Abstract
PII is a protein allosteric effector in Escherichia coli and other bacteria that indirectly regulates glutamine synthetase at the transcriptional and post-translational levels in response to nitrogen availability. Data supporting the notion that plants have a nitrogen regulatory system(s) includes previous studies showing that the levels of mRNA for plant nitrogen assimilatory genes such as glutamine synthetase (GLN) and asparagine synthetase (ASN) are modulated by carbon and organic nitrogen metabolites. Here, we have characterized a PII homolog (GLB1) in two higher plants, Arabidopsis thaliana and Ricinus communis (Castor bean). Each plant PII-like protein has high overall identity to E. coli PII (50%). Western blot analyses reveal that the plant PII-like protein is a nuclear-encoded chloroplast protein. The PII-like protein of plants appears to be regulated at the transcriptional level in that levels of GLB1 mRNA are affected by light and metabolites. To initiate studies of the in vivo function of the Arabidopsis PII-like protein, we have constructed transgenic lines in which PII expression is uncoupled from its native regulation. Analyses of these transgenic plants support the notion that the plant PII-like protein may serve as part of a complex signal transduction network involved in perceiving the status of carbon and organic nitrogen. Thus, the PII protein found in archaea, bacteria, and now in higher eukaryotes (plants) is one of the most widespread regulatory proteins known, providing evidence for an ancestral metabolic regulatory mechanism that may have existed before the divergence of these three domains of life.
As the pathways of carbon and nitrogen metabolism are highly coordinated in higher plants, it is likely that crosstalk between signals derived from carbon and nitrogen metabolites may exist in plants, as they do in bacteria. In plants, supplementation of growth media with sucrose and organic nitrogen can cause changes in the expression of some nitrogen assimilatory genes, such as those encoding the enzymes nitrate reductase (NR) (1, 2), glutamine synthetase (GLN) (2–4), and asparagine synthetase (ASN) (4, 5). Whereas mechanisms for metabolite sensing are known in bacteria, the means by which these signals are sensed in plants have yet to be discovered. The ability of plants to sense changes in the levels of carbon and nitrogen metabolites is reminiscent of a nitrogen regulatory system (Ntr) in Escherichia coli. A component of the Ntr system in E. coli is a protein called PII (encoded by the glnB gene), which interacts with other regulatory proteins to regulate the activity of the glutamine synthetase (GS) at the post-translational level and the transcription of the gene encoding GS (glnA) (6–8). Under nitrogen-limiting conditions, the PII protein is uridylylated by uridylyltransferase and the PII-UMP then interacts with an adenylyltransferase to deadenylylate and activate the GS enzyme (6, 7). By contrast, under conditions of nitrogen excess, PII-UMP is deuridylylated allowing it to interact with adenylyltransferase to cause the adenylylation and inactivation of the GS enzyme. PII can also interact with a two-component system (NtrB/NtrC) to decrease glnA transcription whereas PII-UMP appears to have no effect (7–13). Thus, the protein allosteric effector PII works in concert with other proteins, including uridylyltransferase, adenylyltransferase, NtrB, and NtrC, to regulate E. coli GS at multiple levels in response to the status of carbon and nitrogen metabolites (6–15).
Here, we report the identification and characterization of a plant gene GLB1 encoding a PII homolog found in Castor bean and Arabidopsis. Before this report, the only glnB gene identified in a eukaryote was shown to be bacterial origin because it was located on the chloroplast genome of a red alga, Porphyra purpurea (16). We demonstrate that the Arabidopsis GLB1 gene is nuclear whereas the PII-like protein it encodes is chloroplast-localized. Levels of GLB1 mRNA are regulated by light and metabolites. To test the in vivo function of the Arabidopsis PII-like protein, we created transgenic plants that express GLB1 constitutively. Analysis of these PII overproducing Arabidopsis supports the notion that the plant PII-like protein may be one of the components involved in signaling the status of carbon and organic nitrogen in higher plants.
MATERIALS AND METHODS
Plant Materials.
The plant tissues used in all experiments were from A. thaliana Columbia ecotype. Plants were grown on MS agar plates [MS salts (GIBCO/BRL), pH adjusted to 5.7 with potassium hydroxide, 0.8% (wt/vol) agar] containing 3% sucrose. The concentrations of Asn, Gln, and Glu supplied in the media were as previously described (5). MS basal medium without a nitrogen source used in Fig. 6 was from GIBCO/BRL (formula no. 97–5068EC). Plants were grown in EGC growth chambers at 45–60 μE m−2 sec−1 on a 16-hr light/8-hr dark cycle, unless otherwise noted.
Figure 6.
