Abstract
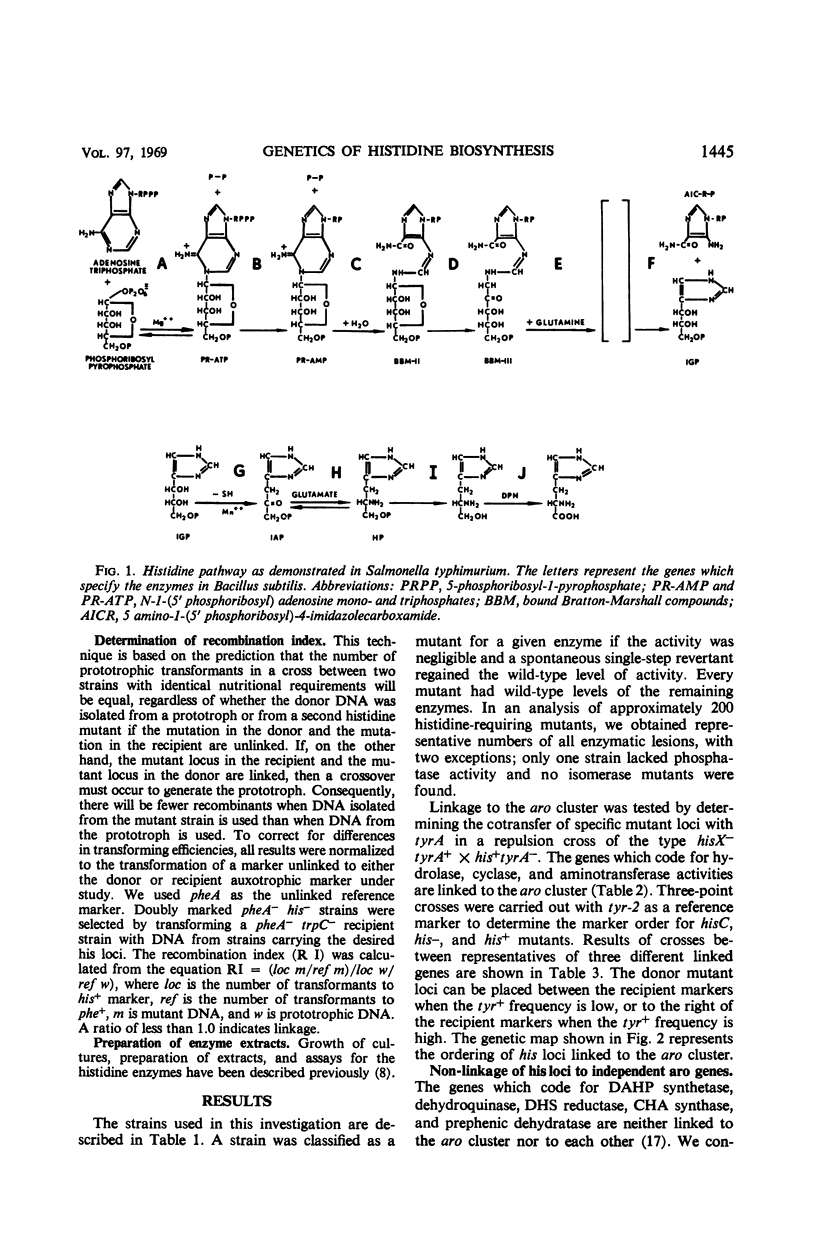
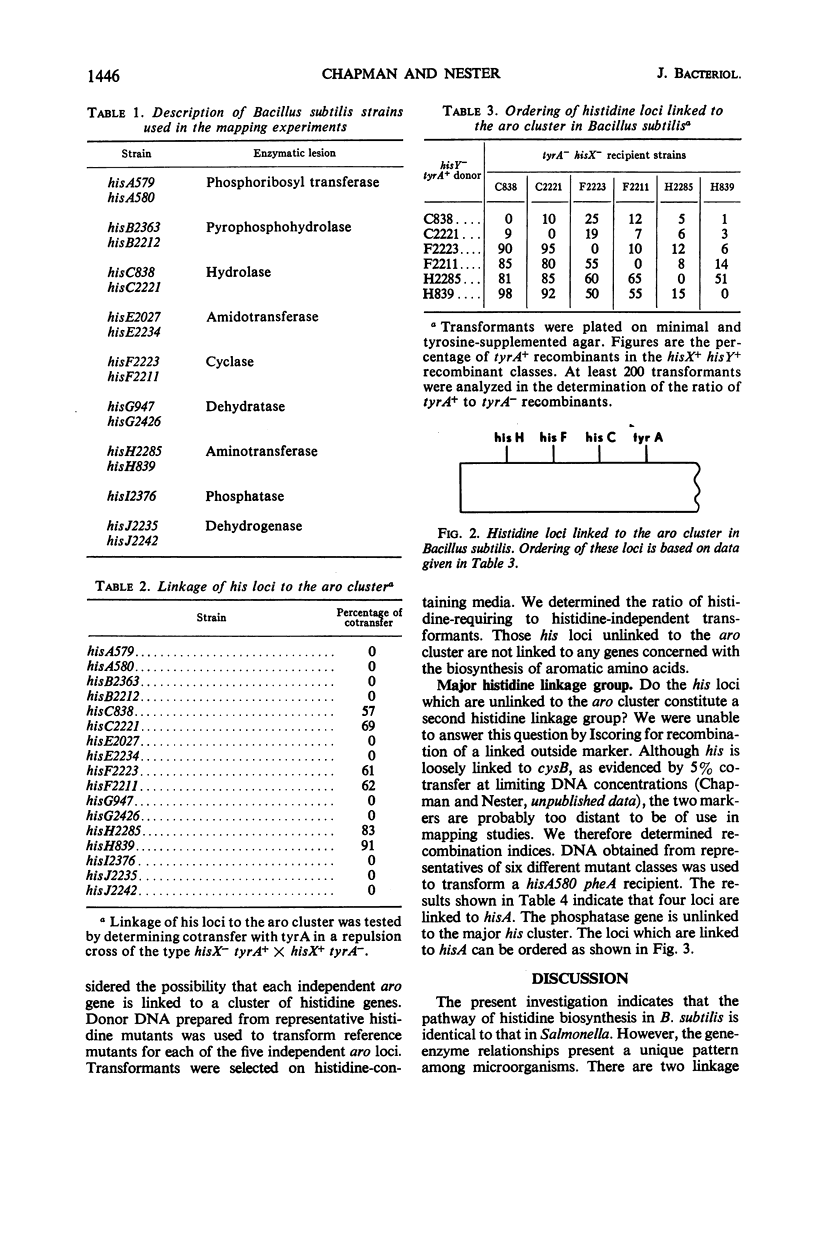
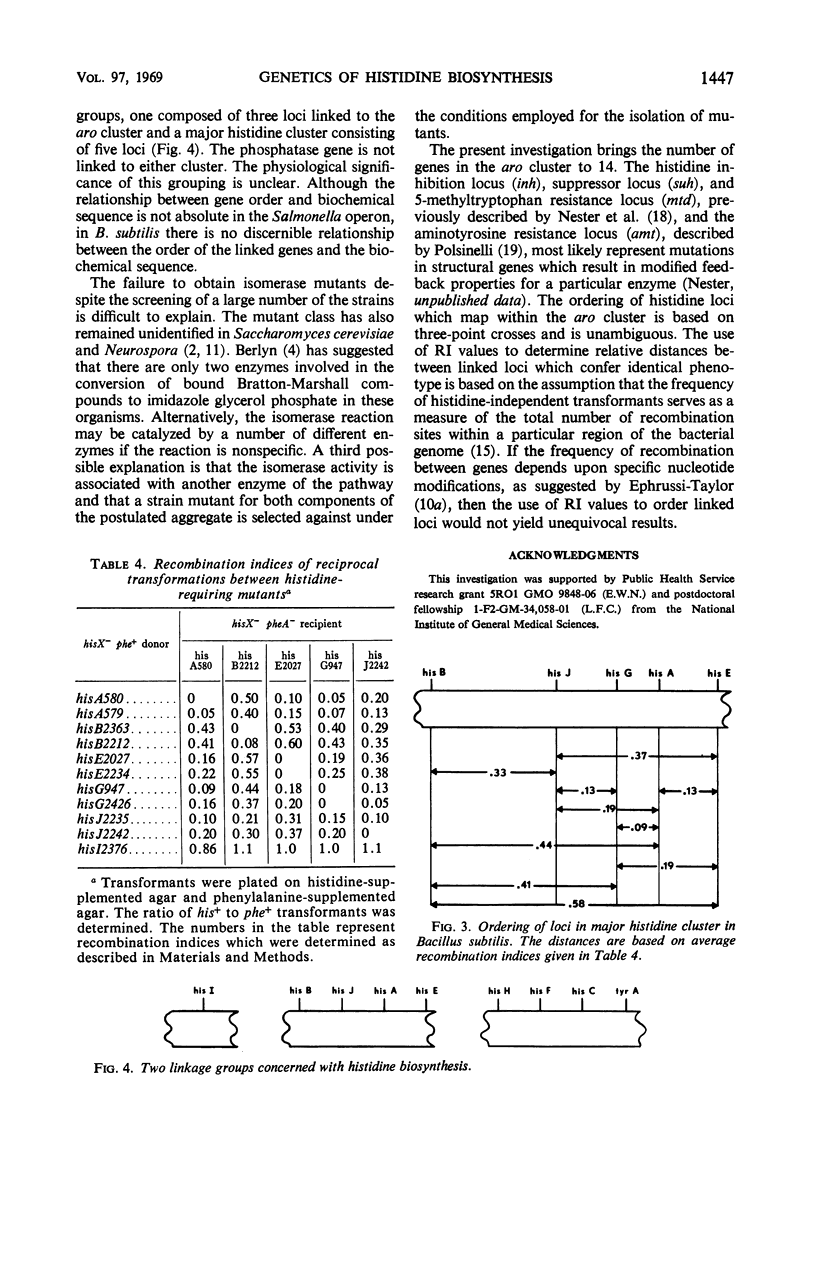
Mutants for 9 of the 10 steps in histidine biosynthesis have been isolated and identified by enzyme assay. Each locus has been mapped in relation to the aro cluster and to other histidine loci by deoxyribonucleic acid-mediated transformation. The genes which code for enzymes 3, 6, and 8 of the pathway are linked to the aro cluster. A major histidine linkage group is composed of the genes which specify enzymes 1, 2, 5, 7, and 10. The locus which codes for step 9 of the pathway is unlinked to any other identified his loci. The major histidine cluster is loosely linked to cysB and is unlinked to any of the loci concerned with aromatic amino acid biosynthesis.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- AHMED A., CASE M. E., GILES N. H. THE NATURE OF COMPLEMENTATION AMONG MUTANTS IN THE HISTIDINE-3 REGION OF NEUROSPORA CRASSA. Brookhaven Symp Biol. 1964 Dec;17:53–65. [PubMed] [Google Scholar]
- BURTON K. A study of the conditions and mechanism of the diphenylamine reaction for the colorimetric estimation of deoxyribonucleic acid. Biochem J. 1956 Feb;62(2):315–323. doi: 10.1042/bj0620315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlyn M. B. Gene-enzyme relationships in histidine biosynthesis in Aspergillus nidulans. Genetics. 