco
| Plant genotype | 35SPro[s]S4 Pro[a/s]
|
35S Pro[s]35S Pro[a/s]
|
35S Pro[s]
|
35S Pro[a/s]
|
||||
|---|---|---|---|---|---|---|---|---|
| Copy no. | T1 I:S | Copy no. | T1 I:S | Copy no. | T1 I:S | Copy no. | T1 I:S | |
| Immune plant 1 | 1 | 15:5 | 1 | 16:4 | 6 | 17:3 | 8 | 2:18 |
| Immune plant 2 | 1 | 12:8 | 2 | 15:5 | 1 | 2:18 | ||
| Immune plant 3 | 1 | 14:6 | 2 | 16:4 | ||||
| Immune plant 4 | 1 | 15:5 | 2 | 16:4 | ||||
| Susceptible plant 1 | 8 | 0:20 | 1 | 0:20 | 1 | 0:20 | 1 | 0:20 |
| Susceptible plant 2 | 2 | 0:20 | 1 | 0:20 | 1 | 0:20 | 8 | 0:20 |
I:S, number of immune plants/number of susceptible plants in 20 T1 progeny; Copy no., number of transgene copies as determined by Southern blot analysis.
PVY Immunity in Progeny from Crosses Between Susceptible Pro[s] and Pro[a/s] Lines.
Transgenes integrating into the plant genome as inverted repeats have been correlated with the induction of PTGS in plants (18), and it has been suggested that this is caused by the structure of the transgene DNA formed within the plant genomic DNA. The sense plus antisense constructs described above have inverted repeat configurations. Therefore, we questioned whether the DNA structure or the production of an RNA duplex was responsible for the increased frequency of producing immune plants. To investigate this, five different lines of PVY-susceptible Pro[s] plants were crossed with three different lines of PVY-susceptible Pro[a/s] plants (Table 1), and at least 20 progeny plants from each cross were inoculated with PVY. Progeny of the selfed parents and Pro[s] × Pro[s] and Pro[a/s] × Pro[a/s] crosses were also tested. The Pro[s] parent plants each contained a single copy of the Pro[s] gene, the Pro[a/s]1 and the Pro[a/s]5 plants each contained a single Pro[a/s] gene, and the Pro[a/s]4 plant contained two Pro[a/s] genes. Some of the progeny plants of the Pro[s] × Pro[a/s] crosses should contain both a Pro[s] gene and a Pro[a/s] gene but at different locations in the genome. These transgenes should be no more likely to show ectopic pairing or interacting structures than would occur between transgenes in the progeny of the crosses between two independent lines of Pro[s] plants or between two independent Pro[a/s] plants. The results showed that a proportion of the progeny of each Pro[s] × Pro[a/s] cross was immune to PVY (Table 3), whereas all of the progeny plants of selfed Pro[s], selfed Pro[a/s], Pro[s] × Pro[s], or Pro[a/s] × Pro[a/s] were susceptible to PVY. This finding suggests that it was the expression of the sense and antisense mRNAs together in the same plant, and not the genomic arrangement of the transgenes, that was inducing the PVY immunity.
Table 3.
PVY resistance in progeny plants resulting from crosses between susceptible Pro[s] plant lines and Pro[a/s] plant lines
| Pro[s]♂
|
Pro[a/s]♂
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 1 | 4 | 5 | ||
| 1 | 0/20* | 8/20 | 11/20 | ||||||
| 2 | 0/20 | 0/20 | 3/20 | 4/20 | |||||
| Pro[s] | 2 | 0/20 | 0/20 | 3/20 | |||||
| ♀ | 3 | 0/20 | 6/20 | 3/20 | |||||
| 4 | 0/20 | 3/20 | 2/20 | ||||||
| 5 | 0/20 | 12/20 | 3/20 | ||||||
| 1 | 4/20 | 5/20 | 0/20 | 0/20 | |||||
| Pro[a/s] | 1 | 7/20 | 6/20 | 0/20 | |||||
| ♀ | 4 | 8/20 | 0/20 | ||||||
| 5 | 0/20 | 0/20 | |||||||
Number of immune or resistant plants/number of progeny plants inoculated.