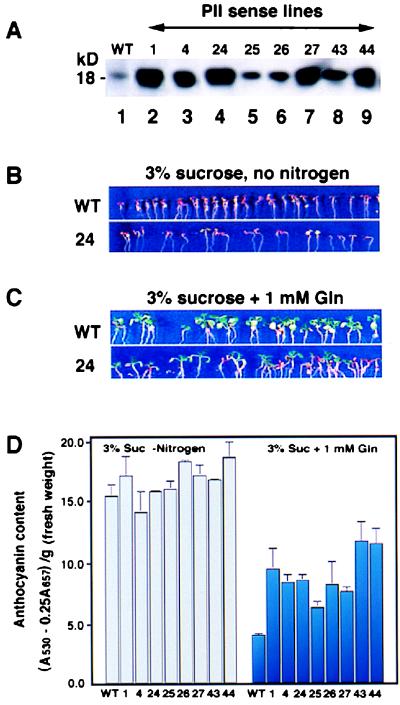
Phenotypic analysis of Arabidopsis PII overexpressors. (A) Western blot analysis of 50 μg of total proteins extracted from wild-type (WT, lane 1) or PII overexpressors (T3 homozygous lines 1, 4, 24, 25, 26, 27, 43, and 44; lanes 2–9). (B) The 10-day-old Arabidopsis wild-type (WT) and the representative PII overexpressor (line 24) grown on MS basal medium containing 3% sucrose without a nitrogen source accumulate high levels of anthocyanin. (C) Difference in accumulation of anthocyanin between 10-day-old Arabidopsis WT and PII-24 overexpressor. Plants were grown on MS basal medium containing 3% sucrose and 1 mM Gln as the only nitrogen source. (D) Anthocyanin content of WT and PII overexpressors (lines 1, 4, 24, 25, 26, 27, 43, and 44) grown on MS basal medium containing 3% sucrose, no nitrogen (gray bars) or on 3% sucrose plus 1 mM glutamine as the only nitrogen source (blue bars). Results are shown as means ± SEM from anthocyanin determination of four independent pools of 30 seedlings/bar.
Isolation of Castor Bean and Arabidopsis PII cDNA Clones.
A λZAPII library containing cDNAs from developing endosperm and embryos of castor bean (Ricinus communis) seeds was used for mass sequencing (17). One of the clones sequenced showed similarity to the bacterial nitrogen-regulatory protein PII. The GLB1 cDNA clone of castor bean is not full length, in that the derived peptide is missing methionine at the N terminus (Fig. 1C). This 0.85-kb cDNA was used to screen an Arabidopsis genomic library at a reduced stringency as follows. The hybridization was performed with QuickHyb (Stratagene) at 50°C for 1 hr, washing was with 2× SSC and 0.1% SDS, 5 min twice at room temperature followed by 0.5× SSC and 0.1% SDS at 50°C for 30 min. One Arabidopsis genomic GLB1 clone with a 3-kb DNA insert was isolated. A 1.1-kb NcoI–NcoI DNA fragment of the genomic clone was used to screen an Arabidopsis silique (pod) cDNA library. Under high stringency hybridization conditions, two Arabidopsis cDNA clones were obtained from 1 × 106 plaques. These Arabidopsis GLB1 cDNAs were sequenced by using the Sequenase method (United States Biochemical).
Figure 1.
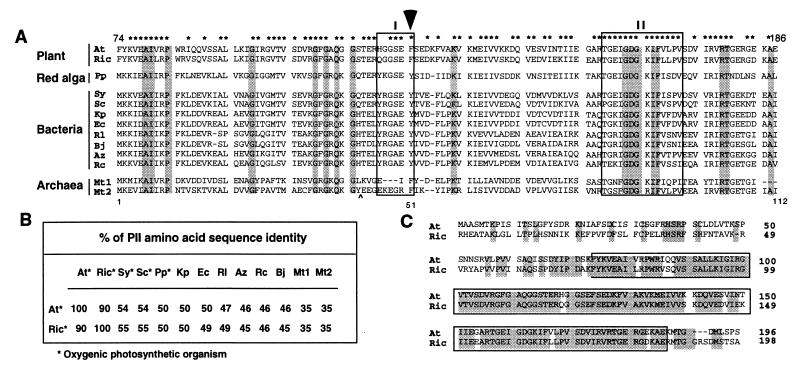
(A) Comparison of the deduced amino acid sequences of plant and microbial PII-like polypeptides. The amino acid residues conserved in all PII-like proteins are shaded. The asterisks (∗) indicate residues that are shared between the plant sequences and any prokaryotic PII-like proteins. Box I and box II refer to PII signature domains I and II. The position of the tyrosine residue that is uridylylated in E. coli is indicated by an arrow. Abbreviations and sequence references: At, A. thaliana; Ric, R. communis; Pp, P. purpurea (16), Sy, Synechocystis sp. PCC 6803 (29); Sc, Synechococcus strain PCC 7942 (30); Kp, K. pneumoniae (31); Ec, E. coli (28); Rl, R. leguminosarum (32); Bj, B. japonicum (33); Az, A. brasilense (34); Rc, R. capsulatus (35); and Mt1 and Mt2, M. thermolithotrophicus PII-like protein 1 and 2 (36). For best alignment, the last two residues GN and a peptide sequence FSANLPEIVDIQKII, which is indicated by “∧”, are deleted in Mt2. The Arabidopsis PII-like protein sequence shown starts with amino acid residue 74 and ends with residue 186. The numbers 1, 51, and 112 indicate the residue position of the E. coli PII protein (product of the glnB gene). (B) The identity of the deduced amino acid sequences of plant and prokaryotic PII proteins. (C) Comparison of the deduced amino acid sequences of plant PII proteins. At, A. thaliana; Ric, R. communis. The identical amino acid residues are shaded. The boxed region corresponds to full-length prokaryotic PII protein.
Antibody Preparation and Western Blot Analyses.
The in-frame Arabidopsis full-length PII cDNA was cloned into a pMALc2 vector (New England Biolabs) to generate the maltose-binding protein (MBP)-PII fusion protein. Induction, extraction, and purification of the MBP-PII fusion protein were performed as described (18). The affinity-purified MBP-PII protein was separated by 12% (wt/vol) SDS/PAGE according to Laemmli (19). Coomassie blue R-250 dissolved in water was used to visualize the protein bands (20). The MBP-PII bands identified by Coomassie blue staining were cut out of the gels, bulked, and eluted. The antibody preparation and Western blot analyses were performed as described (21). The enhanced chemiluminescence (ECL) system was used for detection in Western blot analyses and was performed according to the manufacturer’s instructions (Amersham).
