Abstract
In mammals, one of the major actions of insulin-like growth factor I (IGF-I) is to increase skeletal growth by stimulating new cartilage formation. IGF-I stimulates chondrocytes in vitro to synthesize new cartilage matrix, measured by enhanced uptake of 35S-sulfate, but the addition of insulin does not produce a similar effect except when added at high concentrations. However, recent studies have shown that, in teleosts, both insulin and IGF-I are potent activators of 35S-sulfate uptake in gill cartilage. To further characterize the growth-promoting activities of these hormones in fish, we have used reverse transcriptase-linked PCR to analyze the expression of insulin receptor family genes in salmon gill cartilage. Partial cDNA sequences encoding the tyrosine kinase domains from six distinct members of the IR gene family were obtained, and sequence comparisons revealed that four of the cDNAs encoded amino acid sequences that were highly homologous to human IR whereas the encoded sequences from two of the cDNAs were more similar to the human type I IGF receptor (IGF-R). Furthermore, a comparative reverse transcriptase-linked PCR assay revealed that the four putative IR mRNAs expressed in toto in gill cartilage were 56% of that found in liver whereas the expressed amount of the two IGF-R mRNAs was 9-fold higher compared with liver. These results suggest that the chondrogenic actions of insulin and IGF-I in fish are mediated by the ligands binding to their cognate receptors. However, further studies will be required to characterize the binding properties and relative contribution of the individual IR and IGF-R genes.
Keywords: somatomedins, teleost, development
Linear skeletal growth in vertebrates is dependent on the synthesis of new cartilage, which is a complex process involving proliferation and differentiation of chondrocytes, cellular hypertrophy, and formation of extracellular matrix (1). Growth hormone is a major regulator of this process, but it is now established that the action of growth hormone on chondrocytes is mediated, at least in part, by insulin-like growth factor I (IGF-I) (reviewed in refs. 2 and 3). Salmon and Daughaday (4) first reported that growth hormone increased production of plasma somatomedin (IGF-I), which stimulated 35S-sulfate uptake by rat cartilage in vitro. In contrast, the addition of insulin, a hormone with close structural homology to IGF-I, had no effect except at high nonphysiological concentrations (4, 5). More recently, transgenic mice with a null mutation in the IGF-I gene have been generated, and these animals exhibited a growth retardation phenotype with a clear defect in cartilage synthesis (6, 7). Taken together, these results demonstrate that IGF-I plays a major role in regulating chondrogenesis in mammals.
Teleosts also contain IGF-I (and -II) genes (8–10), and it has been shown that IGF-I stimulated 35S-sulfate uptake by isolated gill cartilage (11–14). It is of interest, however, that several studies have reported that, in fish, insulin is also a potent activator of sulfate uptake by gill cartilage in vitro (14–16). Using a highly purified preparation of eel insulin, Duan and coworkers (17) showed that eel insulin was equipotent with bovine IGF-I in stimulating sulfate uptake in eel ceratobranchial cartilage. These findings are intriguing, but because of the close structural similarity between insulin and IGF-I, it is well known that these hormones can cross-bind to each other’s receptor although usually with a lowered affinity. Thus, to further characterize the chondrogenic actions of insulin and IGF-I, it is important to determine which members of the IR tyrosine kinase family genes are expressed in fish cartilage. Using reverse transcriptase-PCR (RT-PCR), we report here that six distinct partial cDNA sequences were obtained: Four clones encoded primary sequences that are more similar to the mammalian IR, and the remaining two cDNA clones encoded sequences more similar to the IGF receptor (IGF-R). Our results suggest that both insulin and IGF-I stimulate chondrogenesis in fish by binding to their own receptors. Our results also indicate that teleosts contain two IR genes per diploid genome due to gene duplication.
METHODS
Materials.
Radioisotopes were obtained from Amersham. Recombinant human insulin was purchased from Sigma. Oligonucleotides were synthesized on a Applied Biosystems 380B DNA synthesizer. Plasmid pGEM4Z was purchased from Promega. Restriction enzymes and other DNA-modifying enzymes were molecular biology grade and were purchased from several sources.
Gill Cartilage Sulfate Uptake Assay.
