Abstract
Leucine-rich nuclear export signals (NESs) are recognized by the NES receptor exportin 1 and are central to the export of multiple shuttling proteins and RNAs. The export of messenger RNA in vertebrates was, however, thought to occur by a different pathway, because inhibition by injection of a synthetic Rev NES conjugate could not be demonstrated. Here we find that peptide conjugates composed of the NES of either protein kinase A inhibitor protein (PKI) or the HIV-1 Rev protein, when coupled to human serum albumin, are potent inhibitors of mRNA and small nuclear RNA export. These results provide direct evidence that mRNA export in vertebrates depends on interactions between an NES and its cognate NES receptors. PKI NES conjugates are significantly more efficient at inhibiting RNA export than are REV NES conjugates, indicating that different NESs may have different abilities to promote protein and RNA export. Surprisingly, an expected control conjugate containing the mutant Rev NES sequence M10 strongly inhibited the export of intronless dihydrofolate reductase mRNA. Nuclear injection of NES peptide conjugates led to mislocalization to the nucleus of 10–20% of the cytoplasmic Ran GTPase-binding protein (RanBP1) indicating that RanBP1 shuttles between the nucleus and the cytoplasm via an NES pathway. These results demonstrate that in vertebrates the export of mRNA, like that of small nuclear RNA, 5S rRNA, and transport factors such as RanBP1, employs NES-mediated molecular machinery.
Keywords: protein kinase inhibitor, REV, exportin 1
Nuclear export of several proteins is directed by nuclear export signals (NESs) that consist of short amino acid sequences rich in leucine or other aliphatic amino acids plus acidic residues. Whereas NES is a general term for a nuclear export signal, throughout this paper NES refers specifically to the class of leucine-rich NESs, and peptides containing these sequences are named in uppercase letters. Such signals were first identified in the Rev protein of the HIV type 1 (HIV-1) and in the protein kinase A inhibitor protein (PKI) (1–6). Similarly, two yeast proteins that are essential for export of poly(A)+ RNA, Gle1p and Mex67p, also contain functional NES sequences (7, 8).
In yeast two-hybrid experiments, NESs have been shown to interact, perhaps indirectly, with a variety of proteins such as the Rev-interacting proteins (hRip/Rab) (9–12) and several yeast and vertebrate nucleoporins (nuclear pore proteins; refs. 11, 13, 14). In vertebrates, at least one nucleoporin, Nup98, plays an important role in the export of mRNAs, small nuclear RNAs (snRNAs), 5S rRNA, and large rRNAs (15), and two others, Nup153 and Nup214/CAN, may be involved in the export of poly(A)+ RNAs (16, 17).
NES-mediated export is saturable and therefore dependent on one or more limiting cell factors (3, 18). By coupling many copies of the Rev NES peptide CLPPLERLTL onto BSA, Fischer et al. (3) generated an inhibitor of RNA export; as a control they used a mutant form of Rev NES, M10 (1, 19). Injection of these conjugates into Xenopus laevis oocyte nuclei showed that the Rev NES conjugate inhibited Rev–RRE-mediated RNA export (RRE, Rev response element), as well as snRNA and 5S rRNA export. This inhibition was attributed to titration of a putative NES receptor. However, their Rev NES conjugate did not cause inhibition of mRNA export; this finding is surprising because several yeast proteins involved in mRNA export contain NES sequences essential for their action. More recently, a conjugate containing the NES of the yeast protein Mex67p was also reported to inhibit the export of snRNAs and 5S RNA, but not mRNA (8). The lack of effect of these NES conjugates on the export of mRNA was interpreted as indicating that mRNA uses a factor with a signal that is recognized by a different type of receptor.
Recent work has identified the highly conserved protein CRM1 as a major receptor for the NESs of Rev and PKI (and by implication for the NES of other proteins) in both yeast and vertebrates (12, 20, 21). The CRM1 protein, renamed exportin 1, forms a complex with the NES of a “cargo” protein, but does so only in the presence of RanGTP (20), the nuclear form of the Ras-like GTPase Ran (22). This NES cargo–exportin 1–RanGTP trimeric complex is thought to interact directly or indirectly with proteins of the nuclear pore, to facilitate translocation of the NES cargo through the nuclear pore (reviewed in ref. 23). In one study, exportin 1 was found to be required for the export of poly(A)+ nuclear RNAs in yeast (21), making it curious that conjugates containing the Rev NES failed to inhibit the export of mRNA in vertebrates (3).
To analyze the role of NES in RNA export more closely, we tested conjugates containing the NES from PKI for their abilities to affect export from Xenopus oocyte nuclei. Surprisingly, we found that the PKI conjugate strongly inhibited the export of mRNA, in addition to snRNA. This led us to test Rev NES conjugates for comparison, and we found that Rev NES conjugates also inhibited mRNA export, albeit less well than PKI NES conjugates. We find a hierarchy of inhibition, ranging from a very strong inhibition by PKI NES to a moderate inhibition by our Rev NES conjugates, with even the Rev mutant NES M10 conjugate inhibiting export to a slight but significant degree. A scrambled PKI NES sequence gave no inhibition of export. Nuclear injection of the normal PKI NES conjugate also led to accumulation in the nucleus of the NES-containing Ran-binding protein 1 (RanBP1), raising the possibility that mislocalization of this normally cytoplasmic Ran-binding protein contributes to the persistent inhibition of RNA export that we observe at later time points.
MATERIALS AND METHODS
DNA Templates and in Vitro Transcription.
