Abstract
Nuclear hormone receptors are potent repressors of transcription in the unliganded state. We describe here the cloning of a nuclear receptor corepressor that we call SUN-CoR (Small Unique Nuclear receptor CoRepressor), which shows no homology to previously described nuclear hormone receptor corepressors, N-CoR, or SMRT. SUN-CoR is a highly basic, 16-kDa nuclear protein that is expressed at high levels in adult tissues and is induced during adipocyte and myogenic differentiation. SUN-CoR potentiates transcriptional repression by thyroid hormone receptor and RevErb in vivo, represses transcription when fused to a heterologous DNA binding domain, and interacts with RevErb as well as with thyroid hormone receptor in vitro. SUN-CoR also interacts with N-CoR and SMRT in vitro and with endogenous N-CoR in cells. We conclude that SUN-CoR is a corepressor and may function as an additional component of the complex involved in transcriptional repression by unliganded and orphan nuclear hormone receptors.
Gene expression is regulated at the transcriptional level by sequence-specific transcriptional activators and repressors. The mechanism by which transcriptional repressors modulate gene expression is beginning to be elucidated. One mechanism involves protein–protein interaction with the basal transcription apparatus preventing recruitment of general transcription factors or maintaining the complex in a conformation that is inactive for transcriptional initiation. Many activation and repression domains have been shown to interact directly with components of the basal transcription apparatus, whereas others interact indirectly by first contacting an intermediate protein termed a coactivator or corepressor (reviewed in ref. 1). Another mechanism involves modification of chromatin structure in the vicinity of the promoter. There is convincing evidence that packaging of DNA into nucleosomes represses gene expression by inhibiting transcriptional initiation (2, 3). Also, hyperacetylation of histone tails has been associated with actively transcribed DNA, and hypoacetylation is associated with inactive heterochromatin (reviewed in ref. 4). Thus, transcriptional regulatory proteins may function by modifying nucleosome positioning, structure, or functional groups to modulate gene expression either directly or by the recruitment of proteins and enzymatic activities that mediate these effects.
One class of repressor proteins is the nuclear hormone receptors (NHRs) such as thyroid hormone receptor (TR) and retinoic acid receptor, which repress basal transcription in the absence of ligand (5). The cloning of two NHR corepressors has begun to shed light on the mechanism by which these receptors silence transcription. The corepressors, called N-CoR and SMRT, are large (270 and 168 kDa, respectively) proteins that share significant homology to each other (6, 7). It is hypothesized that in the absence of ligand, TR and retinoic acid receptor bind to a member of this family of corepressors and, upon ligand binding, the corepressor is released, relieving repression and permitting transcriptional activation. The AF2 amphipathic α-helix present in the C terminus of many NHRs is necessary for releasing corepressor (7–9) as well as subsequently activating transcription (10–12).
RevErb is an orphan receptor encoded on the noncoding strand of the thyroid receptor α gene (13, 14). It is induced during adipocyte differentiation (15) and is down-regulated during muscle differentiation (16). RevErb has no known ligand, lacks the AF2 domain, and constitutively represses transcription when bound as a dimer to a RevDR2 site (17). The C terminus of RevErb, including the hinge region and heptad repeats, has been shown to be sufficient for repression when bound to the Gal4 heterologous DNA binding domain (17). The mechanism of repression by RevErb has also begun to be elucidated. We have shown that N-CoR interacts with the RevErb repression domain and functions as a corepressor for the orphan receptor (18) by interacting with receptor dimers on DNA (19). Despite its homology to N-CoR, SMRT is not a corepressor for RevErb (19).
Recently, it has become evident that repression by NHRs involves recruitment of a complex of proteins that mediates its repressive effects. N-CoR and SMRT have been shown to interact with Sin3, which recruits the histone deacetylase HDAC1 (20–22). Sin3 coimmunopurifies with HDAC1, HDAC2, the Rb-associated proteins RbAP48 and RbAP46, and proteins of apparent molecular mass of 30 and 18 kDa called SAP30 (Sin3-associated polypeptide) and SAP18, respectively (23). SAP18 has been cloned and enhances Sin3-mediated repression (23). The Rb-associated proteins have been implicated in targeting histone-modifying enzymes to core histones. This recruitment of histone deacetylase activity may contribute to a repressed state of local chromatin. Thus, transcriptional repressors may function by recruiting large multiprotein complexes that target several downstream factors to mediate their transcriptional effects.
