Abstract
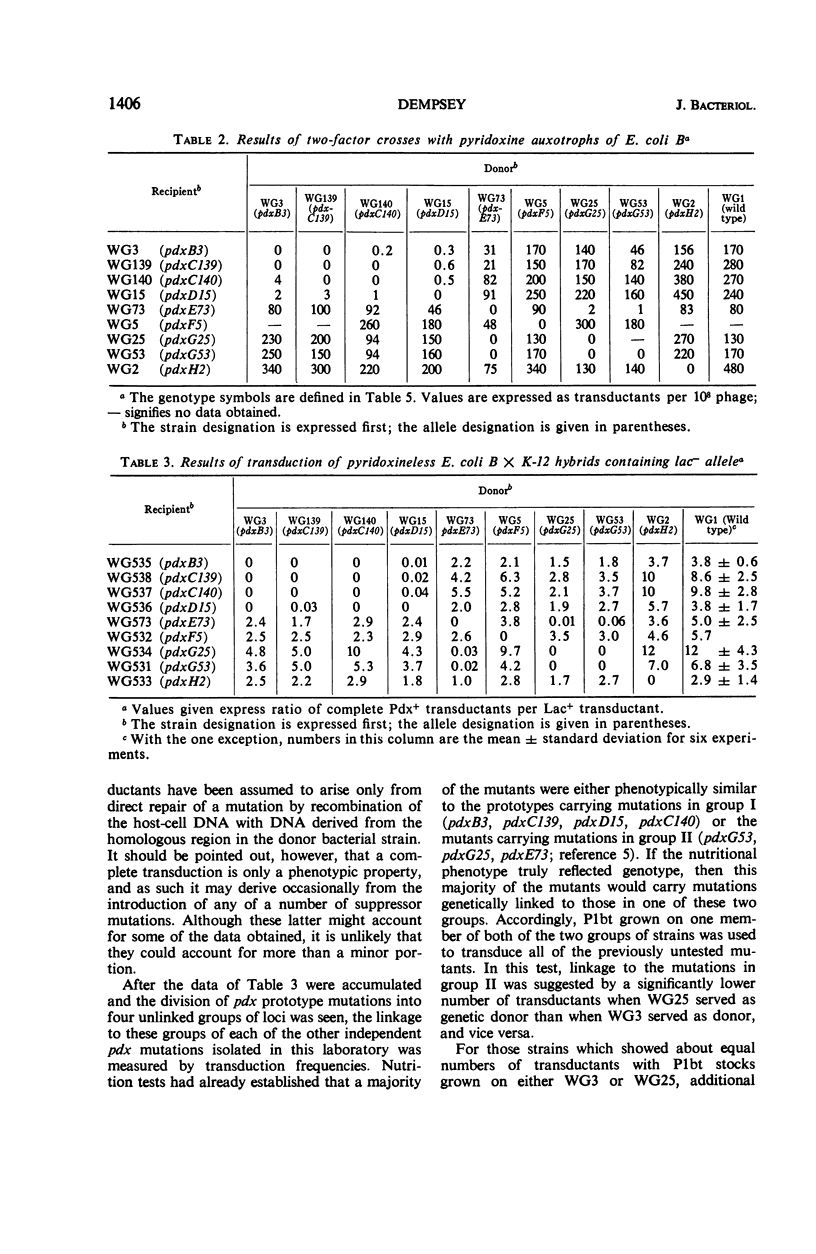
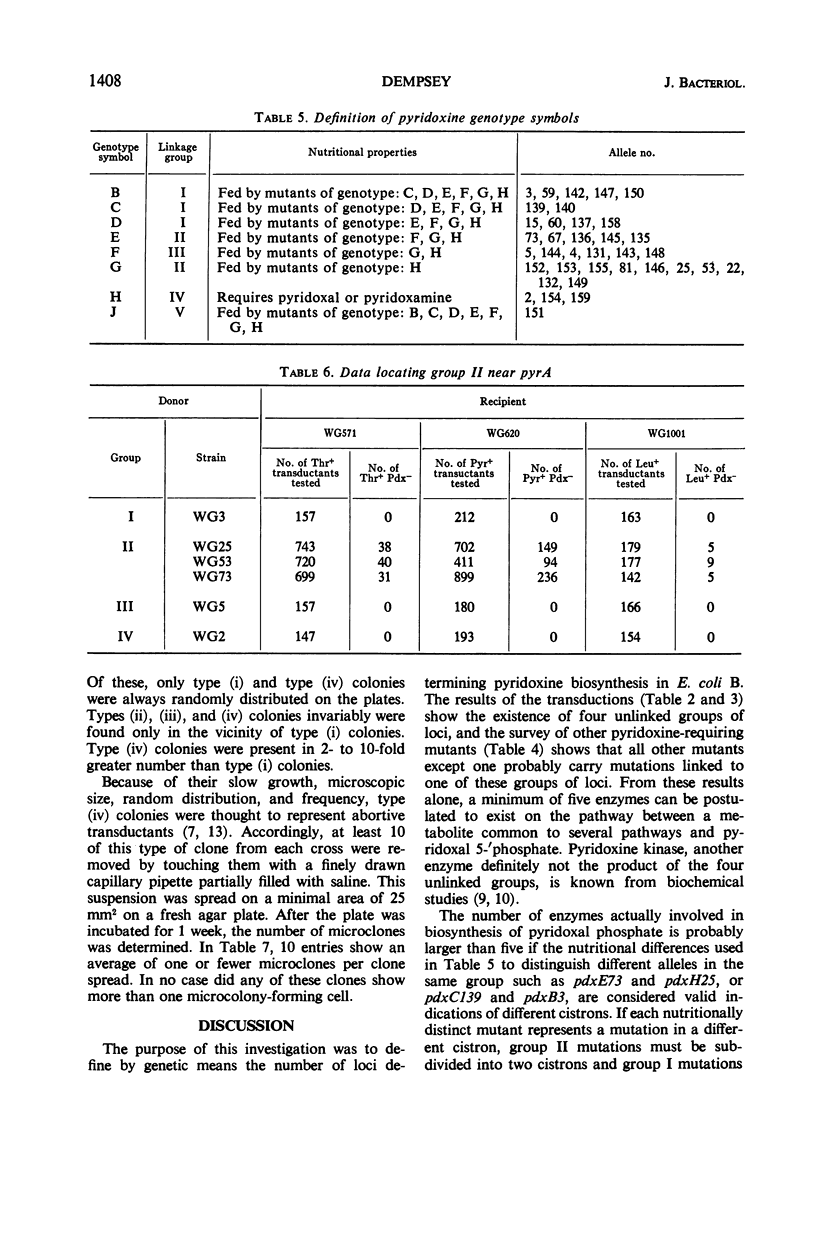
Pyridoxine mutants of Escherichia coli B, previously divided into a minimum of six groups by cross-feeding tests, were characterized by transduction studies performed with phage P1bt. The results of these studies allowed division of pyridoxine mutants into five unlinked groups and set the minimum number of enzymes between pyridoxal phosphate and a metabolite common to other pathways at six or seven, with the probable maximum at ten. One group was shown to be linked to thr, leu, and pyrA.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BERTANI G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951 Sep;62(3):293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demerec M., Adelberg E. A., Clark A. J., Hartman P. E. A proposal for a uniform nomenclature in bacterial genetics. J Gen Microbiol. 1968 Jan;50(1):1–14. doi: 10.1099/00221287-50-1-1. [DOI] [PubMed] [Google Scholar]
- Dempsey W. B., Pachler P. F. Isolation and characterization of pyridoxine auxotrophs of Escherichia coli. J Bacteriol. 1966 Feb;91(2):642–645. doi: 10.1128/jb.91.2.642-645.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey W. B. Synthesis of Pyridoxine by a Pyridoxal Auxotroph of Escherichia coli. J Bacteriol. 1966 Aug;92(2):333–337. doi: 10.1128/jb.92.2.333-337.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GROSS J., ENGLESBERG E. Determination of the order of mutational sites governing L-arabinose utilization in Escherichia coli B/r bv transduction with phage Plbt. Virology. 1959 Nov;9:314–331. doi: 10.1016/0042-6822(59)90125-4. [DOI] [PubMed] [Google Scholar]