DNA templates for transcription of U3 (24), U1Sm− (25), and U5 snRNAs (26) were generated by PCR amplification of RNA coding regions by using appropriate primers as described previously. The templates for transcription of dihydrofolate reductase (DHFR) mRNA (an intronless mRNA; ref. 27), pre-adenovirus major late (AdML) mRNA (an adenovirus intron-containing pre-mRNA substrate; refs. 15 and 28), and tRNAiMet (ref. 29; E.L. and J.E.D., unpublished data) were generated by linearization of previously described plasmids. In vitro transcription of radiolabeled RNA was performed as described (26, 30) by using [α-32P]GTP and SP6 RNA polymerase (for U3, U1Sm−, U5, and pre-AdML RNAs) or T7 RNA polymerase (for DHFR mRNA and tRNA) according to manufacturer’s conditions (Promega). All RNAs were synthesized with m7GpppG caps (25), except for tRNA, which was made as uncapped RNA.
Peptide Conjugate Preparation.
Peptides of the sequences given in Table 1 were synthesized, and their structures were confirmed by mass spectroscopy (Research Genetics, Huntsville, AL). A cysteine was included at the N terminus of each peptide for use in crosslinking. For preparation of conjugates, human serum albumin (HSA; Calbiochem) was dissolved in 0.1 M sodium bicarbonate, pH 8.5 (conjugation buffer 1), and mixed with the bifunctional crosslinker, m-maleimidobenzoyl-N-hydroxysuccinimide ester (MBS; Pierce) (10 mg/ml in dimethylformamide), to give final concentrations of 10 mg/ml HSA and 2.5 mg/ml MBS. Following a 1-hr incubation at room temperature, the free crosslinker was removed by using a Bio-Gel P6-DG column (Bio-Rad) equilibrated in 0.1 M sodium phosphate, pH 6.0 (conjugation buffer 2). HSA/MBS was pooled, and 5 mg was mixed with 2.5 mg of peptide (10 mg/ml in conjugation buffer 2) and incubated for 1 hr at room temperature. Free peptide was removed by using a Bio-Gel P6-DG column equilibrated in phosphate-buffered saline. Conjugate-containing fractions were pooled and adjusted to the indicated concentrations by using a Microcon 30 concentrator (Amicon). For production of biotinylated conjugates, HSA (10 mg in 1 ml of conjugation buffer 1) was first incubated with 0.4 mg of N-hydroxysuccinimide-LC-biotin II (Pierce) for 1 hr at 4°C. Biotinylated HSA was separated from free biotin by several rounds of concentration with a Centriprep 10 (Amicon). Peptide conjugation was then performed as described above. The number of peptides per conjugate was estimated following gel electrophoresis and comparison with HSA/MBS. Protein concentrations were determined by the Bio-Rad protein assay. Once conjugated to HSA, the peptide sequences of the REV, M10, and PKI peptides (Table 1) were again confirmed by 10 rounds of Edman sequencing on an Applied Biosytems Procise Microsequencer.
Table 1.
Amino acid content of peptide conjugates
| Conjugate | Peptide |
|---|---|
| PKI | CNELALKLAGLDINKT |
| MIX | CTKNILDGALKLALNE |
| REV | CLPPLERLTLD |
| BSA-R | CLPPLERLTL |
| M10 | CLPPDLRLTLD |
Peptides PKI, MIX, REV, and M10 were conjugated to HSA and used in injection experiments as described in the text. BSA-R refers to the Rev-NES peptide conjugate used by Fischer et al. (3). Leucine residues shown to be critical for function of the nuclear export signals (1, 2, 4) are in bold. Mix is a scrambled PKI NES.
Oocyte Injections and Analysis of RNA and Protein Transport.
Preparation and injection of X. laevis oocytes were performed as previously described (24–26). To analyze the effect of peptide conjugates on the export of specific RNAs, 1–5 fmol of individual 32P-labeled RNAs was mixed with the peptide conjugates in 9 mM NaCl/0.6 mM DTT/0.25 unit/μl RNasin and injected in the amounts indicated in the figure legends. The mixtures were injected into the nuclei of oocytes, and at the times indicated in the figure legends, the oocytes were dissected under oil (31). The nucleocytoplasmic distributions of the RNAs were analyzed by polyacrylamide gel electrophoresis, as previously described (26, 32). U3 snRNA, which normally is not exported from the nucleus (24), and blue dextran (29) were included in all injection experiments as controls for the accuracy of nuclear injection and oocyte dissection.
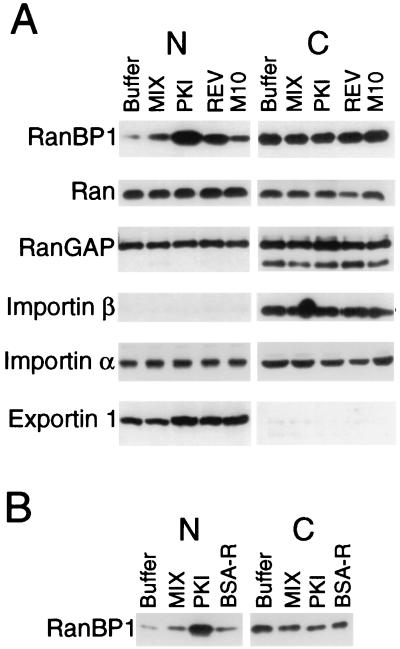
To analyze the effects on the intracellular localization of endogenous transport factors, the peptide conjugates were injected into the nuclei of oocytes in the amounts indicated in the legend to Fig. 4. The oocytes were subsequently dissected into nuclear and cytoplasmic fractions, and the nuclear proteins were solubilized directly in 1× SDS gel loading buffer. Cytoplasms were homogenized in 10 mM Mops, pH 7.2/75 mM KCl/25 mM NaCl/2 mM DTT/1 μg/ml leupeptin/1 μg/ml pepstatin/2 μg/ml aprotinin. After removal of the yolk from the cytoplasmic extract by centrifugation at 14,000 rpm in a microcentrifuge for 4 min, the cleared extract was mixed 1:1 with 2× SDS gel loading buffer before resolution by gel electrophoresis. Proteins from the nuclear and cytoplasmic fractions, as indicated in the legend to Fig. 4, were separated in SDS-containing 10% polyacrylamide gels (33). The distributions of specific proteins were analyzed by immunoblotting by using appropriate antibodies to the different transport factors and ECL according to the manufacturer’s protocol (Amersham). Rabbit polyclonal antibodies were the generous gifts of M. Dasso (National Institute of Child Health and Human Development, Bethesda, MD) [anti-Ran GTPase-activating protein (RanGAP), anti-Ran, and anti-RanBP1], M. Moore (Baylor College of Medicine, Houston, TX) (anti-importin β), F. Grosveld (St. Jude’s Hospital, Memphis, TN) [anti-CRM1 (exportin 1)], and D. Görlich (ZMB Universität, Heidelberg, Germany) (anti-importin α) and were used for immunoblotting at the following dilutions: anti-Ran, 1:1,000; anti-RanGAP, 1:2,000; anti-RanBP1, 1:2,000; anti-importin α, 1:10,000; anti-importin β, 1:750; and anti-CRM1, 1:2,000.