Using a yeast two-hybrid screen to identify other proteins involved in the RevErb repression, we have cloned a small protein we call SUN-CoR (Small Unique Nuclear hormone receptor CoRepressor). This protein shows no homology to N-CoR or SMRT. It interacts with both RevErb and TR in vitro, it represses transcription when fused to a heterologous DNA binding domain (DBD), and it potentiates transcriptional repression by TR and RevErb in vivo. SUN-CoR also interacts with N-CoR and SMRT in vitro and associates with endogenous N-CoR in cells. We conclude that SUN-CoR is an additional factor that could be involved in transcriptional repression by unliganded and orphan NHRs.
MATERIALS AND METHODS
Two-Hybrid Screen.
RevErb amino acids 376–614 were used as bait to screen a 17-day mouse embryo library in the yeast dihybrid system (CLONTECH) as previously described (18).
Northern Analysis.
Northern analysis was performed on total cellular RNA as previously described (24) using cDNA probes for SUN-CoR and N-CoR. The full-length SUN-CoR probe is the 500-bp EcoRI fragment containing only the SUN-CoR coding region. The N-CoR probe consisted of the 2400-bp SacI fragment of the mouse N-CoR cDNA. Probes were labeled with 32P using random primers (Boehringer Mannheim).
Immunofluorescence.
293T cells grown on glass coverslips were transfected with 20 μg of vector containing hemagglutinin (HA)-tagged SUN-CoR by the calcium phosphate precipitation method. Two days posttransfection, the cells were fixed for 30 min in 4% paraformaldehyde, permeabilized with 0.1% Triton X-100 in PBS, and blocked with 1% nonfat dry milk in PBS. Cells were incubated with a 1:75 dilution of anti-HA (monoclonal 12CA5) and anti-SUN-CoR (polyclonal rabbit serum raised against GST-SUN-CoR1–141) antibodies for 1 h in a humid chamber at 37°C. After washing with PBS, rhodamine conjugated donkey anti-rabbit and fluorescein-conjugated donkey anti-mouse secondary antibodies were applied at 1:200 dilution for 1 h at 37°C. Cells were washed in PBS and mounted prior to photographing.
Coimmunoprecipitation and Western Analysis.
C2C12 cells were harvested on days 0–4 of differentiation, and equal quantities of protein were separated by SDS-PAGE and transferred to nitrocellulose. Immunoblotting was performed with polyclonal anti-SUN-CoR antibody at a 1:400 dilution followed by detection with the ECL reagent system (Amersham).
Immunoprecipitation assays were performed in HeLa, 293, and CV-1 cells transfected with both HA-SUN-CoR and RevErb (10 μg per 150-mm plate) or HA-SUN-CoR alone (for N-CoR immunoprecipitation) and lysed with NETN buffer (300 mM NaCl/0.5% Nonidet P-40) containing protease inhibitors. Whole cell extracts were clarified by centrifugation, and equal quantities of protein were immunoprecipitated at 0°C for 1 h with 5 μl of anti-RevErb (25) or anti-N-CoR (20) antibodies or 2 μl of monoclonal anti-HA antibody (12CA5). Immunoprecipitation reactions were then incubated with protein-A/G agarose beads (Santa Cruz Biotechnology) for 1 h at 4°C. Samples were washed with NETN buffer, boiled, separated by SDS-PAGE, and transferred to nitrocellulose. Immunoblotting was performed with anti-HA antibody at 1:500 dilution or anti-RevErb monoclonal antibody at 1:400 followed by detection by ECL reagent.
Plasmid Constructs for GST Interaction Assays.