- HARTMAN P. E., HARTMAN Z., SERMAN D. Complementation mapping by abortive transduction of histidine requiring Salmonella mutants. J Gen Microbiol. 1960 Apr;22:354–368. doi: 10.1099/00221287-22-2-354. [DOI] [PubMed] [Google Scholar]
- HURWITZ J. The enzymatic phosphorylation of pyridoxal. J Biol Chem. 1953 Dec;205(2):935–947. [PubMed] [Google Scholar]
- Helling R. B. The effect of arabinose-specific enzyme synthesis on recombination in the arabinose genes of Escherichia coli. Genetics. 1967 Nov;57(3):665–675. doi: 10.1093/genetics/57.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny K. V., Dempsey W. B. Phosphorylation of pyridoxine by Escherichia coli B. Nature. 1967 Feb 25;213(5078):830–830. doi: 10.1038/213830b0. [DOI] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- LURIA S. E., ADAMS J. N., TING R. C. Transduction of lactose-utilizing ability among strains of E. coli and S. dysenteriae and the properties of the transducing phage particles. Virology. 1960 Nov;12:348–390. doi: 10.1016/0042-6822(60)90161-6. [DOI] [PubMed] [Google Scholar]
- SWANSTROM M., ADAMS M. H. Agar layer method for production of high titer phage stocks. Proc Soc Exp Biol Med. 1951 Nov;78(2):372–375. doi: 10.3181/00379727-78-19076. [DOI] [PubMed] [Google Scholar]
- Taylor A. L., Trotter C. D. Revised linkage map of Escherichia coli. Bacteriol Rev. 1967 Dec;31(4):332–353. doi: 10.1128/br.31.4.332-353.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]