Figure 4.
RanBP1 mislocalizes to the nucleus in the presence of NES peptide conjugates. (A) The PKI and REV NES peptide conjugates affect the intracellular distribution of RanBP1 but not other tested components of the transport machinery. The indicated peptide conjugates (≈115 ng per nucleus) were injected into oocyte nuclei, and 6.5 hr later the oocytes were dissected. The protein extracts were separated in SDS-containing 10% polyacrylamide gels, and the distributions of specific proteins were analyzed by immunoblotting by using appropriate antibodies and ECL. Oocyte equivalents of the nuclear (N) and cytoplasmic (C) extracts varied for the different proteins: RanBP1, importin β, importin α, and exportin, 3.0 (N) and 0.1 (C); Ran, 1 (N) and 0.5 (C); and RanGAP, 0.5 (N) and 0.1 (C). (B) The PKI NES conjugate, but not the BSA-R conjugate, causes prolonged mislocalization of RanBP1. Oocyte nuclei were each injected with ≈130 ng (MIX and PKI) or ≈190 ng (BSA-R) of peptide conjugates or buffer. Nineteen hours later the oocytes were dissected into nuclear (N) and cytoplasmic (C) fractions. The distribution of RanBP1 in 2 (N) and 0.1 (C) oocyte equivalents was analyzed by immunoblotting as in A.
RESULTS
Inhibition of mRNA Export by a PKI NES Conjugate.
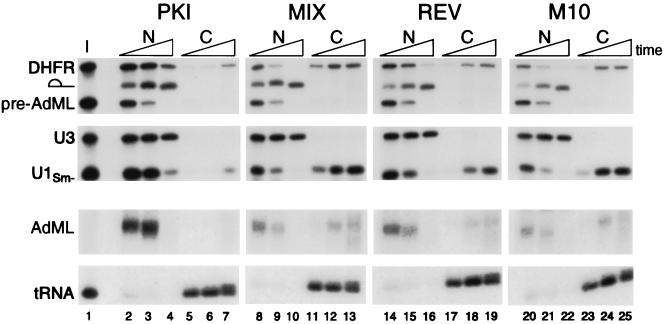
We asked whether a PKI NES conjugate could competitively inhibit the export of snRNA from oocyte nuclei, as had been previously reported for Rev and Mex67p NES conjugates (3, 8). PKI NES peptides conjugated to HSA (Table 1) did indeed inhibit the export of U1 snRNA (Fig. 1, lanes 2–7). Surprisingly, the PKI conjugate was also a very effective inhibitor of the export of DHFR mRNA and spliced AdML mRNA (lanes 2–7). The export of tRNA was unaffected by the PKI conjugate, demonstrating that the inhibition was specific. A control conjugate, containing a peptide with the same amino acids as PKI NES, but arranged in a mixed order that does not resemble an NES sequence (termed MIX conjugate; Table 1) had no effect on the export of any class of RNA (Fig. 1, lanes 8–13).
Figure 1.
Inhibition of mRNA and snRNA export by the PKI NES conjugate. The PKI and REV NES conjugates differ in their abilities to inhibit RNA export. One to three femtomoles of each 32P-labeled DHFR mRNA, AdML pre-mRNA, U3 snRNA, U1Sm− snRNA, and tRNAiMet were coinjected into oocyte nuclei together with ≈20 ng per nucleus of the PKI NES (lanes 2–7), MIX (lanes 8–13), REV NES (lanes 14–19), or M10 (lanes 20–25) peptide conjugates (see Table 1). The oocytes were dissected 1, 4, and 20 hr after injection, and RNA was extracted from the nuclear (N) and cytoplasmic (C) fractions; 0.5 oocyte equivalent of RNA was resolved on denaturing 8% polyacrylamide gels and visualized by autoradiography. The AdML mRNA and tRNAiMet panels were exposed twice as long as the upper panels, to allow for detection of AdML mRNA export. The triangles at the top of the lanes indicate increasing times since injection. U3 snRNA, which normally is not exported from the nucleus (24), served as a marker for the accuracy of nuclear injections and oocyte dissections. The RNA injection mixture (I) is shown in lane 1. Pre-AdML RNA is the injected form of the intron-containing pre-mRNA; the lariat symbol represents the excised AdML intron, and AdML represents the spliced mRNA, which normally is exported from the nucleus.