Branchial cartilages were dissected from fasted 1-yr-old coho salmon and washed in Eagle’s medium. Cartilages (two per well) were incubated in a 24-well plate containing 1 ml of Eagle’s medium and 1 uCi 35S-sulfate in the absence or presence of insulin (10–500 ng) for 48 h at 15°C. Incorporation of 35S-sulfate was determined as described (18).
RT-PCR and cDNA Cloning.
Total RNA was isolated from frozen salmon gill cartilage and liver by homogenization in guanidinium thiocyanate and ultracentrifugation through a 5.7-M CsCl cushion as described (19). The RNAs were quantitated based on absorbance at 260 nm and appeared undegraded on a ethidium bromide-stained 1.0% denaturing agarose gel. RNA was reverse-transcribed into cDNA using oligo(dT) 12–18 (5 μg/ml) or random hexamers (1 μg/ml) as primer and Superscript II (Life Technologies, Gaithersburg, MD) under conditions recommended by the manufacturer. The cDNA product from 1.0 μg RNA was subjected to 30–40 cycles of PCR, each cycle 94 × 1 min, 52 × 1 min, 70 × 1 min, in 100-μl volumes containing 10 mM Tris⋅HCl (pH 8.3), 1.5 mM MgCl2, 50 mM KCl, 100 pmol each of sense and antisense primers (see Figs. 2, 4, and 5 for primer sequences), and 2.5 U Taq polymerase. The RT-PCR products were analyzed on a 1.4% agarose gel; Southern blotting was performed using Hybond-N membranes (Amersham), and hybridization with radiolabeled oligonucleotide probes was performed using standard methods (20).
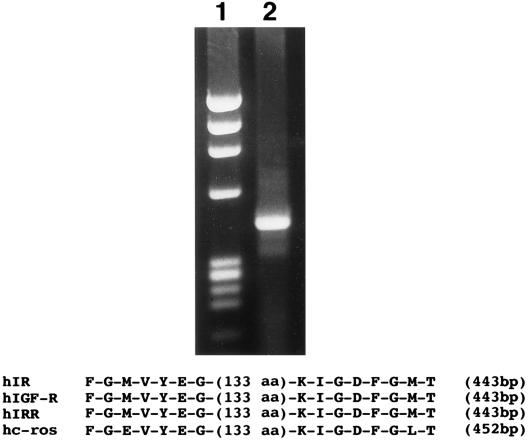
Figure 2.
RT-PCR amplification of IR superfamily genes in salmon gill cartilage. Degenerate oligonucleotide primers were synthesized, which encoded conserved amino acid sequences in the tyrosine kinase domains of human IR, IGF-R, IRR, and c-ros as shown. Primer IR-8(sense) = 5′-TTQGGNATGGTNTAQGAPGG; primer IR-3(antisense) = 5′-GTCATNCCPAAPTCNCCNATQTT with N = G+A+T+C, P = G+A, Q = T+C. These primers were used to amplify 100-ng coho cartilage RNA that had been reverse-transcribed into cDNA for 40 cycles PCR, and the RT-PCR product was analyzed on a 1.4% agarose gel (lane 2). Lane 1 = QX174/HaeIII markers.
Figure 4.
Expanded partial sequence of SIR-1 cDNA. The sequence was obtained by performing RT-PCR on salmon gill cartilage RNA with primers IR-10 (sense sequence = 5′-AAPCCNTGGACNCAPTAQGC) and IR-3; a 1800-bp DNA fragment was isolated by agarose gel electrophoresis, subcloned into pGEM4Z/SmaI, and sequenced. The 36-bp sequence homologous to exon 11 found in the hIR gene is boxed, and the tetrabasic cleavage site separating the a and B subunits are underlined.
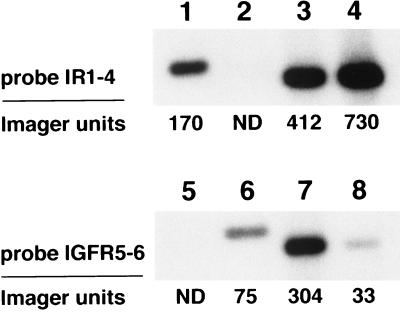
Figure 5.