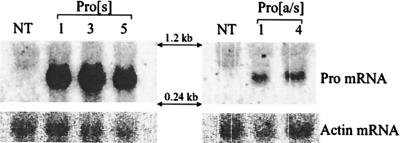
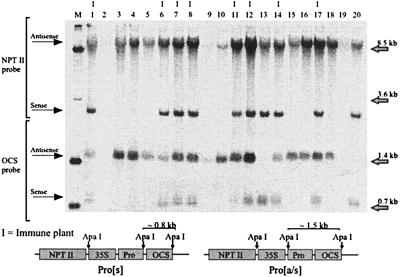
The parent plants for the Pro[s]1 × Pro[a/s]1, Pro[s]1 × Pro[a/s]4, and Pro[a/s]4 × Pro[s]3 crosses and their progeny were examined in more detail. Northern blot analysis showed that each of the parent plants was expressing readily detectable levels of the appropriate Pro transgene mRNA (Fig. 3), with higher levels in the Pro[s] plants than in the Pro[a/s] plants. Southern blot analysis of the progeny plants showed that every plant that contained both a Pro[s] and a Pro[a/s] gene was immune to PVY, whereas all plants containing neither gene, Pro[s] alone, or Pro[a/s] alone were susceptible to PVY (Fig. 4; Table 4). The Pro[a/s]4 plant has two independently segregating Pro[a/s] genes that either alone or together, when associated with a Pro[s] gene, conferred PVY immunity.
Figure 3.
Pro transgene mRNA expression in Pro[s] and Pro[a/s] plants. (Left) Total RNA from nontransgenic (NT), Pro[s]1, Pro[s]3, and Pro[s]5 tobacco plants probed for Pro sense mRNA. (Right) Nontransgenic (NT), Pro[a/s]1, and Pro[a/s]4 tobacco plants probed for antisense Pro mRNA. Both filters were also probed for actin mRNA (see Materials and Methods) for details.
Figure 4.
Southern blot analysis of ApaI-digested genomic DNA (15 μg per lane) from 20 randomly selected progeny plants (lanes 1–20) from a Pro[s]1 × Pro[a/s]1 cross. Lanes marked with “I” or unmarked contained extracts from plants subsequently shown to be immune or susceptible to PVY infection, respectively. Lane M, Mr standard. The top portion of the filter was probed with a neomycin phosphotransferase (NPTII) sequence-specific probe to show transgene copy/locus number. The bottom portion was probed with an octopine synthase (OCS) terminator sequence-specific probe to show the presence or absence of Pro[s] and/or Pro[a/s] transgenes.
Table 4.
Genotypes of individual progeny from Pro[s] × Pro[a/s] crosses and their reaction to PVY inoculation
| Cross ♀ × ♂ | S | S A/S1 | S A/S2 | S A/S1 A/S2 | A/S1 A/S2 | A/S1 | A/S2 | Null | ND | Total no. of plants |
|---|---|---|---|---|---|---|---|---|---|---|
| Pro[s]1 × Pro[a/s]1 | 8 | 8 | — | — | — | 2 | — | 2 | 20 | |
| Pro[a/s]4 × Pro[s]3 | 1 | 5 | 2 | 1 | 4 | 2 | 4 | 1 | 20 | |
| Pro[s]1 × Pro[a/s]4 | 5 | 2 | 4 | 9 | 20 | |||||
| Reaction | Sus | Imm | Imm | Imm | Sus | Sus | Sus | Sus | Sus |
Sus, Susceptible to PVY; Imm, immune to PVY infection; ND, not determined.
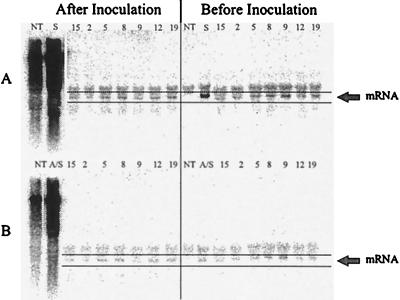
The transgene mRNA levels were analyzed in the progeny that were showing PVY immunity from the Pro[s]1 × Pro[a/s]4 cross (Fig. 5). Both the Pro[s] and Pro[a/s] mRNAs were detected in the plants before and 14 days after inoculation with PVY. PVY viral RNA was detected in the inoculated parental lines (Pro[s]1 and Pro[a/s]4) and in nontransgenic control plants, but none was detected in the symptomless progeny. This result suggests that the Pro[s] and Pro[a/s] mRNAs formed a duplex that induced the sequence-specific degradation to give PVY immunity but the duplex was resistant to the degradation.
Figure 5.