The amount of PKI conjugate used in the experiments shown above (≈20 ng per nucleus) was low, only about one-seventh as much as Fischer et al. (3) used in studies with their Rev NES conjugate (designated BSA-R in Table 1). To compare the NES of PKI and Rev more closely, we synthesized a Rev NES peptide (designated REV in Table 1); this conjugated peptide contains at its C terminus an aspartic acid residue that is not present in BSA-R but is present in Rev protein. When a low amount of our REV conjugate (≈20 ng) was injected into oocytes, very little inhibition of U1 RNA or mRNA export was observed (Fig. 1, lanes 14–19), consistent with the previous study of Fischer et al. (3). Only at early time points were the levels of exported U1 RNA different in oocytes that had been injected with the REV conjugate vs. a mutant REV conjugate, M10 (Table 1), in which two amino acids were changed (Fig. 1, lanes 20–25). Thus, the PKI conjugate is a much better inhibitor of RNA export than is the REV conjugate. This same low level of the PKI conjugate was also a very effective inhibitor of RNA export mediated by Rev-RRE (data not shown), indicating that the PKI conjugate most likely titrates export factors utilized by the Rev NES. Finally, even the low level of the PKI conjugate used in Fig. 1 was in excess over what was needed to interfere with RNA export because injection of 5- to 10-fold lower amounts of the PKI conjugate retarded the export of both snRNAs and mRNAs (data not shown).
Inhibition of mRNA Export by a REV NES Conjugate.
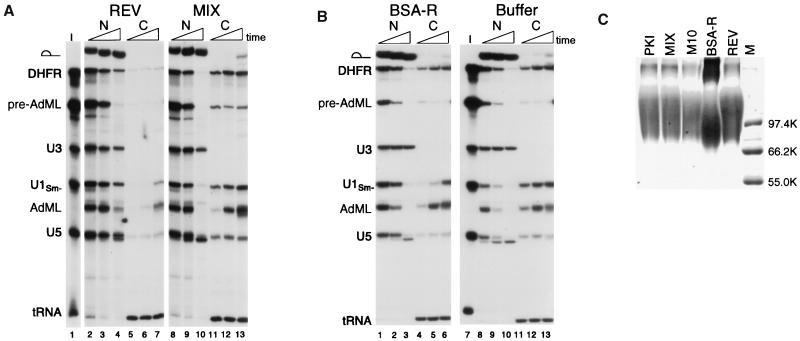
When higher amounts of our REV conjugate were injected at a level comparable with that used by Fischer et al. (3) (≈130 ng per nucleus), the export of snRNAs was effectively blocked (U1 and U5; Fig. 2A, lanes 2–7), as had been observed previously. However, our Rev conjugate also inhibited the export of both DHFR mRNA and spliced AdML mRNA (compare REV to MIX, Fig. 2A). The REV conjugate did not affect export of tRNAs, again showing that the inhibition of export was specific. Thus, both the PKI conjugate and the REV conjugate that we tested blocked export of mRNAs, as well as snRNAs. However, the REV conjugate was less potent, requiring a higher injected concentration for export inhibition. Because these results differed from those of Fischer et al. (3) with regard to inhibition of mRNA export, we repeated our experiments by using BSA-R, a REV NES conjugate provided by U. Fischer (IMT, Philipps-Universität, Marburg, Germany). In that case, we obtained results identical to those previously reported by him for this conjugate: inhibition of snRNA export at early time points, relief of inhibition of snRNA export at late time points, and no inhibition of mRNA export (Fig. 2B, lanes 1–6). Thus, the conjugates used by Fischer et al. and by us differ in activity.
Figure 2.
Inhibition of mRNA export by the REV NES conjugate. (A) The REV NES conjugate inhibits mRNA and snRNA export when present in high amounts. Mixtures containing 32P-labeled DHFR mRNA, AdML pre-mRNA, U3 snRNA, U1Sm− snRNA, U5 snRNA, and tRNAiMet (I, lane 1) were coinjected into oocyte nuclei together with ≈130 ng per nucleus of the REV (lanes 2–7) or the MIX (lanes 8–13) peptide conjugates. Oocytes were dissected 1.5, 4, and 21 hr after injection, and RNA export was analyzed as in Fig. 1. The export of a small amount of pre-AdML RNA is because of saturation of the splicing machinery by the bolus of injected RNA. (B) The BSA-R conjugate fails to inhibit mRNA export even when present in high amounts. Mixtures containing 32P-labeled DHFR mRNA, AdML pre-mRNA, U3 snRNA, U1Sm− snRNA, U5 snRNA, and tRNAiMet (I, lane 7) were coinjected into oocyte nuclei together with ≈190 ng per nucleus of the BSA-R (lanes 1–6) conjugate or with buffer alone (lanes 8–13). Oocytes were dissected 1.5, 4, and 18.5 hr after injection, and RNA export was analyzed as in Fig. 1. (C) SDS/PAGE analysis of peptide conjugates. Ten micrograms of each of the indicated peptide conjugates (Table 1) were subjected to electrophoresis in an SDS-containing 8% polyacrylamide gel, and the proteins were detected by Coomassie blue staining. The marker (M) lane contains 0.5 μg of each protein molecular weight marker (Promega).
Compositions of the Conjugates.