Comparative RT-PCR of salmon gill cartilage RNA and liver RNA. Duplicate southern blots were prepared that contained the RT-PCR products from 100-ng gill cartilage RNA (lanes 3 and 7) or liver RNA (lanes 4 and 8). The RNAs were reverse-transcribed into cDNA using random hexamers as primer, subjected to 30 cycles PCR with primer set IR-8/IR-3, electrophoresed on a 1.4% agarose gel, and blotted onto Hybond-N membrane. The upper blot was hybridized with radiolabeled probe IR1–4 (sequence = 5′-CCAAGGACATCGTPAAGGG); the lower blot was probed with IGFR5–6 (sequence = 5′-GPTGACAGTTGAACTCCTTC). As positive controls, lanes 1 and 5 contained 50 ng each pSIR-1/pSIR-2/pSIR-3/pSIR-4 plasmid DNA digested with EcoRI + HindIII; lanes 2 and 4 contained 100 ng each pSIR-5/pSIR-6 plasmid DNA digested with EcoRI + PstI. The blots were exposed to x-ray film for 1 h and were scanned on a Molecular Dynamics PhosphoImager. ND, not detected.
For subcloning, the RT-PCR product was made blunt-ended with T4 DNA polymerase, phosphorylated with T4 polynucleotide kinase, ligated to pGEM4z linearized with SmaI, and transformed into competent DH5a Escherichia coli. Isolation of plasmid DNA and dideoxysequencing were performed as described (20).
RESULTS
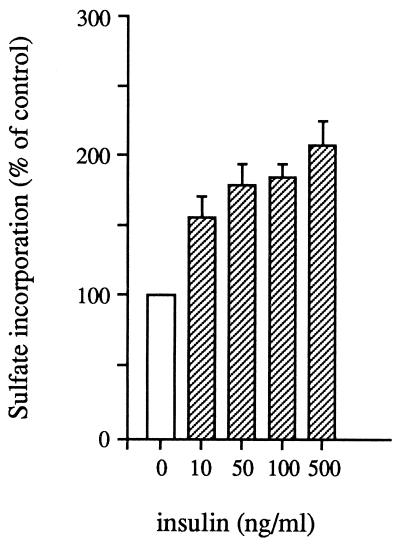
Before performing the RT-PCR analysis, we checked on the sensitivity of isolated coho gill cartilage to respond to insulin in the 35S-sulfate uptake assay. The results obtained from a representative experiment are shown in Fig. 1. The addition of insulin in vitro stimulated 35S-sulfate uptake ≈2-fold above control (P < 0.05), and the effect was detectable at a 10 ng/ml insulin concentration. This is within the physiological range of insulin found in salmon plasma (21). These results confirm that physiological concentrations of insulin can stimulate sulfate uptake by salmon cartilage in vitro.
Figure 1.
Stimulation of sulfate uptake in salmon gill cartilage by insulin. Equivalent amounts of gill cartilage pieces were incubated in Eagle’s medium containing 35S-sulfate and indicated concentrations of human insulin for 48 h at 15°C. Incorporation of 35S-sulfate into cartilage matrix was measured as described in Materials and Methods. Average stimulation above control ± SE is shown.
Total RNA was isolated from coho salmon branchial cartilage and reverse transcribed into cDNA. For RT-PCR, degenerate oligonucleotide primers corresponding to highly conserved amino acid sequences in the IR tyrosine kinase domain were used. These primers were designed to amplify all members of the IR family including IR, IGF-R, and insulin receptor-related receptor (IRR) (Fig. 2).
Analysis of the coho salmon cartilage RT-PCR product revealed a single band of the expected size, which was gel purified and subcloned into pGEM4Z (Fig. 2). We isolated plasmid DNA from 64 colonies, from which 27 clones were sequenced and from these 6 distinct cDNA sequences, labeled SIR-1 to SIR-6, were obtained (Fig. 3A). At least two independent clones for each cDNA species were sequenced, and the clone sequences were essentially identical. (We occasionally found 1–4 nucleotide changes in some clones, which we attributed to allelic variation or cloning error.)
Figure 3.
Nucleotide and deduced amino acid sequences of SIR cDNAs. Six distinct cDNA sequences were obtained by subcloning the 443-bp fragment amplified by RT-PCR from salmon gill cartilage as described in the text. Nucleotides which are identical in all six cDNAs are boxed. (Lower) The deduced amino acid sequences are compared with human IR, IGF-1R, and IRR. Amino acids that are identical in all nine sequences are boxed.