Protein Extraction and Chloroplast Isolation.
Rosette leaves from 4-week-old Arabidopsis plants grown in soil were used for total protein extraction and chloroplast isolation. Total proteins were extracted from Arabidopsis leaves with SDS extraction buffer as described (21). Chloroplasts were isolated with discontinuous Percoll gradients, and the isolated chloroplasts were treated with protease as described (22).
Analyses of DNA and RNA.
Arabidopsis genomic DNA was isolated as described (18). RNA was isolated by using a phenol extraction protocol (23). Southern and Northern blotting were performed as described (24). For detection of GLB1 mRNA, membranes were hybridized with single-stranded digoxigenin-labeled probes made from GLB1 cDNA by using PCR (25). To generate probes ≈540 nucleotides in length covering the PII-coding region and part of the 3′-noncoding region, primers MH5 (GAAACCAAACACAGACTCC) and MH6 (CCGAGTAATAACAGTCGTC) were used. The ASN1 single-stranded digoxigenin-labeled probe was generated by PCR using oligonucleotide primers 5′-AACTCCGATAGCGGCTC-3′ and 5′-CTCTATTTCCACAAGGCACC-3′ (5). The control digoxigenin-labeled probe for 18S rRNA was made by random priming. Northern blots were hybridized at 42°C in 50% formamide hybridization solution overnight and washed with 0.5× SSC, 0.1% (wt/vol) SDS at 65°C for 30 min. Detections were performed according to the Boehringer Mannheim Genius System user’s guide. All Northern blots presented herein were repeated at least three times with comparable results.
Analyses of Arabidopsis PII Transgenic Plants.
The full-length Arabidopsis GLB1 cDNA was subcloned into a plant expression vector (pTEV) (26) in the sense or antisense orientation. After vacuum infiltration, 81 sense transformants were recovered. T1 progeny test revealed that 46 lines harbored a single transgene insertion as determined by the ratio of 3:1 (KanR:KanS). The overexpression of GLB1 in these lines was confirmed by Northern blot analyses (data not shown). Ten single insertion lines were randomly selected to test for homozygocity in the T2 generation. The ectopic expression of PII in the T3 homozygous lines was further confirmed by both Northern and Western blot analyses. These lines were used for carbon/nitrogen sensing test. Results from eight independent lines, PII-1, -4, -24, -25, -26, -27, -43, and -44, are shown in Fig. 6. The extraction and quantitation of anthocyanin was performed as described (27). Of >100 antisense GLB1 lines generated, none showed significantly reduced levels of GLB1 mRNA or PII-like protein.
RESULTS
Cloning and Identification of cDNAs for PII Homologs from Arabidopsis and Castor Bean.
The predicted full-length PII polypeptide encoded by the Arabidopsis GLB1 cDNA is 196 aa with a molecular weight of 21,375 daltons. So far, glnB or glnB-like genes have been identified in a broad set of microorganisms, including enteric bacteria, cyanobacteria, and various archaea. Representative bacterial and archael PII sequences were aligned with plant PII sequences and the results are shown as Fig. 1A. The region of homology to prokaryotic PII encompasses the entire microbial PII protein and occurs between amino acids 74 and 186 of the Arabidopsis PII-like protein (Fig. 1A). In this region, the overall identity between the encoded plant PII-like protein and E. coli PII is 50% (28). Within the region corresponding to E. coli PII, the deduced amino acid sequences of plant PII share high overall identities with other representative microbial glnB genes including: Synechosystis sp. PCC 6803 (54%) (29), Synechococcus sp. PCC 7942 (54%) (30), Klebsiella pneumoniae (50%) (31), Rhizobium leguminosarum (47%) (32), Bradyrhizobium japonium (46%) (33), Azospirillum brasilense (46%) (34), Rhodobacter capsulatus (46%) (35), and Methanococcus glnB-like protein 1 (35%) (36) (Fig. 1B).
In addition to the high overall identity between various PII proteins of prokaryotes and the PII-like proteins in plants, extremely high local identities occur over signature domains, especially in signature domain II of the prokaryotic PII protein (79% identity, Fig. 1A) and in signature domain I (Y-R-G-[AS]-E-Y), which contains the Tyr-51 residue that is post-translationally modified by uridylylation in enteric bacteria (6, 8). In both Arabidopsis and Castor bean PII-like proteins, the corresponding Tyr-51 in signature domain I is replaced by a phenylalanine (Fig. 1A). This suggests that the plant PII-like protein may not be modified by uridylylation in the corresponding signature domain I. Interestingly, even though the PII protein identified in cyanobacteria contains the conserved Tyr-51 residue in signature domain I, it is not modified by uridylylation and is instead modified by phosphorylation at the Ser-49 residue (37). Both plant PII-like proteins contain this conserved Ser-49 residue. The conserved signature domain II, T-x(3)-G-D-G-[KR]-I-F-[LIVM]-x(2)-[LIVM], common to all prokaryotic PII proteins, is extremely well conserved within Arabidopsis and Castor bean PII-like proteins (Fig. 1C). The high conservation of signature domain II among archael, bacterial, and eukaryotic PII proteins suggests that this domain may play an important, as yet unknown, function in PII proteins across all three domains of life.