We questioned how the preparations of Rev NES conjugates used by Fischer et al. (3) and by us might differ. The presence of biotin on our HSA carrier protein is unlikely to be significant, because control experiments performed with biotin-free REV conjugate gave similar results (data not shown); moreover, the MIX conjugate, which was biotinylated, did not inhibit export even when injected at high concentrations (Fig. 2A and Fig. 3B). Previous studies have shown that the presence of biotin does not affect the export of conjugates from vertebrate nuclei (18). The use of HSA, rather than BSA, as the carrier protein is not the critical difference, because the MIX-HSA conjugate has no effect on export. Sequencing of our REV and M10 peptide conjugates confirmed that their sequences were as presented in Table 1 (data not shown). One obvious difference common to both our REV conjugate and M10 conjugate is the presence of an aspartic acid residue at the C terminus of the peptide on each conjugate (Table 1). However, when we resynthesized and tested the REV and M10 peptides in the form of nonbiotinylated conjugates with or without this aspartic acid, we observed no detectable difference in the pattern of RNAs whose export was inhibited, although there was a modest but reproducible decrease in the ability of the shorter conjugates to inhibit RNA export (data not shown).
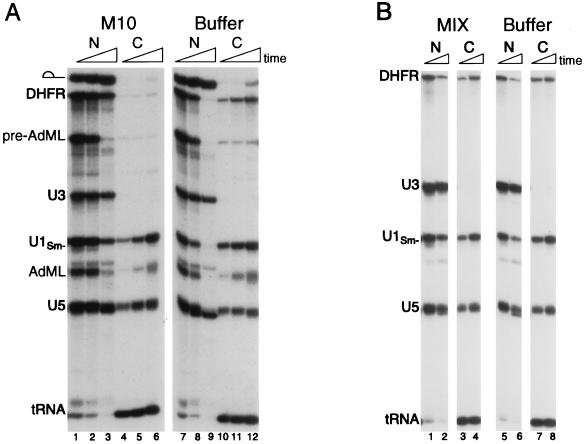
Figure 3.
The M10 but not the MIX peptide conjugate can inhibit RNA export. (A) The M10 conjugate is a potent inhibitor of DHFR mRNA export. Mixtures containing 32P-labeled DHFR mRNA, AdML pre-mRNA, U3 snRNA, U1Sm− snRNA, U5 snRNA, and tRNAiMet were coinjected into oocyte nuclei with ≈130 ng per nucleus of the M10 (lanes 1–6) peptide or with buffer only (lanes 7–12). Oocytes were dissected 1.5, 4, and 18 hr after injection, and RNA export was analyzed as in Fig. 1. (B) The MIX conjugate does not interfere with RNA export, even when present in high amounts. Mixtures containing 32P-labeled DHFR mRNA, U3 snRNA, U1Sm− snRNA, U5 snRNA, and tRNAiMet were coinjected into oocyte nuclei with ≈130 ng per nucleus of the MIX (lanes 1–4) peptide or with buffer alone (lanes 5–8). Oocytes were dissected 1.5 and 4 hr after injection, and RNA export was analyzed as in Fig. 1.
It is possible that the conditions under which the peptides were conjugated to the protein carriers are responsible for the differences in the abilities of the conjugated proteins to inhibit mRNA export. We carried out peptide conjugation for 1 hr, whereas Fischer et al. (3) carried out the peptide conjugation reaction for ca. 16 hr. By polyacrylamide gel electrophoresis (Fig. 2C), we estimate that our conjugates and the BSA-R conjugate furnished to us by Fischer have about the same average number of peptides per carrier (≈15–20). However, the electrophoretic profiles of the two conjugates indicated that a significant proportion of the BSA-R conjugate is in very large complexes, possibly reflecting extensive crosslinking between the conjugates.
Inhibition of DHFR mRNA Export by the M10 Conjugate.
When comparing the inhibitory activities of the different NES conjugates relative to their controls, we noted a difference between the M10 and MIX control conjugates. The M10 conjugate itself, even when injected at the relatively low amount in Fig. 1, appeared to inhibit export of DHFR mRNA slightly (Fig. 1, compare lanes 8–13 and 20–25). Therefore, we asked whether the M10 conjugate, at higher levels, might interfere with RNA export. Indeed, at higher concentrations, M10 conjugate strongly inhibited the export of DHFR mRNA and, to a lesser extent, the export of spliced AdML mRNA and snRNAs (Fig. 3A, lanes 1–6 compared with lanes 7–12); however, this M10 conjugate did not block Rev-mediated export of an RRE-containing RNA (data not shown). In contrast, even high amounts of the control MIX conjugate had no effect on RNA export when compared with injected buffer alone (Fig. 3B).
Protein and RNA Import in the Presence of the PKI NES.
To test whether the efficient and broad inhibition of mRNA and snRNA export by the PKI NES conjugate reflected nonspecific effects on nuclear pore function, we monitored protein import from the cytoplasm in the presence of the conjugate. As expected, the PKI conjugate caused only a slight reduction in the rate of import of radioactively labeled nuclear proteins that had been injected into the cytoplasm (data not shown).
Although the import of proteins does not appear to be inhibited by the NES conjugates, the import of certain RNAs was significantly retarded. Injection of the PKI conjugate into the nucleus inhibited by ≈2- to ≈3-fold the rate of import of both U6 RNA and NL-15 RNA, an RNA whose import into the nucleus requires interaction with the nuclear antigen La (34) (data not shown). However, the PKI conjugate did not affect the import of U1 or U5 small nuclear ribonucleoproteins into the nucleus, which has been proposed to occur by a pathway different than that used by U6 small nuclear ribonucleoproteins (35, 36). The results reported here suggest that import of U6 and NL-15 RNAs depends on the presence of shuttling proteins whose export from the nucleus to the cytoplasm is affected by the NES conjugates (37).
Nuclear Mislocalization of RanBP1.