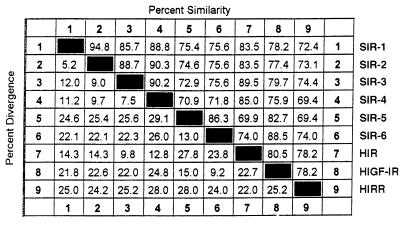
The nucleotide sequence identities between the 6 SIR cDNAs ranged from 69.7 to 90.8%, and comparisons of the translated protein sequences revealed that they were all members of the IR family. Fig. 3B shows that 87/132 amino acids in the tyr kinase domain are completely conserved in all SIR clones. However, a sequence identity/divergence matrix (Table 1) clearly shows that SIR-1, SIR-2, SIR-3, and SIR-4 were related most closely to hIR (83.5–89.0% identity) whereas SIR-5 and SIR-6 shared greatest amino acid identity (82.7% and 88.5%, respectively) with hIGF-R. None of the clones that we analyzed exhibited maximal homology to hIRR.
Table 1.
Comparison of deduced amino acid sequences from the cloned SIR cDNAs with human IR, IGF-R, and IRR
Percent similarity/divergence values were calculated using the megalign program from DNAstar (Madison, WI) and inputting the amino sequences shown in Fig. 3.
A further examination of the salmon IR sequences suggested that they could also be divided into 3 subsets. Clones SIR-1 and SIR-2 shared 94.8% identity and formed one possible subset. Similarly, SIR-3 and SIR-4 were 90.2% identical to each other. The two putative salmon IGF-R clones, SIR-5 and SIR-6, shared 86.3% identity.
Although the above comparisons suggested that four IR and two IGF-R sequences have been cloned from coho gill cartilage, because of the relatively short lengths that were compared and the high sequence conservation in the tyr kinase domain, it was possible that some of the sequence identities were attributable to fortuitous mutations. To test this, we used RT-PCR to obtain an expanded cDNA sequence for SIR-1, and the results corroborated the identification of this clone as an IR. The expanded SIR-1 cDNA sequence, shown in Fig. 4, encoded 580 amino acids, which consisted of the COOH-terminal region of the α-subunit and most of the β-subunit and exhibited 61.5%, 50%, and 46.5% identity with hIR, hIGF-R, and hIRR, respectively. The SIR-1 cDNA also contained a 36-bp (12-aa) sequence homologous to exon 11 in the hIR gene, which is not present in either hIGF-R or hIRR genes (22–24). These results provide strong evidence that SIR-1 and likely SIR-2, SIR-3, and SIR-4 indeed represent salmon IR genes.
To assess the relative expression of the putative IR and IGF-R genes, a comparative RT-PCR assay was performed with primer set IR-8/IR-3 and by using gill cartilage RNA and liver RNA as the substrates. We used salmon liver because insulin binding has been shown to be abundant in this tissue (25, 26). RT-PCR was performed for 30 cycles, which preliminary experiments established was within the linear response range with RNA from either tissue, and duplicate Southern blots of the amplified cDNA products were prepared. One blot was hybridized with radiolabeled oligonucleotide IR1–4, which was designed to specifically bind SIR-1/SIR-2/SIR-3/SIR-4 cDNAs (Fig. 5, lanes 1 and 2); the second blot was hybridized to oligonucleotide IGFR-5–6, which specifically bound SIR-5/SIR-6 cDNAs (Fig. 5, lanes 5 and 6). The blots were scanned and quantitated using a Molecular Dynamics PhosphoImager, and the results showed that, with probe IR1–4, the gill cartilage RT-PCR product yielded 56% of the signal obtained with liver (Fig. 5). In contrast, with probe IGFR5–6, the signal obtained from gill cartilage was 9-fold higher than that from liver. These results suggest that gill chondrocytes express approximately half the level of IR mRNA as liver but a much higher amount of IGF-R mRNA.
DISCUSSION
Gill cartilage from teleosts, including salmon, responds to physiological concentrations of insulin with increased synthesis of extracellular matrix. Because of possible cross-binding of insulin and IGFs to each other’s receptor, however, it was not known whether the insulin action was mediated by activation of its own receptor or the IGF-R. Our results here indicate that the former case is more likely. Using RT-PCR, we have shown that chondrocytes from salmon gill cartilage expressed at least six distinct genes from the IR superfamily, and sequence comparisons showed that four of these shared highest amino acid sequence identity with mammalian IR.