The derived sequences of plant PII proteins from Arabidopsis and Castor bean are 90.3% identical to each other within the region corresponding to microbial PII (Fig. 1C). Beyond this region, the identity between these two plant PII-like proteins decreases dramatically, especially in the N-terminal region (amino acid residues 1–55) (Fig. 1C). This nonconserved N-terminal region in the plant PII-like protein (which is not present in the microbial PII protein) most likely serves as a chloroplast localization signal (see below).
The Arabidopsis PII-Like Protein Is Localized in Isolated Chloroplasts.
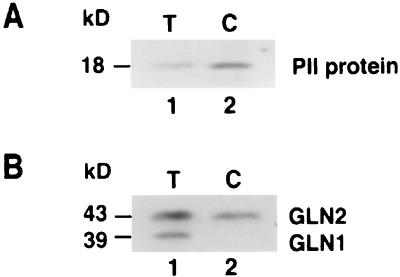
The molecular mass of the Arabidopsis PII-like protein deduced from the GLB1 cDNA clone is ≈21.4 kDa. However, Western blot analysis of total leaf proteins indicates the Arabidopsis PII-like protein is ≈18 kDa (Fig. 2A, lane 1). This size discrepancy suggests that the Arabidopsis PII-like protein may be processed inside the plant cells, most likely in chloroplasts. This is a likely site of localization because the glnB gene of a red alga, P. purpurea, is located in the chloroplast genome (16). To determine whether the plant PII-like protein is localized in chloroplasts, the proteins from isolated, protease-treated, Arabidopsis chloroplasts were probed by Western blot analysis. The PII-like protein detected was 18 kDa in size (Fig. 2A, lane 2). As a positive control for chloroplast purity, antibodies that recognize both cytosolic and chloroplastic isoforms of GS were used to probe the same protein samples by Western blot analysis. Both cytosolic GS (39 kDa) and chloroplastic GS (43 kDa) were detected in the Arabidopsis total proteins (Fig. 2B, lane 1). By contrast, only chloroplastic GS was detected in the protein fraction from isolated chloroplasts (Fig. 2B, lane 2), thus confirming the purity of the chloroplast preparation. Therefore, the PII-like protein of higher plants appears to be a chloroplast protein that is nuclear encoded.
Figure 2.
The Arabidopsis PII-like protein is localized in chloroplasts. Western blot analyses of Arabidopsis PII-like protein (A) and GS (B). The Arabidopsis PII-like protein can be detected in leaf total proteins (T) or in chloroplastic proteins (C) as shown in A. As a control, cytosolic GS (GLN1) was detected in leaf total proteins (T) but not in the chloroplastic proteins (C). By contrast, chloroplastic GS (GLN2) was detected in both total proteins (T) and chloroplastic proteins (C) as shown in B.
The Arabidopsis GLB1 Gene Maps to the Top of Chromosome IV.
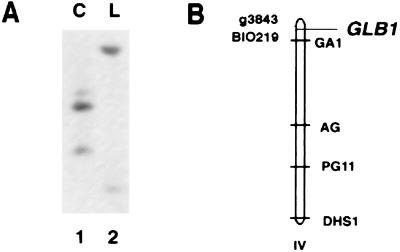
To prove that the Arabidopsis GLB1 cDNA is a bona fide plant gene, we first performed genomic Southern analyses at both high and low stringencies. The results indicate that GLB1 appears to be a single or low copy number gene in Arabidopsis (data not shown). We also mapped the GLB1 gene to a location on the Arabidopsis chromosomes by using recombinant inbred lines (38). The GLB1 gene mapped at 10.8 cM from the top of chromosome IV with neighboring physical markers g3843 and BIO219 (Fig. 3B).
Figure 3.
The Arabidopsis GLB1 gene maps to the top of chromosome IV. Restriction enzyme BglII was used to digest Arabidopsis genomic DNA to determine an restriction fragment length polymorphism between the Columbia (C) and Landsberg (L) ecotype as shown in A. This restriction fragment length polymorphism was used to determine the genotype in 32 recombinant inbred lines developed for C and L ecotypes as described (38). The map position of GLB1 gene is shown relative to its physical flanking markers g3843 and BIO219.
Levels of Arabidopsis GLB1 mRNA Are Induced by Light.
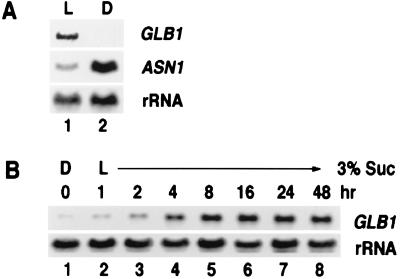
Because light appears to coordinate the expression of nitrogen assimilatory genes such as NR, GLN2, and GLU1 (glutamate synthase) with that of photosynthetic genes such as rbcS (Rubisco small subunit) (4), we examined whether light also could regulate the accumulation of Arabidopsis GLB1 mRNA. Northern blot analyses showed that GLB1 mRNA accumulates preferentially in light-grown plants compared with plants dark-adapted for 48 hr (Fig. 4A, lanes 1 and 2). Because levels of GLB1 mRNA appeared to be induced by light, we conducted a time course of light induction of the expression of the GLB1 gene to see whether it occurs rapidly. Levels of GLB1 mRNA, which are extremely low in dark-adapted plants (Fig. 4B, lane 1), are rapidly induced after only 1 hr of light treatment and increase to maximal levels at 8–16 hr of light treatment (Fig. 4B, lanes 2–6). Levels of GLB1 mRNA remain high even after 24- or 48-hr light treatment (Fig. 4B, lanes 7 and 8).