Northern blot analysis of PVY immune plants generated by crossing Pro[s]1 with Pro[a/s]4. Total RNA was extracted from nontransgenic tobacco plants (NT), immune progeny plants 2, 5, 8, 9, 12, 15, and 19, and susceptible parent plants Pro[s]1 (S) and Pro[a/s]1 (A/S) before PVY inoculation (right) and 14 days after inoculation (left). The RNAs were run on 0.9% agarose gels, transferred to membranes, and probed for Pro sense mRNA (A) and Pro antisense mRNA (B). Equal loading of samples is apparent by the similar levels of ribosomal trapping of the probe above the mRNA bands.
Silencing of a Reporter Gene by Using a Single, Self-Complementary mRNA.
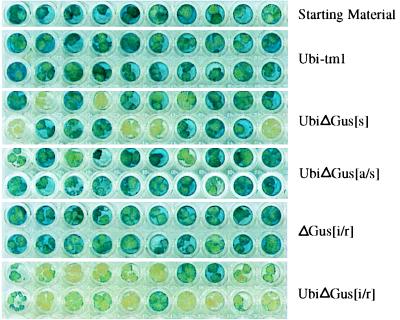
The effectiveness of an RNA duplex, formed from a self-complementary mRNA transcript, to induce silencing of an endogenous reporter gene was investigated. Transgenic rice tissue constitutively expressing GUS from a single transgene was supertransformed by using vectors that contained the bar gene conferring bialaphos resistance and various sense, antisense, and self-complementary (sense + antisense) constructs derived from a crippled GUS gene (Fig. 1C). The supertransformed tissue was maintained on bialaphos selection medium for 3 weeks and then analyzed for GUS activity (Fig. 6). A crippled GUS gene (ΔGUS), with a 231-bp internal deletion, was used so that its activity would not be superimposed on the endogenous GUS activity. Calli supertransformed with the binary vector containing the bar gene and promoter-terminator cassette without the ΔGUS gene gave no silencing of the endogenous GUS activity. Supertransformation with ΔGUS in a sense or antisense orientation showed some silencing of the endogenous GUS activity. However, supertransformation with a construct expressing the ΔGUS gene mRNA designed such that 558 bases of its 3′ end could form a duplex with the 5′ end of the transcript, to give a “panhandle” structure, gave almost 90% silencing of the endogenous GUS activity. Supertransformation with the panhandle construct from which the promoter had been deleted resulted in very little silencing. This result suggests that mRNA with the panhandle structure is an effective trigger for inducing silencing and operates in a way similar to dsRNA formed from two independent molecules.
Figure 6.
Analysis of GUS expression of supertransformed rice callus. Transgenic rice tissue containing a single Gus transgene supertransformed with UbiΔGus[s], UbiΔGus[a/s], UbiΔGus[i/r], ΔGus[i/r], and the binary vector containing the bar gene but not the ΔGus gene. For more details see Materials and Methods and Fig. 1.
DISCUSSION
Extreme virus resistance conferred by a virus-derived transgene has been shown in a number of cases to be mediated by a mechanism of PTGS (2–4, 18). There have been a number of models proposed for the induction and operation of PTGS involved with antisense, cosuppression, and virus resistance (4, 5, 19, 20), but none completely fits the observed results. All models propose that gene silencing and virus immunity involve the rapid degradation of RNA molecules that have high levels of sequence homology with the silencing transgene. In cases in which the silencing transgene is the same sense as the target endogenous gene transcripts or viral genomic RNA, it has been suggested (2, 3, 19) that a plant-encoded RNA-dependent RNA polymerase (6, 7) makes complementary strands from the transgene mRNA and that these small cRNAs potentiate the degradation of the target RNA. Indeed, Dougherty and Parks (19) suggested that, even in antisense plants, cRNAs are synthesized to mediate the degradation. In most models, the antisense RNA or cRNA is proposed to hybridize with the target RNA in some way marking it for degradation (4, 5, 19). However, there are conflicting ideas about the induction of the degradation system. Lindbo and Dougherty (2, 3) proposed that the system was triggered by high levels of transcription and that this correlated with high gene copy number; yet Van Blokland et al. (21) observed silencing of chalcone synthase that could not be correlated with the level of transgene transcription. PTGS is often associated with multicopy T-DNA loci (5, 22–25), and Stam et al. (5) considered that the repetitive nature of a locus was essential for eliciting PTGS and suggested two possible mechanisms: the arrangement leads to the production of aberrant RNA by read-through transcription, abnormal processing, or transcription of a methylated template and the aberrant RNA activates the cRNA mediated RNA degradation process; and ectopic pairing between the transgene DNA and the target endogenous gene DNA impairs the processing and/or transport of the endogenous gene RNA transcript; such RNA may be intrinsically unstable and rapidly degraded or may act as aberrant RNA causing degradation of homologous RNAs. The ectopic pairing between different transgene copies could also produce aberrant RNA that induces the degradation mechanism.