To examine whether endogenous transport factors might be sequestered in the nucleus as a consequence of competition for NES receptors by the NES conjugates, we analyzed the intracellular distribution of the transport-related factors Ran, RanBP1, RanGAP, importin β, importin α, and exportin 1 by immunoblotting. For the majority of the proteins, no significant change in the nucleocytoplasmic distribution was evident (Fig. 4A). However, ≈10–20% of the RanBP1 was observed to accumulate in the nucleus in the presence of the PKI or REV conjugates but not in the presence of the MIX or M10 conjugates. This result supports the proposal that RanBP1, which has an essential NES, normally cycles between the nucleus and cytoplasm (38, 39), as blockage of its export by the NES conjugate would result in its accumulation in the nucleus. Aberrant localization of RanBP1 protein was observed for at least 19 hr after injection of the PKI conjugate (Fig. 4B), consistent with the persistent inhibition of RNA export (Fig. 1). In contrast, at this same time point we detected no significant nuclear accumulation of RanBP1 in oocytes injected with BSA-R; that may account for the eventual alleviation of inhibition of snRNA export by this conjugate (Fig. 2B; ref. 3).
DISCUSSION
The export of RNAs from the nucleus is believed to be mediated by specific RNA binding proteins (reviewed in ref. 40). A well known example of such a protein is the HIV factor Rev, in which a specific NES sequence has been characterized (reviewed in ref. 41). Similar leucine-rich NES sequences have been found in PKI and in several factors implicated in poly(A)+ RNA export in yeast. A potential NES receptor, exportin 1, has recently been identified in both yeast and higher eukaryotes (reviewed in ref. 23).
Peptides consisting of an NES sequence coupled to a carrier protein such as serum albumin act as competitive inhibitors of Rev-mediated export and of cellular snRNA and 5S rRNA export, presumably by titration of an NES receptor. The inhibition of export of several RNAs—but not of mRNA—by the REV NES conjugate used by Fischer et al. (3) led to the proposal that distinct signals and receptors are used for the export of the different classes of RNA. Specifically, they proposed that Rev, via its NESs, makes use of a receptor normally used for the export of snRNAs and 5S rRNA but that mRNA uses a different system (3). The recent identification of exportin 1 as a receptor for Rev NES and PKI NES (20, 21) is not inconsistent with this possibility. However, export of poly(A)+ RNA in yeast depends on NES-containing proteins as well as exportin 1 and related proteins (7, 8, 42), making it likely that similar factors also function in the export of mRNAs in higher eukaryotes. Paradoxically, before this report no NES conjugate had been shown to be capable of inhibiting mRNA export in vertebrates (3, 8).
The experiments presented here show that in higher eukaryotes mRNA export is effectively blocked by competition with NES conjugates containing either PKI or REV NES. It is unclear why this inhibition of mRNA export by NES conjugates has escaped notice until now. Possibly, most experiments of this type used conjugates that were much more extensively crosslinked, as a consequence of carrying out the conjugation reaction for 16–18 hr rather than the 1 hr used here (see Materials and Methods). The extra time of incubation with the crosslinking reagent apparently allowed for formation of a significant proportion of larger aggregates between several protein molecules (Fig. 2C), which might differ in their interactions with NES receptors or receptor complexes.
Our REV conjugate and the BSA-R conjugate used by Fischer et al. (3) differ from each other functionally in their abilities to inhibit mRNA export, although the NES peptides differ chemically only by the presence of the aspartic acid residue in our conjugate (Table 1). This extra residue cannot be responsible for the difference in activities because we resynthesized a new peptide conjugate identical in amino acid sequence to the peptide in BSA-R and found that this minimal Rev NES conjugate inhibited both snRNA and mRNA export (data not shown). Regardless of the differences between the REV and BSA-R conjugates, the fact remains that the REV NES conjugate inhibits the export of mRNA, as well as snRNA.
Whereas Rev is a bifunctional export factor that binds directly to RNA and to exportin 1, it is possible that comparable cellular mRNA export factors have these functions on separate polypeptides. Thus, an RNA binding protein may gain access to exportin 1 or a similar receptor through protein–protein interactions with an NES-containing protein. Therefore the factor(s) responsible for cellular mRNA export are not necessarily bifunctional RNA binding proteins that contain an NES (7).
Unexpectedly, we found that the M10 conjugate could also inhibit RNA export to some extent, being most effective on DHFR mRNA export and less so on AdML mRNA or snRNA export. This pattern of inhibition mimics the inhibition seen after nuclear injection of an excess of heterogeneous nuclear ribonucleoprotein (hnRNP) A1 protein (43, 44) or the DHFR mRNA itself (3, 32). The inability of the M10 conjugate to interfere with Rev-mediated export (data not shown) or the shuttling of RanBP1 (Fig. 4) indicates that it targets a specific receptor. Thus, the M10 peptide conjugate may affect interactions such as those between hnRNP A1 protein and its cognate nuclear export receptor. The inhibition of export of certain mRNAs by M10 conjugate observed here might also explain why expression of M10 mutant Rev protein is toxic in stably transformed CMT3 cells (M.-L. Hammarskjöld, personal communication) and in Saccharomyces cerevisiae (C. M. Hammell and C. N. Cole, unpublished results cited in ref. 45). Because of the ability of this conjugate to inhibit mRNA export, its use as an “inactive” control for Rev NES conjugates in other studies may be problematic.
