Abstract
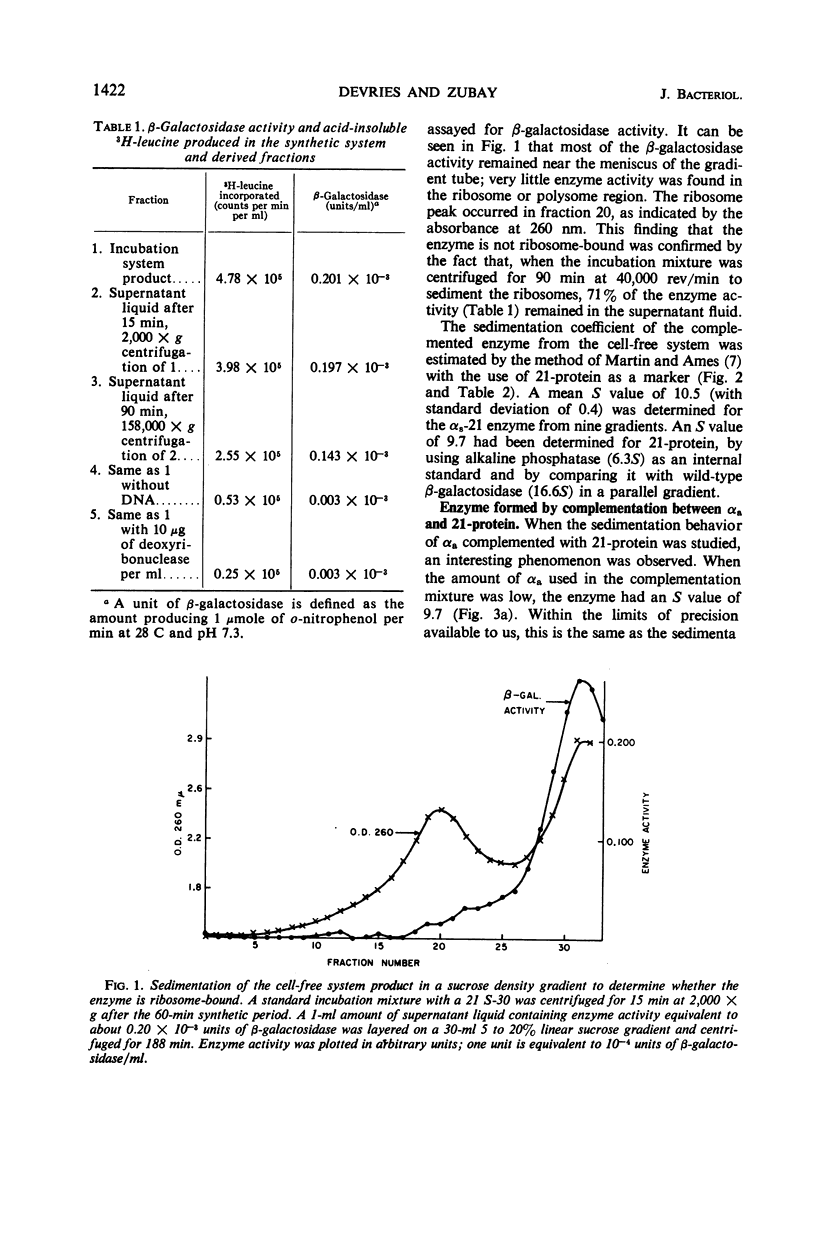
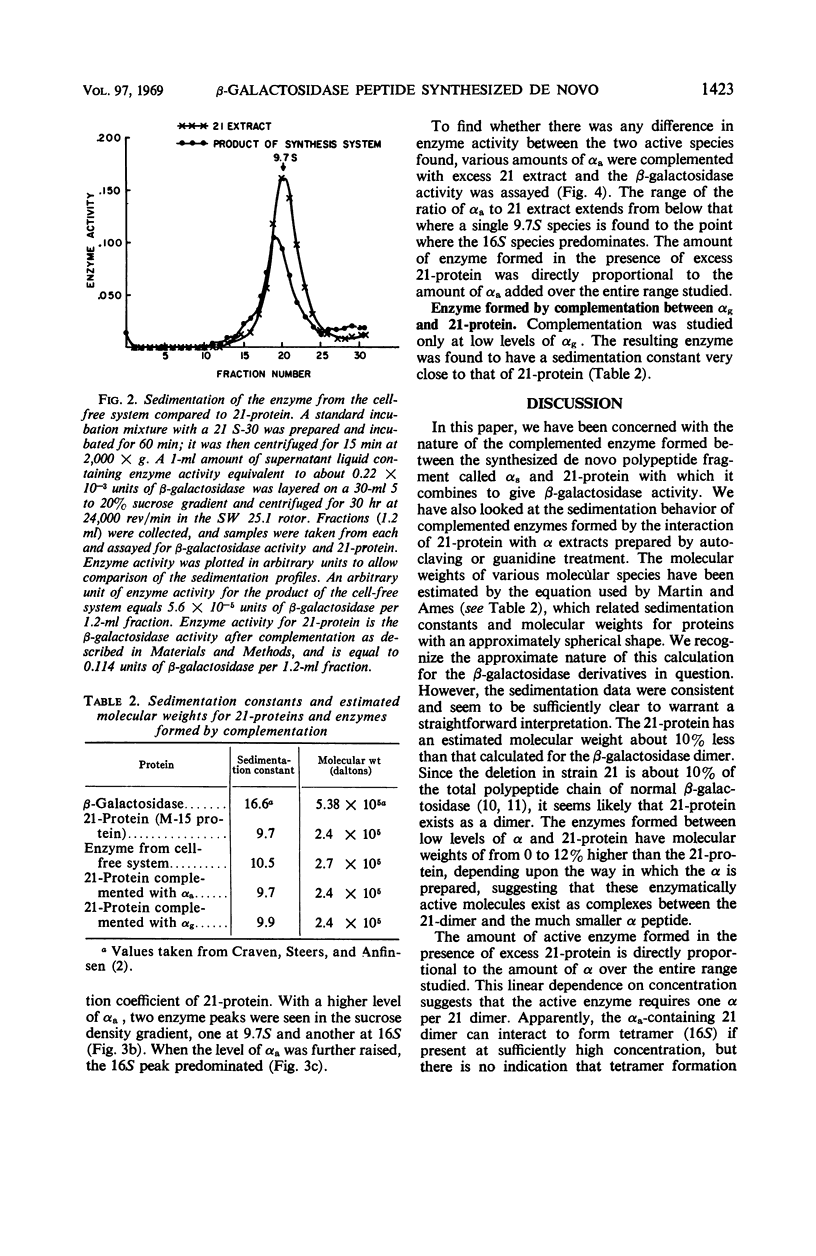
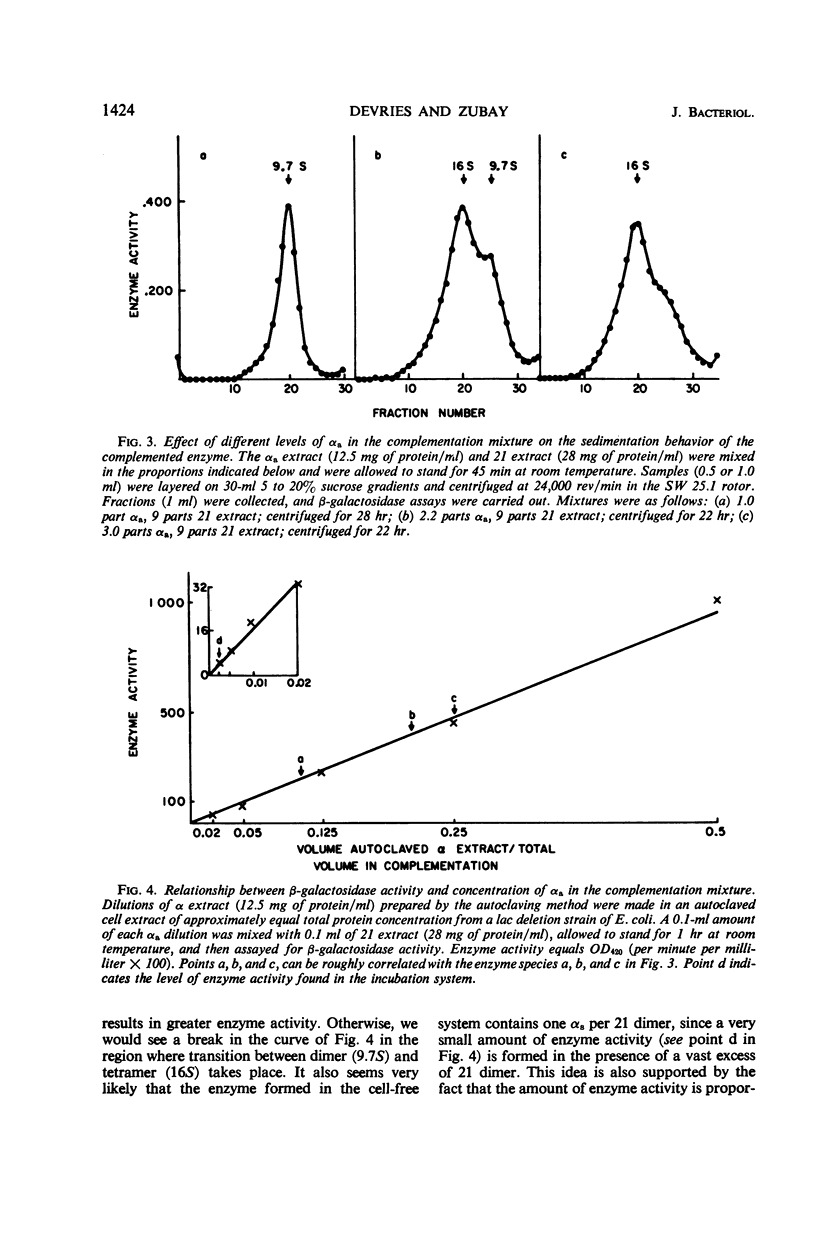
In a cell-free system, φ80dlac can be transcribed, and the resulting ribonucleic acid can be translated to yield a product which interacts with an enzymatically inactive z protein to produce active enzyme. The inactive z protein is produced by Escherichia coli strain 21, which contains a deletion in the first part of the gene for β-galactosidase and appears to exist as a dimer. The enzyme formed in the cell-free system appears to be composed of one strain 21 z protein dimer and one newly synthesized polypeptide chain with a molecular weight of about 3 × 104. The estimated size of this complementing segment is in good agreement with Ullmann, Jacob, and Monod's estimate of the size of the α region of β-galactosidase. Using α fragments produced by autoclaving or guanidine treatments, we found that the active portion of α seems to be smaller than the full α region. We also found, using α produced by the autoclaving technique, that active dimer undergoes conversion to tetramer as the amount of α is increased. Evidently, the binding of α favors this conversion, but it is unlikely that the conversion of dimer to tetramer per se results in increased enzyme activity.