Our results with sense or antisense gene constructs derived from the Pro gene of PVY confirm the findings of others that such genes, when transformed into plants, can confer immunity to the virus from which the transgene was derived. The results also demonstrate a correlation between high transgene copy number and virus immunity and that the immunity is not mediated by the transgene protein. By analogy with other studies (2–5, 18, 25), the PVY immunity we observed is mediated by a sequence-specific degradation of the Pro sequence within the genome of the challenging PVY. Our experiments show that coexpression of sense and antisense Pro mRNAs, from a single T-DNA construct or by introduction through crossing, was much more effective at inducing PVY immunity than by transforming plants with only Pro[s] or Pro[a/s] genes. Furthermore, the immunity was not correlated with the presence of multiple loci or multiple transgenes within a locus and was conferred in some plant lines by a single T-DNA locus containing one Pro[s] gene and one Pro[a/s] gene. This result is contrary to the results expected by using current models. Such models predict that immunity is mediated by the hybridization of the Pro[a/s] or cRNA molecules to the challenging PVY genomic RNA. Therefore, coexpressed Pro[s] and Pro[a/s] mRNAs would be expected to hybridize with each other and the Pro[s]-derived cRNA, thus reducing the amount of free Pro[a/s] mRNA or cRNA available to hybridize with the PVY genome. The increased efficiencies (5- to 10-fold) of the Pro[s] plus Pro[a/s] constructs over simple Pro[s] or Pro[a/s] constructs and the 100% efficiency of Pro[s] plus Pro[a/s] achieved by crossing conflict with this prediction.
In the crossing experiments, plants containing a single Pro[s] gene together with a single Pro[a/s] were immune to PVY, whereas the progeny of their selfed parents or of different Pro[s] × Pro[s] or Pro[a/s] × Pro[a/s] crosses were susceptible to PVY. It seems improbable that one sense and one antisense gene at two different loci are more likely to ectopically pair (and induce PTGS) than two loci of same sense genes. These data, and the lack of silencing obtained by the promoterless GUS panhandle construct, suggest that the PTGS is not mediated by ectopic pairing and that transcription is essential.
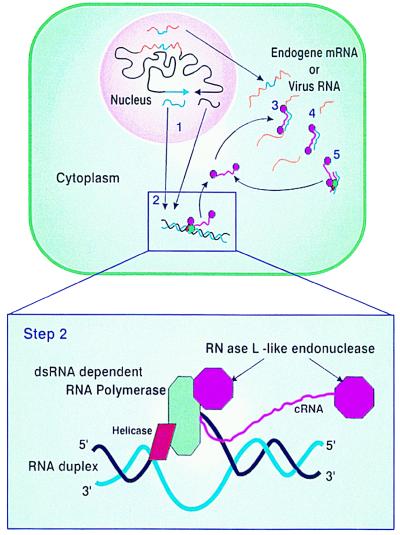
To explain our results and those of others, we propose a model (Fig. 7) that incorporates parts of previously proposed models, most notably that of Dougherty and Parks (19), but has some important modifications. The cornerstone of our model is that PTGS is induced by dsRNA and that this is mediated by an RNA-dependent RNA polymerase, present in the cytoplasm, that requires a dsRNA template. This specificity for dsRNA template is the key to producing sequence-specific degradation. The dsRNA is formed by hybridization of complementary transgene mRNAs or complementary regions of the same transcript. In our experiments, these RNAs were deliberately designed, but in cases of cosuppression or antisense with single sense transgenes, we suggest that multicopy transgenes are required in an inverted repeat arrangement so that transcriptional read-through will produce mRNAs with self-complementarity. In rare cases, dsRNA could also be produced by a single transgene being integrated into the plant genome such that its 3′ end is adjacent to an endogenous promoter, thus producing an antisense mRNA that could hybridize with the sense transgene mRNA. This would explain the characteristics of 35S Pro-immune plant 2 (Table 2). The dsRNA-dependent RNA polymerase produces cRNA to which RNase molecules are attached. These cRNA-RNase molecules hybridize to the endogene mRNA or viral RNA and cleave the single-stranded RNA adjacent to the hybrid. The cleaved single-stranded RNAs are further degraded by other host RNases because one will lack a capped 5′ end and the other will lack a poly(A) tail. If the transgene is derived from a viral sequence, this will provide sequence-specific cleavage of viral genomic and subgenomic RNAs, thus preventing viral infection. If the transgene is derived from an endogene, this will lead to cleavage and degradation of the endogene mRNA. The model predicts that the dsRNA formed by the cRNA and the target RNA will remain and act as a template for the dsRNA-dependent