Full-length RevErb (17), TRβ1 (19), N-CoR (6), SMRT (7), and Luciferase (Promega), vectors for in vitro transcription/translation, were previously described. All GST fusion constructs were cloned into the BamHI site of the pGEX2T vector (Pharmacia). The cloning of GST-N-CoR amino acids 1944–2453 (18) and GST-SMRT amino acids 982-1495 (19) were previously described. SUN-CoR amino acids 1–141 (no. 1 and no. 4), 1–100 (no. 1 and no. 3), 1–49 (no. 1 and no. 2), 50–141 (no. 5 and no. 4), and 101–141 (no. 6 and no. 4) were generated with BamHI restriction sites (indicated in bold) by PCR using the indicated primers as described: no. 1, 5′-gccggatccatggcaggtgaagaaatgaa-3′; no. 2, 5′-ggcggatcctgggtccaacttctgcaaca-3′; no. 3, 5′-cccggatcctatttctttaactctgttca-3′; no. 4, 5′-cccggatccttagtgtttgcttttccctt-3′; no. 5, 5′-gccggatccttggaacaagcaaaggtgga-3′; no. 6, 5′-gcgggatccacagacaagaagaaggctgc-3′. All PCR products were confirmed by sequencing.
Protein Binding Assays Using GST Fusion Proteins.
GST fusion proteins were prepared as previously described (18). GST beads (50 μl) containing the fusion protein were incubated at room temperature in a buffer containing 50 mM KCl, 20 mM Hepes, pH 7.9, 2 mM EDTA, 0.1% Nonidet P-40, 10% glycerol, 0.5% nonfat dry milk, and 5 mM DTT. Five microliters of in vitro translated RevErb, TRβ1, N-CoR, SMRT, or Luciferase was added to the beads (input was 1 μl). Binding was allowed to proceed for 1 h, and the beads were washed four times in the same buffer. The bound proteins were eluted by boiling in 30 μl of SDS-PAGE loading buffer and resolved by electrophoresis. The GST fusion proteins were stained with Coomassie to ensure equal loading, and the bound proteins were visualized by autoradiography.
Plasmid Constructs for Transfection.
The cloning of the reporter vectors Gal4X5-SV40-Luciferase and RevDR2-SV40-Luciferase (17) as well as Gal4-RevErb (17), Gal4-TRβ1-LBD (19), and Gal4-KRAB (18) were previously described. Full-length SUN-CoR used for potentiation experiments and immunofluorescence was generated by subcloning the pGAD10 insert isolated in the dihybrid screen (using BamHI and Bgl2) into a BamHI site in frame with an HA tag in the CMX vector (18). Gal4-SUN-CoR used in Fig. 3B was generated by filling in the above pGAD10 insert using Klenow enzyme followed by ligation into a filled in EcoRI site of the CMX vector containing the Gal4-DBD. The Gal4-SUN-CoR deletion mutants were made by subcloning the GST constructs described above into the CMX-Gal4 vector using BamHI. The reading frames of all fusion junctions were confirmed by sequencing.
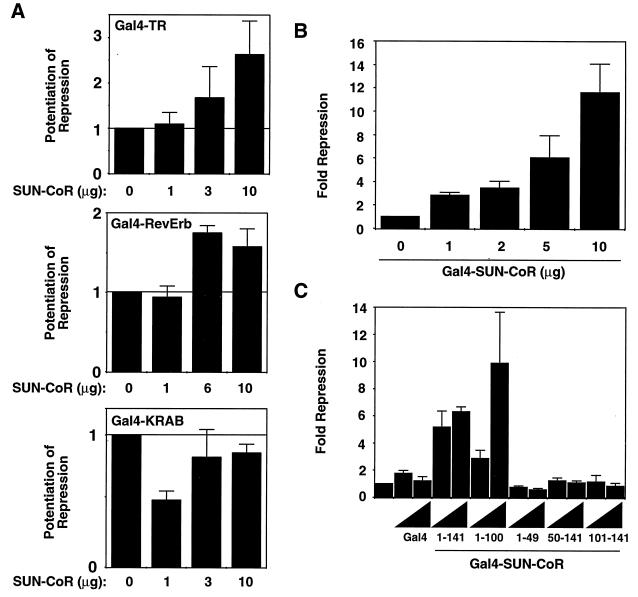
Figure 3.