We have not yet performed a ribonuclease protection assay for each of the six SIR mRNAs that will be necessary to quantitate the expression level of each gene. However, our results obtained in the comparative RT-PCR analysis indicate that the putative IR mRNAs (SIR-1, SIR-2, SIR-3, and SIR-4) are expressed at readily detectable levels in gill cartilage. Compared with liver, a known insulin responsive tissue, gill cartilage RNA yielded 56% of the IR-specific signal obtained with liver RNA (Fig. 5). The RT-PCR assay also demonstrated that gill cartilage expressed much higher levels of the IGF-R mRNAs, i.e., SIR-5 and SIR-6, than liver, and this is consistent with the known major action of IGF in this tissue.
Because of the tetraploid nature of the salmon genome (27), we had anticipated obtaining partial cDNA sequences for two IR and two IGF-R if these genes were expressed in cartilage. Consistent with this expectation, we obtained two clones (SIR-5 and SIR-6) that exhibited high sequence similarity to IGF-R. However, four salmon IR cDNA sequences were identified, and this could be due to either allelic variation or duplication of the IR gene. To resolve this issue, we recently have cloned partial sequences for two distinct IR cDNAs from another teleost species, the sea bream, which has a diploid genome (S.J.C., B. Funkenstein, and D.F.S., unpublished results). These results suggest that a duplication of the IR gene has occurred during evolutionary development in the teleosts.
Although the RT-PCR analyses demonstrate that all six SIR mRNAs are expressed in salmon cartilage, it is important to note that further experiments will be necessary to characterize the receptor proteins encoded by these mRNAs, including measurements of their binding affinities for insulin and/or IGF and intracellular signaling properties. In particular, because of the heterodimeric structure of the IR family, it is possible that multiple hybrid forms of the receptors can be synthesized that exhibit differences in biological activity. Knowledge of the partial cDNA sequences reported here should facilitate these studies through use of the deduced peptides as antigens to generate specific antibodies.
The finding that both putative IR and IGF-R genes are expressed in salmon cartilage makes it likely that both insulin and IGF may play a role in fish development by stimulating chondrocyte proliferation and skeletal growth. In mammals, insulin has both growth-promoting and metabolic regulatory activities, but it is the metabolic regulatory function of the hormone to maintain glucose homeostasis that predominates. However, in teleosts, it has been shown that other nutrients, including amino acids such as arginine, are more potent than glucose in stimulating insulin secretion (21, 28, 29). This suggests that, in fish, insulin may supplement the action of IGF-I by increasing growth under conditions of nutrient excess. This dual control would provide teleosts with greater flexibility in controlling their rate of growth in response to changing genetic and environmental factors.
Acknowledgments
We thank Dr. William H. Daughaday for critically reading this manuscript and Paul Gardner and Jeff Stein for expert technical assistance. This work was supported by the Howard Hughes Medical Institute and by a grant from the National Institute of Health (DK 13914).