The stronger inhibitory activity of the PKI NES peptide conjugate relative to the Rev NES correlates well with the greater ability of the PKI NES to support the formation of a trimeric complex of NES peptide, exportin 1, and RanGTP in vitro (20). The differences between the NESs could result from inherent affinities of their “minimal” NES sequences for a receptor or from differences in the contexts in which they are presented to a receptor. Context certainly seems to be important, as a version of the PKI NES that is longer than the one used here (GSNELALKLAGLDINKTGGC; cf. ref. 4) is an even more effective inhibitor of RNA export (data not shown). Similarly, the addition of an aspartic acid residue to the C terminus of the Rev NES modestly increased the ability of that NES conjugate to inhibit RNA export, in keeping with the finding that this residue is important for the function of Rev protein in the expression of nonspliced HIV pre-mRNA in mammalian cells (1, 46). However, these changes in overall efficiency of inhibition do not alter the spectrum of targets affected. For example, addition of the extra aspartic acid residue did not differentially affect the ability of the REV NES conjugate to block mRNA export relative to snRNA export. Similarly, a shorter version of the PKI NES (LALKLAGLDI) inhibited the export of mRNAs and snRNAs comparably (data not shown).
The NES conjugates also affected the localization of at least one transport factor. Injection of either the PKI or the Rev NES conjugate led to nuclear accumulation of 10–20% of the normally cytoplasmic RanBP1. Because RanBP1 contains a functional NES, it has been proposed to shuttle between the nucleus and cytoplasm (38, 39); inappropriate accumulation of this protein in the nucleus might also contribute to the overall inhibition of nuclear export. In contrast, the distributions of several other transport factors such as importin α, importin β, exportin 1, Ran, and RanGAP were not significantly affected by the conjugates. The distribution of importin β, which appears to have an NES (47), is not affected by the NES conjugate, indicating that this NES may not be required for its recycling in vertebrate cells. Importin α and Ran are exported by using other export receptors (48), which might not be sensitive to the inhibitory actions of the NES conjugates.
We have shown here that mRNA export in vertebrates can be blocked by NES conjugates, indicating that it utilizes NES-containing proteins and thus resembles mRNA export in yeast. Our results do not yet resolve the question as to whether all NES-dependent export uses a single NES receptor (e.g., exportin 1) or multiple receptors; however, the selective inhibition of the export of certain RNAs by the M10 and the BSA-R conjugates is consistent with the latter possibility. In either case, the finding that mRNA export occurs in a manner similar to that of snRNAs, 5S rRNA, and HIV Rev-mediated RNA export demonstrates that general features exist and that these are likely to be mechanistically similar in yeast and vertebrates.
Acknowledgments
We thank Ariane Grandjean and Susanne Imboden for excellent technical assistance and Dr. Frank Masiarz, Chiron Corporation, for N-terminal protein sequencing of the peptide conjugates. We thank Drs. Katharine Ullman, Christian Grimm, and Jeannine Petersen for stimulating discussions. We also thank Dr. Susan Taylor (University of California, San Diego) for the long version of the PKI NES peptide and Dr. Utz Fischer for providing the BSA-R REV peptide conjugate. We thank Drs. Mary Dasso, Frank Grosveld, Dirk Görlich, and Mary Moore for generously providing us with antibodies. This work was supported by National Institutes of Health Grant GM30220 (to J.E.D.) and National Institutes of Health Grant GM33279 and American Cancer Society Grant CB199 (to D.F.). A.E.P. was supported in part by a gift from the Lucille Markey Foundation and a National Institutes of Health predoctoral training grant.
ABBREVIATIONS
- NES
nuclear export signal
- PKI
protein kinase A inhibitor protein
- RanBP1
Ran-binding protein 1
- snRNA
small nuclear RNA
- RRE
Rev response element
- DHFR
dihydrofolate reductase
- AdML
adenovirus major late
- HSA
human serum albumin
- MBS
m-maleimidobenzoyl-N-hydroxysuccinimide ester
- RanGAP
Ran GTPase-activating protein
References
- 1.Malim M H, McCarn D F, Tiley L S, Cullen B R. J Virol. 1991;65:4248–4254. doi: 10.1128/jvi.65.8.4248-4254.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyer B E, Malim M H. Genes Dev. 1994;8:1538–1547. doi: 10.1101/gad.8.13.1538. [DOI] [PubMed] [Google Scholar]
- 3.Fischer U, Huber J, Boelens W C, Mattaj I W, Lührmann R. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 4.Wen W, Meinkoth J L, Tsien R Y, Taylor S S. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 5.Bogerd H P, Fridell R A, Benson R E, Hua J, Cullen B R. Mol Cell Biol. 1996;16:4207–4214. doi: 10.1128/mcb.16.8.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim F J, Beeche A A, Hunter J J, Chin D J, Hope T J. Mol Cell Biol. 1996;16:5147–5155. doi: 10.1128/mcb.16.9.5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murphy R, Wente S R. Nature (London) 1996;383:357–360. doi: 10.1038/383357a0. [DOI] [PubMed] [Google Scholar]