SUN-CoR is a corepressor for TR and RevErb. (A) SUN-CoR potentiates TR and RevErb but not KRAB domain repression. Gal4-TR, Gal4-RevErb, or Gal4-KRAB repression domain expression plasmid was transfected into 293T cells along with a (Gal4)5-SV40 luciferase reporter plasmid and increasing amounts of SUN-CoR expression plasmid as indicated. (B) SUN-CoR contains an intrinsic repression domain. Increasing concentrations of Gal4-SUN-CoR expression plasmid were transfected into 293T cells along with a (Gal4)5-SV40 luciferase reporter plasmid. (C) The SUN-CoR N terminus contains the repression domain. Two concentrations (2 and 5 μg) of the indicated Gal4-SUN-CoR fusion construct were transfected into 293T cells.
Cell Culture and Transfection.
293T cells were maintained, transfected, lysed, and assayed for β-galactosidase and luciferase activity as previously described (26). Cells were transfected with 1 μg of luciferase reporter, 0.5 μg of β-galactosidase expression vector, receptor or corepressor expression vector in quantities indicated in figure legends, and empty expression vector (CDM or CMX) to equalize the total amount of transfected DNA. The measured relative light units were normalized to β-galactosidase activity, which served as an internal control for transfection efficiency. Each experiment was repeated two to five times, figures show the results of representative experiments in which individual data points were assayed in duplicate, and the range of the results is shown.
RESULTS
Cloning of SUN-CoR.
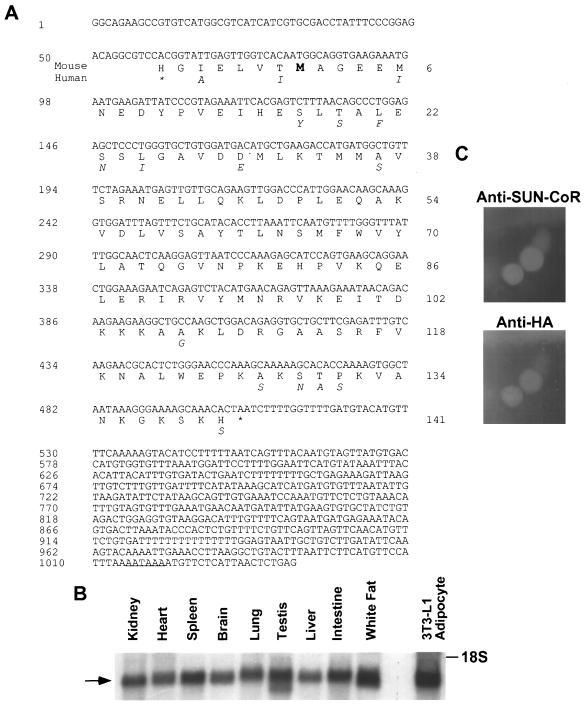
The RevErb C terminus has been shown previously to include a potent transcriptional repression domain (17). A yeast two-hybrid screen was used to identify putative RevErb corepressors. Using amino acids 376–614 of RevErb fused to the Gal4 DBD, 4 million clones from a 17-day mouse embryo library were screened, yielding 84 interacting clones. Five of these clones included a 1041-bp insert (Fig. 1A) containing an ORF that encodes a basic protein (pKa 8.9) of 141 amino acids with a predicted molecular mass of 16 kDa. We call this protein SUN-CoR for Small Unique Nuclear hormone receptor CoRepressor, as it shows no homology to the previously cloned NHR corepressors, which are considerably larger. The initiator methionine codon (ATG) at bp 80–82 is in the appropriate context for translational initiation, and a polyadenylation signal (underlined in Fig. 1A) is located at bp 1015–1021. When cloned into a vector suitable for in vitro transcription and translation in rabbit reticulocyte lysate, the SUN-CoR cDNA produced a protein of the predicted size using its own translational start site (data not shown).
Figure 1.