ABBREVIATIONS
- IGF
insulin-like growth factor
- RT-PCR
reverse transcriptase–PCR
- IGF-R
IGF receptor
- IRR
insulin receptor-related receptor
Footnotes
References
- 1.Gilbert S F. Developmental Biology. Sunderland, MA: Sinauer; 1991. pp. 209–214. [Google Scholar]
- 2.Stewart C E H, Rotwein P. Physiol Rev. 1996;76:1005–1026. doi: 10.1152/physrev.1996.76.4.1005. [DOI] [PubMed] [Google Scholar]
- 3.Daughaday W H. Perspec Biol Med. 1989;32:194–211. doi: 10.1353/pbm.1989.0006. [DOI] [PubMed] [Google Scholar]
- 4.Salmon W E, Jr, Daughaday W H. J Lab Clin Med. 1957;49:825–836. [PubMed] [Google Scholar]
- 5.Salmon W D, Jr, DuVall M R. Endocrinology. 1970;87:1168–1180. doi: 10.1210/endo-87-6-1168. [DOI] [PubMed] [Google Scholar]
- 6.Liu J P, Baker J, Perkins A S, Robertson E J, Efstratiadis A. Cell. 1993;75:59–72. [PubMed] [Google Scholar]
- 7.Baker J, Liu JP, Robertson E J, Efstratiadis A. Cell. 1993;75:73–82. [PubMed] [Google Scholar]
- 8.Cao Q P, Duguay S J, Plisetskaya E, Steiner D F, Chan S J. Mol Endocrinol. 1989;3:2005–2010. doi: 10.1210/mend-3-12-2005. [DOI] [PubMed] [Google Scholar]
- 9.Shamblott M J, Chen T T. Proc Natl Acad Sci USA. 1992;89:8913–8917. doi: 10.1073/pnas.89.19.8913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duguay S J, Lai-Zhang J, Steiner D F, Funkenstein B, Chan S J. J Mol Endocrinol. 1996;16:123–132. doi: 10.1677/jme.0.0160123. [DOI] [PubMed] [Google Scholar]
- 11.Duan C, Inui Y. Gen Comp Endocrinol. 1990;79:326–331. doi: 10.1016/0016-6480(90)90118-6. [DOI] [PubMed] [Google Scholar]
- 12.Duan C, Hirano T. J Exp Zool. 1990;256:347–350. [Google Scholar]
- 13.McCormick S D, Tsai P I, Kelley K M, Nishioka R S, Bern H A. J Exp Zool. 1992;262:166–171. doi: 10.1002/jez.1402620206. [DOI] [PubMed] [Google Scholar]
- 14.Cheng C M, Chen T T. J Endocrinol. 1995;147:67–73. doi: 10.1677/joe.0.1470067. [DOI] [PubMed] [Google Scholar]
- 15.Duan C, Hirano T. J Endocrinol. 1992;133:211–219. doi: 10.1677/joe.0.1330211. [DOI] [PubMed] [Google Scholar]
- 16.Plisetskaya E M, Bondareva V M, Duan C, Duguay S J. Gen Comp Endocrinol. 1993;91:74–80. doi: 10.1006/gcen.1993.1106. [DOI] [PubMed] [Google Scholar]
- 17.Duan C, Noso T, Moriyama S, Kawauchi H, Hirano T. J Endocrinol. 1992;133:221–230. doi: 10.1677/joe.0.1330221. [DOI] [PubMed] [Google Scholar]
- 18.Duan C. In: Biochemistry and Molecular Biology of Fishes. Hochachka P W, Mommsen T P, editors. Vol. 3. Amsterdam: Elsevier Science; 1994. pp. 525–533. [Google Scholar]
- 19.Chirgwin J M, Przbyla A E, MacDonald R J, Rutter W J. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 21.Plisetskaya E M, Mommsen T. Rev Aquatic Sci. 1991;4:225–259. [Google Scholar]
- 22.Seino S, Seino M, Nishi S, Bell G I. Proc Natl Acad Sci USA. 1989;86:114–118. doi: 10.1073/pnas.86.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abbott A M, Bueno R, Pedrini M T, Murray J M, Smith R J. J Biol Chem. 1992;267:10759–10763. [PubMed] [Google Scholar]
- 24.Shier P, Watt V M. J Biol Chem. 1989;264:14605–14608. [PubMed] [Google Scholar]
- 25.Plisetskaya E M, Fabbri E, Moon T W, Gutierrez J, Ottolenghi C. Fish Physiol Biochem. 1993;11:401–409. doi: 10.1007/BF00004590. [DOI] [PubMed] [Google Scholar]
- 26.Gutierrez J, Parrizas M, Maestro M A, Navarro I, Plisetskaya E M. J Endocrinol. 1995;146:35–44. doi: 10.1677/joe.0.1460035. [DOI] [PubMed] [Google Scholar]
- 27.Allendorf F W, Thorgaard G H. In: Evolutionary Genetics of Fishes. Turner B J, editor. New York: Plenum; 1984. pp. 1–53. [Google Scholar]
- 28.Ronner P, Scarpa A. Gen Comp Endocrinol. 1987;65:354–362. doi: 10.1016/0016-6480(87)90120-1. [DOI] [PubMed] [Google Scholar]
- 29.Plisetskaya E M, Buchelli-Narvàez L I, Hardy R W, Dickhoff W W. Comp Biochem Physiol. 1991;98:165. (abstr.). [Google Scholar]