RNA polymerase to generate more cRNA. We detected accumulation of sense and antisense RNAs with the potential to form dsRNAs in our PVY immune plants. Similar molecules were identified by Metzlaff et al. (20) in plants with cosuppressed chalcone synthase. The RNase in our model may be something akin to RNase L, which is a latent single-stranded RNA endonuclease that is endogenous and ubiquitous in mammalian cells and activated by 5′-phosphorylated 2′-5′-linked oligoadenylates. It has been shown that this RNase can be directed to give site-specific cleavage by conjugation to an antisense oligonucleotide (26). A variation of our model could replace the RNase L-like endonuclease with a dsRNase. This enzyme would not require direct attachment to the cRNA and would specifically degrade the RNA/RNA duplex formed by the cRNA and its target. This would degrade the target RNA, causing PTGS or viral immunity. However, it would also remove the dsRNA from acting as a template for further cRNA production; thus, the mechanism would not be self-perpetuating. We suggest that the dsRNA-dependent RNA polymerase in our model is the same as that found in healthy plants (6, 7, 27, 28). The highly purified RNA-dependent RNA polymerase of Schiebel et al. (6, 7) from healthy plants was reported to be active on single-stranded RNA but not on dsRNA templates in vitro. We suggest that this enzyme is associated with a helicase in vivo that was lost during purification because the less purified RNA-dependent RNA polymerase preparations of Ikegami and Fraenkel-Conrat (27) showed activity with dsRNA templates.
Figure 7.
A model for dsRNA-induced PTGS. The transgenes transcribe both sense and antisense mRNA (step 1), which are exported to the cytoplasm where they hybridize to form a duplex. The duplex is recognized by a complex of dsRNA-dependent RNA polymerase, associated helicase, and RNase L-like molecules (step 2). The complex transcribes cRNA, attaches to it RNase L-like molecules, and releases the cRNA. The cRNA-RNase molecules hybridize to the target endogene or virus RNAs (step 3) and cleave the single-stranded regions adjacent to the hybrids (step 4), which are then degraded by other plant nucleases. This silences the expression of the endogene or provides virus immunity. The remaining RNA duplex is resistant to nonspecific degradation and acts as a template for synthesis of new cRNA-RNase molecules (step 5). Once the cycle is initiated, there may be no further requirement for nuclear-encoded dsRNA. For clarity the model shows cRNA being produced from one strand of the dsRNA duplex, whereas the complex is capable of producing cRNAs from either strand.
Our model provides a mechanism by which dsRNA can direct sequence-specific degradation of RNAs in the cytoplasm. In our preferred version (incorporating and RNase L-like endonuclease), the mechanism, once initiated, will perpetuate and possibly spread independently of nuclear transcription of the transgene. This prediction is compatible with the results of the study of Fire et al. (29) in which a single injection of dsRNA into a nematode directed PTGS, which was then able to spread and perpetuate throughout its body and even into its progeny, presumably by cytoplasmic maternal inheritance. It is also compatible with the recent report that a PTGS signal was graft transmissible in plants (28). However, our model predicts that PTGS should be more readily induced by using simple antisense constructs than is commonly observed. Perhaps, a threshold of dsRNA is needed to induce the system, and this is often not met because the target mRNA is expressed at too low a level or in a different cellular compartment from the antisense RNA.
Irrespective of the mechanism, delivery of RNAs with the potential to form duplexes may be an important new strategy for virus resistance and gene silencing in transgenic plants.
Acknowledgments
We thank Neil Smith for his enthusiasm, dedication, and exceptional technical assistance. We also thank Geoff Ellacott, for performing some of the tobacco transformations, and Jean Finnegan, Liz Dennis, Paul Keese, and Richard Forster for helpful discussions.
ABBREVIATION
- Pro
protease
- PTGS
posttranscriptional gene silencing
- ds
double-stranded
- PVY
potato virus Y
- GUS
β-glucuronidase
Footnotes
A Commentary on this article begins on page 13349.