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BYRNE R., LEVIN J. G., BLADEN H. A., NIRENBERG M. W. THE IN VITRO FORMATION OF A DNA-RIBOSOME COMPLEX. Proc Natl Acad Sci U S A. 1964 Jul;52:140–148. doi: 10.1073/pnas.52.1.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CRAVEN G. R., STEERS E., Jr, ANFINSEN C. B. PURIFICATION, COMPOSITION, AND MOLECULAR WEIGHT OF THE BETA-GALACTOSIDASE OF ESCHERICHIA COLI K12. J Biol Chem. 1965 Jun;240:2468–2477. [PubMed] [Google Scholar]
- DeVries J. K., Zubay G. DNA-directed peptide synthesis. II. The synthesis of the alpha-fragment of the enzyme beta-galactosidase. Proc Natl Acad Sci U S A. 1967 Apr;57(4):1010–1012. doi: 10.1073/pnas.57.4.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederman M., Zubay G. DNA directed peptide synthesis. V. The cell-free synthesis of a polypeptide with beta-galactosidase activity. Biochem Biophys Res Commun. 1968 Aug 21;32(4):710–714. doi: 10.1016/0006-291x(68)90297-0. [DOI] [PubMed] [Google Scholar]
- Lederman M., Zubay G. DNA-directed peptide synthesis. 1. A comparison of T2 and Escherichi coli DNA-directed peptide synthesis in two cell-free systems. Biochim Biophys Acta. 1967 Nov 21;149(1):253–258. doi: 10.1016/0005-2787(67)90706-x. [DOI] [PubMed] [Google Scholar]