- 8.Segref A, Sharma K, Doye V, Hellwig A, Huber J, Lührmann R, Hurt E. EMBO J. 1997;16:3256–3271. doi: 10.1093/emboj/16.11.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bogerd H P, Fridell R A, Madore S, Cullen B R. Cell. 1995;82:485–494. doi: 10.1016/0092-8674(95)90437-9. [DOI] [PubMed] [Google Scholar]
- 10.Fritz C C, Zapp M L, Green M R. Nature (London) 1995;376:530–533. doi: 10.1038/376530a0. [DOI] [PubMed] [Google Scholar]
- 11.Stutz F, Neville M, Rosbash M. Cell. 1995;82:495–506. doi: 10.1016/0092-8674(95)90438-7. [DOI] [PubMed] [Google Scholar]
- 12.Neville M, Stutz F, Lee L, Davis L I, Rosbash M. Curr Biol. 1997;7:767–775. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- 13.Stutz F, Izaurralde E, Mattaj I W, Rosbash M. Mol Cell Biol. 1996;16:7144–7150. doi: 10.1128/mcb.16.12.7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fritz C C, Green M R. Curr Biol. 1996;6:848–854. doi: 10.1016/s0960-9822(02)00608-5. [DOI] [PubMed] [Google Scholar]
- 15.Powers M A, Forbes D J, Dahlberg J E, Lund E. J Cell Biol. 1997;136:241–250. doi: 10.1083/jcb.136.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bastos R, Lin A, Enarson M, Burke B. J Cell Biol. 1996;134:1141–1156. doi: 10.1083/jcb.134.5.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Deursen J, Boer J, Kasper L, Grosveld G. EMBO J. 1996;15:5574–5583. [PMC free article] [PubMed] [Google Scholar]
- 18.Meyer B E, Meinkoth J L, Malim M H. J Virol. 1996;70:2350–2359. doi: 10.1128/jvi.70.4.2350-2359.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malim M H, Böhnlein S, Hauber J, Cullen B R. Cell. 1989;58:205–214. doi: 10.1016/0092-8674(89)90416-9. [DOI] [PubMed] [Google Scholar]
- 20.Fornerod M, Ohno M, Yoshida M, Mattaj I W. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 21.Stade K, Ford C S, Guthrie C, Weis K. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 22.Moore M S, Blobel G. Nature (London) 1993;365:661–663. doi: 10.1038/365661a0. [DOI] [PubMed] [Google Scholar]
- 23.Ullman K S, Powers M A, Forbes D J. Cell. 1997;90:967–970. doi: 10.1016/s0092-8674(00)80361-x. [DOI] [PubMed] [Google Scholar]
- 24.Terns M P, Dahlberg J E. Science. 1994;264:959–961. doi: 10.1126/science.8178154. [DOI] [PubMed] [Google Scholar]
- 25.Terns M P, Dahlberg J E, Lund E. Genes Dev. 1993;7:1898–1908. doi: 10.1101/gad.7.10.1898. [DOI] [PubMed] [Google Scholar]
- 26.Pasquinelli A E, Dahlberg J E, Lund E. RNA. 1995;1:957–967. [PMC free article] [PubMed] [Google Scholar]
- 27.Kambach C, Mattaj I W. J Cell Biol. 1992;118:11–21. doi: 10.1083/jcb.118.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamm J, Mattaj I W. Cell. 1990;63:109–118. doi: 10.1016/0092-8674(90)90292-m. [DOI] [PubMed] [Google Scholar]
- 29.Jarmolowski A, Boelens W C, Izaurralde E, Mattaj I W. J Cell Biol. 1994;124:627–635. doi: 10.1083/jcb.124.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melton D A, Krieg P A, Rebagliati M R, Maniatis T, Zinn K, Green M R. Nucleic Acids Res. 1984;12:7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lund E, Paine P L. Methods Enzymol. 1990;181:36–43. doi: 10.1016/0076-6879(90)81110-g. [DOI] [PubMed] [Google Scholar]
- 32.Pasquinelli, A. E., Ernst, R. K., Lund, E., Grimm, C., Zapp, M. L., Rekosh, D., Hammarskjöld, M.-L. & Dahlberg, J. E. (1997) EMBO J. 16, in press. [DOI] [PMC free article] [PubMed]
- 33.Adolph K W, Cheng S M, Laemmli U K. Cell. 1977;12:805–816. doi: 10.1016/0092-8674(77)90279-3. [DOI] [PubMed] [Google Scholar]
- 34.Grimm C, Lund E, Dahlberg J E. EMBO J. 1997;16:793–806. doi: 10.1093/emboj/16.4.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fischer U, Darzynkiewicz E, Tahara S M, Dathan N A, Lührmann R, Mattaj I W. J Cell Biol. 1991;113:705–714. doi: 10.1083/jcb.113.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michaud N, Goldfarb D S. J Cell Biol. 1991;112:215–223. doi: 10.1083/jcb.112.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dahlberg J E, Lund E. Semin Cell Dev Biol. 1997;8:65–70. doi: 10.1006/scdb.1996.0123. [DOI] [PubMed] [Google Scholar]
- 38.Richards S A, Lounsbury K M, Carey K L, Macara I G. J Cell Biol. 1996;134:1157–1168. doi: 10.1083/jcb.134.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zolotukhin A S, Felber B K. J Biol Chem. 1997;272:11356–11360. doi: 10.1074/jbc.272.17.11356. [DOI] [PubMed] [Google Scholar]
- 40.Görlich D, Mattaj I W. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 41.Gerace L. Cell. 1995;82:341–344. doi: 10.1016/0092-8674(95)90420-4. [DOI] [PubMed] [Google Scholar]
- 42.Seedorf M, Silver P A. Proc Natl Acad Sci USA. 1997;94:8590–8595. doi: 10.1073/pnas.94.16.8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Izaurralde E, Jarmolowski A, Beisel C, Mattaj I W, Dreyfuss G, Fischer U. J Cell Biol. 1997;137:27–35. doi: 10.1083/jcb.137.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saavedra C A, Felber B K, Izaurralde E. Curr Biol. 1997;7:619–628. doi: 10.1016/s0960-9822(06)00288-0. [DOI] [PubMed] [Google Scholar]
- 45.Saavedra C A, Hammell C M, Heath C V, Cole C N. Genes Dev. 1997;11:2845–2856. doi: 10.1101/gad.11.21.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mermer B, Felber B K, Campbell M, Pavlakis G N. Nucleic Acids Res. 1990;18:2037–2044. doi: 10.1093/nar/18.8.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iovine M K, Wente S R. J Cell Biol. 1997;137:797–811. doi: 10.1083/jcb.137.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kutay U, Bischoff F R, Kostka S, Kraft R, Görlich D. Cell. 1997;90:1061–1071. doi: 10.1016/s0092-8674(00)80372-4. [DOI] [PubMed] [Google Scholar]