Figure 4.
Light-induced expression of Arabidopsis GLB1 gene. (A) Ten micrograms of total RNA extracted from 16-day-old Arabidopsis seedlings grown in a normal day/night cycle (16 hr light/8 hr dark) then light-treated (L) or dark-adapted (D) for 48 hr was subjected to Northern blot analysis. As a control for the light vs. dark treatments, ASN1 mRNA also was detected on a separate Northern blot. 18S rRNA was detected on a replicate blot as a loading control. (B) Time course of light induction of GLB1 mRNA in Arabidopsis. Total RNA (10 μg) from 16-day-old Arabidopsis plants grown in media containing 3% sucrose that were dark-adapted for 2 days and then exposed to light for 0 hr (lane 1), 1 hr (lane 2), 2 hr (lane 3), 4 hr (lane 4), 8 hr (lane 5), 16 hr (lane 6), 24 hr (lane 7), and 48 hr (lane 8). As a loading control, 18S rRNA was detected on a replicate blot.
Levels of Arabidopsis GLB1 mRNA Are Induced by Sucrose and Repressed by Amino Acids.
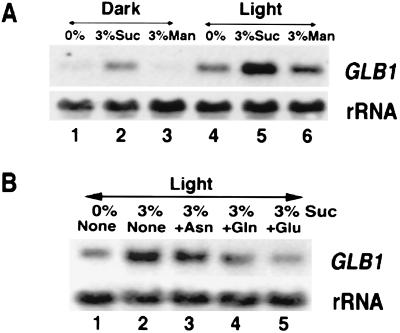
The effects of light on the induction of GLB1 mRNA may be direct via photoreceptors and/or indirect from photosynthetic products. To test the possible involvement of the red light photoreceptor phytochrome, 5-day-old dark-grown seedlings were treated with pulses of light known to activate (red) or inactivate (far-red) phytochrome. The red light pulse or red light followed by a far-red light pulse did not significantly affect levels of GLB1 mRNA (data not shown). These results suggest that phytochrome has no significant effects on the induction of the GLB1 gene. We therefore examined whether sucrose could affect the levels of Arabidopsis GLB1 mRNA. Steady–state levels of GLB1 mRNA are low in dark-adapted plants and are induced by light. (Fig. 5A, cf. lanes 1 and 4). The effects of light on GLB1 mRNA can be mimicked in dark-adapted plants by the addition of 3% sucrose (Fig. 5A, lane 2) but not by mannitol, a nonmetabolizable carbon source (Fig. 5A, lane 3). This control indicates that the induction of GLB1 mRNA by sucrose is not due to osmotic effects. Interestingly, sucrose plus light can induce GLB1 mRNA accumulation beyond light-induced levels (Fig. 5A, lane 5). Again, this additive effect of light and sucrose is not obtained in the control mannitol plus light treatment (Fig. 5A, lane 6). The induction of GLB1 mRNA by sucrose independent of light and the additive induction of GLB1 mRNA by sucrose in the presence of light suggest that the effects of light on GLB1 mRNA induction may derive at least in part from photosynthetic products.
Figure 5.
The expression of Arabidopsis GLB1 gene is regulated by metabolites. (A) Light and sucrose increase levels of GLB1 mRNA. Total RNA (10 μg) from 16-day-old Arabidopsis plants dark-adapted (lanes 1–3) or light-grown (lanes 4–6) for 2 days was used for Northern blot analysis. During the dark or light treatment, plants were grown in media containing 0% sucrose (lanes 1 and 4), 3% sucrose (lanes 2 and 5), or 3% manitol (lanes 3 and 6). As a control, 18 S rRNA was detected on a replicate blot. (B) Amino acids reduce levels of GLB1 mRNA. The 16-day-old Arabidopsis plants grown in a normal day/night cycle were light treated for 48 hr in MS media containing: no sucrose, no amino acid (lane 1); 3% sucrose, no amino acid (lane 2); or 3% sucrose plus 0.4 mM Asn (lane 3), 3.4 mM Gln (lane 4), 3 mM Glu (lane 5). The 18S rRNA was detected on a replicate blot as a loading control.
We have shown that the sucrose effects on the expression of nitrogen assimilatory genes such as AS and GS could be at least partially reversed by the addition of organic nitrogen such as asparagine, glutamine, and glutamate (4, 5). Therefore, we next tested whether these amino acids supplied exogenously also would antagonize the sucrose induction of GLB1 mRNA. Arabidopsis plants grown for 16 days in a normal day/night cycle were light adapted for 48 hr in the absence or presence of 3% sucrose (Fig. 5B, lanes 1 and 2). Levels of GLB1 mRNA are maximally induced under the treatment of light plus sucrose (Fig. 5B, lane 2). The addition of Asn, Gln, or Glu partially reverses the inductive effects of light and sucrose on GLB1 mRNA accumulation albeit to different extents (Fig. 5B, lanes 3–5).
Nitrogen-Related Phenotype of PII Overexpressors.