1967 Nov;57(3):561–570. doi: 10.1093/genetics/57.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton B. C. Fine-structure mapping by transformation in the tryptophan region of Bacillus subtilis. J Bacteriol. 1966 May;91(5):1795–1803. doi: 10.1128/jb.91.5.1795-1803.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton B. C. Transformation mapping of the genes controlling tryptophan biosynthesis in Bacillus subtilis. J Bacteriol. 1967 Sep;94(3):660–665. doi: 10.1128/jb.94.3.660-665.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman L. F., Nester E. W. Common element in the repression control of enzymes of histidine and aromatic amino acid biosynthesis in Bacillus subtilus. J Bacteriol. 1968 Nov;96(5):1658–1663. doi: 10.1128/jb.96.5.1658-1663.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D., Smith I., Marmur J. Gene conservation in Bacillus species. II. The location of genes concerned with the synthesis of ribosomal components and soluble RNA. Proc Natl Acad Sci U S A. 1965 Sep;54(3):724–730. doi: 10.1073/pnas.54.3.724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EPHRATI-ELIZUR E., SRINIVASAN P. R., ZAMENHOF S. Genetic analysis, by means of transformation, of histidine linkage groups in Bacillus subtilis. Proc Natl Acad Sci U S A. 1961 Jan 15;47:56–63. doi: 10.1073/pnas.47.