References
- 1.Powell P A, Sanders P R, Tumer N, Fraley R T, Beachy R N. Virology. 1990;175:124–130. doi: 10.1016/0042-6822(90)90192-t. [DOI] [PubMed] [Google Scholar]
- 2.Lindbo J A, Dougherty W G. Mol Plant–Microbe Interact. 1992;5:144–153. doi: 10.1094/mpmi-5-144. [DOI] [PubMed] [Google Scholar]
- 3.Lindbo J A, Dougherty W G. Virology. 1992;189:725–733. doi: 10.1016/0042-6822(92)90595-g. [DOI] [PubMed] [Google Scholar]
- 4.Baulcombe D C. Plant Cell. 1996;8:1833–1844. doi: 10.1105/tpc.8.10.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stam M, Mol J N M, Kooter J M. Ann Bot. 1997;79:3–12. [Google Scholar]
- 6.Schiebel W, Haas B, Marinkovic S, Klanner A, Sanger H L. J Biol Chem. 1993;268:11851–11857. [PubMed] [Google Scholar]
- 7.Schiebel W, Haas B, Marinkovic S, Klanner A, Sanger H L. J Biol Chem. 1993;268:11858–11867. [PubMed] [Google Scholar]
- 8.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 9.Gleave A P. Plant Mol Biol. 1992;20:1203–1207. doi: 10.1007/BF00028910. [DOI] [PubMed] [Google Scholar]
- 10.Wang M B, Matthews P R, Upadhyaya M N, Waterhouse P M. Acta Hortic. 1998;461:401–405. [Google Scholar]
- 11.Commonwealth Scientific and Industrial Research Organisation and Australian National University (1998) Australian Patent 33,359/95.
- 12.Boevink P, Chu P W G, Keese P. Virology. 1995;207:354–361. doi: 10.1006/viro.1995.1094. [DOI] [PubMed] [Google Scholar]
- 13.Zheng Z, Hayashimoto A, Li Z, Murai N. Plant Physiol. 1991;97:832–835. doi: 10.1104/pp.97.2.832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellis J G, Llewellyn D J, Dennis E S, Peacock W J. EMBO J. 1987;6:11–16. doi: 10.1002/j.1460-2075.1987.tb04711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang M B, Upadhyaya M N, Brettel R I S, Waterhouse P M. J Gen Breed. 1997;51:325–334. [Google Scholar]
- 16.McAlister F M, Jenkins C L D, Watson J M. Aust J Plant Physiol. 1998;25:225–235. [Google Scholar]
- 17.Jefferson R A, Kavanagh T A, Bevan M W. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sijen T, Wellink J, Hiriart J B, Van Kammen A. Plant Cell. 1996;8:2277–2294. doi: 10.1105/tpc.8.12.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dougherty W G, Parks T D. Curr Opin Cell Biol. 1995;7:399–405. doi: 10.1016/0955-0674(95)80096-4. [DOI] [PubMed] [Google Scholar]
- 20.Metzlaff M, O’Dell M, Cluster P D, Flavell R B. Cell. 1997;88:845–854. doi: 10.1016/s0092-8674(00)81930-3. [DOI] [PubMed] [Google Scholar]
- 21.Van Blokland R, Van der Geest N, Mol J N M, Kooter J M. Plant J. 1994;6:861–877. [Google Scholar]
- 22.Hobbs S L A, Warkentin T D, Delong C M O. Plant Mol Biol. 1993;15:851–864. doi: 10.1007/BF00039425. [DOI] [PubMed] [Google Scholar]
- 23.Dehio C, Schell J. Proc Natl Acad Sci USA. 1994;91:5538–5542. doi: 10.1073/pnas.91.12.5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ingelbrecht I, Van Houdt H, Van Montagu M, Depicker A. Proc Natl Acad Sci USA. 1994;91:10502–10506. doi: 10.1073/pnas.91.22.10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.English J J, Mueller E, Baulcombe D C. Plant Cell. 1996;8:179–188. doi: 10.1105/tpc.8.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Torrence P F, Maitra R K, Lesiak K, Khamnei S, Zhou A, Silverman R H. Proc Natl Acad Sci USA. 1993;90:1300–1304. doi: 10.1073/pnas.90.4.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ikegami M, Fraenkel-Conrat H. J Biol Chem. 1979;254:149–154. [PubMed] [Google Scholar]
- 28.Palauqui J C, Elmayan T, Pollien J-M, Vaucheret H. EMBO J. 1998;16:4738–4745. doi: 10.1093/emboj/16.15.4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fire A, Xu S-Q, Montgomery M K, Kostas S A, Driver S E, Mello C C. Nature (London) 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]