Because the expression of the GLB1 gene is regulated by light and by carbon and nitrogen metabolites, we aimed to test the effects of deregulating this controlled expression. To this end, GLB1 mRNA was synthesized constitutively in transgenic plants containing a full-length GLB1 cDNA whose expression was driven by a CaMV 35S promoter. Eight independent transgenic lines (lines 1, 4, 24, 25, 26, 27, 43, and 44) were shown to overexpress the Arabidopsis PII-like protein (Fig. 6A). To test whether these PII overexpressors were affected in carbon/nitrogen sensing, we performed an in planta bioassay. Wild-type (wt) plants grown on media containing 3% sucrose without a nitrogen source accumulate high levels of anthocyanin (Fig. 6B). This sucrose-induced anthocyanin accumulation in wt plants can be relieved by the addition of inorganic nitrogen in the form of ammonium nitrate (data not shown). We extended these studies to the level of organic nitrogen and demonstrated that the sucrose-induced anthocyanin accumulation in wt plants can be relieved by the addition of exogenous Gln (1 mM) (Fig. 6C). Next, we tested whether Gln could reverse sucrose-induced anthocyanin accumulation in PII overexpressors. Like wt plants, PII overexpressors accumulate high levels of anthocyanin when grown in 3% sucrose with no nitrogen source (Fig. 6B, PII-24). Similarly, inorganic nitrogen (ammonium nitrate) can reverse sucrose induced anthocyanin accumulation in PII overexpressing plants (data not shown). However, 1 mM Gln was much less effective in reversing the sucrose-induced levels of anthocyanin in all eight independent PII overexpressing lines compared with wt plants (Fig. 6 C and D). Anthocyanin content in PII overexpressors grown in 3% sucrose plus 1 mM Gln is 60–190% higher than wt (Fig. 6D). This differential effect of Gln on wild type vs. PII overexpressors also was observed at several different concentrations of sucrose and Gln (3% suc + 3 mM Gln, 6% suc + 1 or 3 mM Gln) (data not shown). This in planta phenotype suggests that the PII protein may be involved in sensing the status of carbon and organic nitrogen in higher plants.
DISCUSSION
Unlike the extensive studies of nitrogen regulatory mechanisms in bacteria, very little is known about how plants sense changes in the internal carbon/nitrogen status. Recently, hexokinase has been proposed as a sugar sensor in higher plants (39, 40). It is not clear whether plants have a separate nitrogen sensor or some kind of mechanism involved in sensing the internal status of both carbon and nitrogen. In this report, we identify a plant nuclear gene (GLB1), which encodes a homolog of the prokaryotic PII protein, a component of the bacterial Ntr system. Western blot analysis reveals that the Arabidopsis PII-like protein is localized in the chloroplast (Fig. 2). Thus PII in plants is a nuclear-encoded chloroplast protein. It is noteworthy that the glnB gene encoding the PII protein of a red alga, P. purpurea, is also a chloroplast protein but is encoded by the chloroplast genome (16). The high overall identity of PII-like proteins in prokaryotes and eukaryotes, and the subcellular localization of eukaryotic PII-like proteins in chloroplasts of higher plants and red algae, suggests the genes encoding these proteins in archaea, bacteria, red algae, and higher plants may have evolved from a common ancestral gene.
The Arabidopsis GLB1 gene was expressed in all organs tested (data not shown), and the levels of GLB1 mRNA were regulated by light and metabolites. Thus, it appears that regulation of PII in plants may be transcriptional. In contrast to the glnB gene of E. coli, which is expressed constitutively and regulated post-translationally, the newly identified glnK gene encoding a second PII-like protein in this organism is regulated transcriptionally as well as being post-translationally modified (41). The glnB genes of several bacteria such as B. japonicum, A. brasilense, and R. capsulatus also are regulated at the transcriptional level in response to nitrogen availability (42–44). Recently, it has been reported that the expression of the glnB gene in the cyanobacterium Synechocystis sp. PCC 6803 is regulated in response to the nitrogen source, light–dark transitions and photosynthetic inhibitors (29).
To test the in vivo function of the Arabidopsis PII-like protein, we created transgenic Arabidopsis in which PII expression was constitutive. The phenotype of Arabidopsis PII overexpressors grown under different regimes of carbon/nitrogen supports the notion that the PII protein in plants also may act as a nitrogen sensor. When wt plants are grown in media with high sucrose and no nitrogen, the synthesis of anthocyanins is induced (27, 45). The addition of inorganic nitrogen such as ammonium nitrate can relieve the sucrose-induced accumulation of anthocyanin in these wt plants. Similarly, PII overexpressing plants accumulate high levels of anthocyanin when grown in media containing high sucrose and no nitrogen, and ammonium nitrate can relieve anthocyanin accumulation (data not shown). However, when Gln was used as the only nitrogen source in media with high sucrose, anthocyanin levels in PII overexpressors remained high, compared with wt (Fig. 6 C and D). Thus it appears that PII overexpressors are impaired in their ability to sense or metabolize Gln. It is possible that the over production of the Arabidopsis PII-like protein in chloroplasts may alter the availability or metabolism of Gln as a nitrogen source in the chloroplast. Alternatively, the Arabidopsis PII-like protein may be involved in the regulation of the biosynthesis of anthocyanins. Further studies regarding these hypotheses need to be performed and the components with which the plant PII-like protein interacts need to be identified.
Despite the broad existence of genes encoding PII-like proteins in bacteria and archaea, the functions of PII-like proteins have been defined in only a few bacteria. Among its defined functions, the PII protein is best understood as a protein allosteric effector that controls GS in E. coli and K. pneumoniae, as discussed above. The other proposed functions of the PII protein also are related to nitrogen metabolism. For example, in R. leguminosarum, in addition to regulating glutamine synthetase, PII-UMP has been suggested as a positive effector of nitrate utilization (nas) genes (46). In addition, the R. meliloti PII protein encoded by glnB is involved in infection and nodule development of the host plant alfalfa, and its role in controlling the expression or activity of a bacteroid ammonium transporter has been proposed (47). Moreover, PII is a positive effector of the transcriptional activator NifA for the nitrogen fixation genes in the diazotrophic bacterium A. brasilense (48). In cyanobacteria, no NtrB/NtrC homologs have been identified, and there is no evidence that glutamine synthetase is regulated by adenylylation. However, a glnB gene has been identified in cyanobacteria such as Synechococcus sp. PCC 7942 (30) and Synechocystis sp. PCC 6803 (29). The Synechococcus sp. PCC 7942 PII protein is involved in mediating the coordination between carbon and nitrogen assimilation (49, 50) and in mediating nitrate/nitrite uptake (51).