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FINK G. R. GENE-ENZYME RELATIONS IN HISTIDINE BIOSYNTHESIS IN YEAST. Science. 1964 Oct 23;146(3643):525–527. doi: 10.1126/science.146.3643.525. [DOI] [PubMed] [Google Scholar]
- Fink G. R. A cluster of genes controlling three enzymes in histidine biosynthesis in Saccharomyces cerevisiae. Genetics. 1966 Mar;53(3):445–459. doi: 10.1093/genetics/53.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KLOOS W. E., PATTEE P. A. A BIOCHEMICAL CHARACTERIZATION OF HISTIDINE-DEPENDENT MUTANTS OF STAPHYLOCOCCUS AUREUS. J Gen Microbiol. 1965 May;39:185–194. doi: 10.1099/00221287-39-2-185. [DOI] [PubMed] [Google Scholar]
- KLOSS W. E., PATTEE P. A. TRANSDUCTION ANALYSIS OF THE HISTIDINE REGION IN STAPHYLOCOCCUS AUREUS. J Gen Microbiol. 1965 May;39:195–207. doi: 10.1099/00221287-39-2-195. [DOI] [PubMed] [Google Scholar]
- LACKS S., HOTCHKISS R. D. A study of the genetic material determining an enzyme in Pneumococcus. Biochim Biophys Acta. 1960 Apr 22;39:508–518. doi: 10.1016/0006-3002(60)90205-5. [DOI] [PubMed] [Google Scholar]