- MARTIN R. G., AMES B. N. A method for determining the sedimentation behavior of enzymes: application to protein mixtures. J Biol Chem. 1961 May;236:1372–1379. [PubMed] [Google Scholar]
- OYAMA V. I., EAGLE H. Measurement of cell growth in tissue culture with a phenol reagent (folin-ciocalteau). Proc Soc Exp Biol Med. 1956 Feb;91(2):305–307. doi: 10.3181/00379727-91-22245. [DOI] [PubMed] [Google Scholar]
- Ullmann A., Jacob F., Monod J. Characterization by in vitro complementation of a peptide corresponding to an operator-proximal segment of the beta-galactosidase structural gene of Escherichia coli. J Mol Biol. 1967 Mar 14;24(2):339–343. doi: 10.1016/0022-2836(67)90341-5. [DOI] [PubMed] [Google Scholar]
- Ullmann A., Jacob F., Monod J. On the subunit structure of wild-type versus complemented beta-galactosidase of Escherichia coli. J Mol Biol. 1968 Feb 28;32(1):1–13. doi: 10.1016/0022-2836(68)90140-x. [DOI] [PubMed] [Google Scholar]
- Zubay G., Lederman M., DeVries J. K. DNA-directed peptide synthesis. 3. Repression of beta-galactosidase synthesis and inhibition of repressor by inducer in a cell-free system. Proc Natl Acad Sci U S A. 1967 Oct;58(4):1669–1675. doi: 10.1073/pnas.58.4.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]