To our knowledge, this is the first report describing a protein, the PII-like protein, with a putative role in nitrogen sensing in higher plants. The presence of the PII protein in archaea, bacteria, and now in higher eukaryotes (plants) suggests PII is one of the most widespread regulatory proteins known in these three domains of life. Predictions from x-ray structure and extensive site mutation studies have revealed that the exposed loops of E. coli PII and cleft formed at their interface are the sites of regulatory interactions (14, 15, 52). The primary structure of the plant PII-like protein, especially the signature domain II, is highly homologous to prokaryotic PII. It will be interesting to know whether this highly conserved domain is involved in similar functions. We are in the process of identifying or creating mutants of higher plants that lack PII to examine how the loss of PII affects C/N sensing and to understand the exact mechanism by which this occurs.
Acknowledgments
We would like to thank Dr. C. Somerville for the Castor bean GLB1 cDNA clone, Dr. D. Meinke for the Arabidopsis silique (pod) cDNA library, and Drs. B. Hirel and M. Lara for the GS antibodies. We also thank J. Lee for her technical assistance. This work was supported by National Institutes of Health Grant GM32877 to G.C.
ABBREVIATIONS
- wt
wild type
- Ntr
nitrogen regulatory
- GS
glutamine synthetase
- AS
asparagine synthetase
- UMP
uridylate
- MBP
maltose-binding protein
- MS
Murashige and Skoog
Footnotes
References
- 1.Cheng C-L, Acedo G N, Cristinsin M, Conkling M A. Proc Natl Acad Sci USA. 1992;89:1861–1864. doi: 10.1073/pnas.89.5.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Faure J-D, Jullien M, Caboche M. Plant J. 1994;5:481–491. doi: 10.1046/j.1365-313x.1994.5040481.x. [DOI] [PubMed] [Google Scholar]
- 3.Vincentz M, Moureaux T, Leydecker M-T, Vaucheret H, Caboche M. Plant J. 1993;3:315–324. doi: 10.1111/j.1365-313x.1993.tb00183.x. [DOI] [PubMed] [Google Scholar]
- 4.Lam H-M, Coschigano K, Schultz C, Melo-Oliveira R, Tjaden G, Oliveira I, Ngai N, Hsieh M-H, Coruzzi G M. Plant Cell. 1995;7:887–898. doi: 10.1105/tpc.7.7.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lam H-M, Peng S S-Y, Coruzzi G M. Plant Physiol. 1994;106:1347–1357. doi: 10.1104/pp.106.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown M S, Segal A, Stadtman E R. Proc Natl Acad Sci USA. 1971;68:2949–2953. doi: 10.1073/pnas.68.12.2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Magasanik B. Trends Biochem Sci. 1988;13:475–479. doi: 10.1016/0968-0004(88)90234-4. [DOI] [PubMed] [Google Scholar]
- 8.Magasanik B. J Cell Biochem. 1993;51:34–40. doi: 10.1002/jcb.240510108. [DOI] [PubMed] [Google Scholar]
- 9.Keener J, Kustu S. Proc Natl Acad Sci USA. 1988;85:4976–4980. doi: 10.1073/pnas.85.14.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McFarland N, McCarter L, Artz S, Kustu S. Proc Natl Acad Sci USA. 1981;78:2135–2139. doi: 10.1073/pnas.78.4.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reitzer L J, Magasanik B. Cell. 1986;45:785–792. doi: 10.1016/0092-8674(86)90553-2. [DOI] [PubMed] [Google Scholar]
- 12.Ninfa A J, Magasanik B. Proc Natl Acad Sci USA. 1986;83:5909–5913. doi: 10.1073/pnas.83.16.5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Atkinson M R, Kamberov E S, Weiss R L, Ninfa A J. J Biol Chem. 1994;269:28288–28293. [PubMed] [Google Scholar]
- 14.Jiang P, Zucker P, Atkinson M R, Kamberov E S, Tirasophon W, Chandran P, Schefke B R, Ninfa A J. J Bacteriol. 1997;179:4342–4353. doi: 10.1128/jb.179.13.4342-4353.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang P, Zucker P, Ninfa A J. J Bacteriol. 1997;179:4354–4360. doi: 10.1128/jb.179.13.4354-4360.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reith M, Munholland J. Plant Cell. 1993;5:465–475. doi: 10.1105/tpc.5.4.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van de Loo F, Turner S, Somerville C. Plant Physiol. 1995;108:1141–1150. doi: 10.1104/pp.108.3.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ausubel F M, Brent R, Kingston R E, Moore D D, Seiden J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley Intersciences; 1992. [Google Scholar]
- 19.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Zhao J, Last R L. J Biol Chem. 1995;270:6081–6087. doi: 10.1074/jbc.270.11.6081. [DOI] [PubMed] [Google Scholar]