- Mee B. J., Lee B. T. An analysis of histidine requiring mutants in Pseudomonas aeruginosa. Genetics. 1967 Apr;55(4):709–722. doi: 10.1093/genetics/55.4.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NESTER E. W., LEDERBERG J. Linkage of genetic units of Bacillus subtilis in DNA transformation. Proc Natl Acad Sci U S A. 1961 Jan 15;47:52–55. doi: 10.1073/pnas.47.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasser D., Nester E. W. Aromatic amino acid biosynthesis: gene-enzyme relationships in Bacillus subtilis. J Bacteriol. 1967 Nov;94(5):1706–1714. doi: 10.1128/jb.94.5.1706-1714.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nester E W, Schafer M, Lederberg J. Gene Linkage in DNA Transfer: A Cluster of Genes Concerned with Aromatic Biosynthesis in Bacillus Subtilis. Genetics. 1963 Apr;48(4):529–551. doi: 10.1093/genetics/48.4.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spizizen J. TRANSFORMATION OF BIOCHEMICALLY DEFICIENT STRAINS OF BACILLUS SUBTILIS BY DEOXYRIBONUCLEATE. Proc Natl Acad Sci U S A. 1958 Oct 15;44(10):1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tingle M., Herman A., Halvorson H. O. Characterization and mapping of histidine genes in Saccharomyces lactis. Genetics. 1968 Mar;58(3):361–371. doi: 10.1093/genetics/58.3.361. [DOI] [PMC free article] [PubMed] [Google Scholar]