- 21.Hsieh M H, Chen J T, Jinn T L, Chen Y M, Lin C Y. Plant Physiol. 1992;99:1279–1284. doi: 10.1104/pp.99.4.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartlett S G, Grossman A R, Chua N-H. In: Methods in Chloroplast Molecular Biology. Edelman, editor. Amsterdam, The Netherlands: Elsevier; 1982. pp. 1081–1091. [Google Scholar]
- 23.Jackson A O, Larkins B A. Plant Physiol. 1976;57:5–10. doi: 10.1104/pp.57.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 25.Myerson D. BioTechniques. 1991;10:35–38. [PubMed] [Google Scholar]
- 26.Brears T, Liu C, Knight T, Coruzzi G. Plant Physiol. 1993;103:1285–1290. doi: 10.1104/pp.103.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mita S, Murano N, Akaike M, Nakamura K. Plant J. 1997;11:841–851. doi: 10.1046/j.1365-313x.1997.11040841.x. [DOI] [PubMed] [Google Scholar]
- 28.Son H S, Rhee S G. J Biol Chem. 1987;262:8690–8695. [PubMed] [Google Scholar]
- 29.Garcia-Dominguez M, Florencio F J. Plant Mol Biol. 1997;35:723–734. doi: 10.1023/a:1005846626187. [DOI] [PubMed] [Google Scholar]
- 30.Tsinoremas N F, Castets A M, Harrison M A, Allen J F, Marsac N T. Proc Natl Acad Sci USA. 1991;88:4565–4569. doi: 10.1073/pnas.88.11.4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holtel A, Merrick M. Mol Gen Genet. 1988;215:134–138. doi: 10.1007/BF00331314. [DOI] [PubMed] [Google Scholar]
- 32.Colonna-Romano S A, Riccio A, Guida M, Defez R, Lamberti A, Iaccarino M, Arnold W, Priefer U, Puhler A. Nucleic Acids Res. 1987;15:1951–1964. doi: 10.1093/nar/15.5.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin G B, Thomashow M F, Chelm B K. J Bacteriol. 1989;171:5638–5645. doi: 10.1128/jb.171.10.5638-5645.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Zamaroczy M F, Delorme F, Elmerich C. Mol Gen Genet. 1990;224:421–430. doi: 10.1007/BF00262437. [DOI] [PubMed] [Google Scholar]
- 35.Kranz R G, Pace V M, Caldiott I M. J Bacteriol. 1990;172:53–62. doi: 10.1128/jb.172.1.53-62.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Souillard N, Sibold L. Mol Microbiol. 1989;3:541–551. doi: 10.1111/j.1365-2958.1989.tb00200.x. [DOI] [PubMed] [Google Scholar]
- 37.Forchhammer K, De Marsac N T. J Bacteriol. 1994;176:84–91. doi: 10.1128/jb.176.1.84-91.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lister C, Dean C. Plant J. 1993;4:745–750. [Google Scholar]
- 39.Jang J C, Leon P, Zhou L, Sheen J. Plant Cell. 1997;9:5–19. doi: 10.1105/tpc.9.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jang J C, Sheen J. Trends Plant Sci. 1997;2:208–214. [Google Scholar]
- 41.van Heeswijk W C, Hoving S, Molenaar D, Stegeman B, Kahn D, Westerhoff H V. Mol Microbiol. 1996;21:133–146. doi: 10.1046/j.1365-2958.1996.6281349.x. [DOI] [PubMed] [Google Scholar]
- 42.de Zamaroczy M, Paquelin F A, Elmerich C. J Bacteriol. 1993;175:2507–2515. doi: 10.1128/jb.175.9.2507-2515.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Foster-Hartnett D, Kranz R G. J Bacteriol. 1994;176:5171–5176. doi: 10.1128/jb.176.16.5171-5176.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin G B, Thomashow M F, Chelm B K. J Bacteriol. 1989;171:5638–5645. doi: 10.1128/jb.171.10.5638-5645.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taiz L, Zeiger E. Plant Physiology. CA: Benjamin/Cummings Redwood City; 1991. p. 114. [Google Scholar]
- 46.Amar M, Patriarca E J, Manco G, Bernard P, Riccio A, Lamberti A, Defez R, Iaccarino M. Mol Microbiol. 1994;11:685–693. doi: 10.1111/j.1365-2958.1994.tb00346.x. [DOI] [PubMed] [Google Scholar]
- 47.Arcondeguy T, Huez I, Tillard P, Gangneux C, de Billy F, Gojon A, Truchet G, Kahn D. Genes Dev. 1997;11:1194–1206. doi: 10.1101/gad.11.9.1194. [DOI] [PubMed] [Google Scholar]
- 48.Arsene F, Kaminski P A, Elmerich C. J Bacteriol. 1996;178:4830–4838. doi: 10.1128/jb.178.16.4830-4838.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Forchhammer K, Hedler A. Eur J Biochem. 1997;244:869–875. doi: 10.1111/j.1432-1033.1997.00869.x. [DOI] [PubMed] [Google Scholar]
- 50.Forchhammer K, De Marsac N T. J Bacteriol. 1995;177:2033–2040. doi: 10.1128/jb.177.8.2033-2040.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee H M, Flores E, Herrero A, Houmard J, de Marsac T N. FEBS Lett. 1998;427:291–295. doi: 10.1016/s0014-5793(98)00451-7. [DOI] [PubMed] [Google Scholar]
- 52.Carr P D, Cheah E, Suffolk P M, Vasudevan S G, Dixon N E, Ollis D L. Acta Crystallogr D. 1996;52:93–104. doi: 10.1107/S0907444995007293. [DOI] [PubMed] [Google Scholar]