SUN-CoR sequence, expression, and nuclear localization. (A) Sequence of mouse SUN-CoR cDNA and predicted amino acid sequence. The clone contains a 141-amino acid ORF beginning with an initiation methionine (indicated in bold type) preceded by a well conserved Kozak consensus sequence and ends with a stop codon (indicated by an asterisk). The putative polyadenylation signal is underlined. Amino acids that differ in the human protein are indicated below the mouse protein. (B) SUN-COR tissue distribution. Northern analysis of SUN-CoR mRNA in mouse kidney, heart, spleen, brain, lung, testis, liver, intestine, white fat, and 3T3-L1 adipocytes using the SUN-CoR coding region as probe. (C) SUN-CoR is nuclear. Immunofluorescence of 293T cells transfected with HA-SUN-CoR with polyclonal anti-SUN-CoR antibody and monoclonal anti-HA antibody.
A BLAST database search showed that the SUN-CoR sequence is identical to a previously cloned cDNA known as C1D, whose function is unknown (GenBank accession number 1185119; ref. 27). The mouse and human SUN-CoR/C1D proteins share 90% identity at the amino acid level, indicating that this protein is evolutionarily conserved (amino acid substitutions in the human protein are indicated below the mouse protein sequence in Fig. 1A). The human sequence has an in-frame stop codon located 21 bp upstream of the initiator ATG, strongly suggesting that the mouse clone contains the entire ORF. Also, the cDNA is near the size of the mRNA corresponding to SUN-CoR, as shown below. Indeed, several attempts to screen a testis cDNA library and performance of 5′-rapid amplification of cDNA ends PCR yielded no additional 5′ sequence.
Expression and Cellular Localization of SUN-CoR.
The cDNA encoding the SUN-CoR ORF was used to probe a Northern blot of RNA from mouse kidney, heart, spleen, brain, lung, testes, liver, intestine, and white fat as well as in 3T3-L1 adipocytes in culture (Fig. 1B). This analysis revealed a 1.2-kb mRNA present in all of these differentiated tissues, and it is noteworthy that the size of the mRNA is only ≈100 bp larger than the cloned cDNA, providing additional evidence that the clone is full-length as the additional bases are likely attributable to the poly(A)+ tail of the mRNA.
To determine the cellular localization of SUN-CoR, 293T cells were transiently transfected with HA epitope-tagged SUN-CoR cDNA in a mammalian expression vector lacking a nuclear localization signal. The protein was found in the nucleus as indicated by immunofluorescence (Fig. 1C) using both anti-SUN-CoR and anti-HA antibodies.
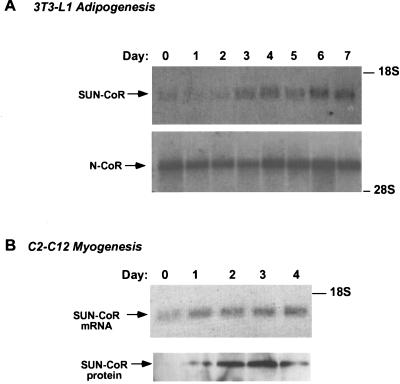
SUN-CoR Expression During Differentiation.
We had found SUN-CoR mRNA to be expressed in all the differentiated tissues tested (Fig. 1B) but to be absent from the immortal cell lines used for transfections (data not shown). Thus, we were interested in the expression pattern of SUN-CoR during differentiation. We chose to look at 3T3-L1 preadipocytes that differentiate into fat cells and C2C12 cells that differentiate into muscle, as RevErb expression is known to be regulated during 3T3-L1 (15) and C2C12 differentiation (16), and TR is known to be involved in muscle differentiation. SUN-CoR expression was up-regulated during differentiation of adipocytes, whereas N-CoR message remained high throughout the process (Fig. 2A). Similarly, SUN-CoR message and protein levels increased with differentiation of C2C12 myocytes (Fig. 2B). All lanes of the Northern and Western blots were evenly loaded as visualized by ethidium bromide and Ponceau-S staining, respectively (data not shown).
Figure 2.
SUN-CoR expression is up-regulated during differentiation. (A) SUN-CoR gene expression is up-regulated during differentiation of 3T3-L1 adipocyte. Northern analysis of 3T3-L1 cells during the 7-day differentiation protocol using the mouse SUN-CoR coding region and the 2400-bp SacI fragment of the mouse N-CoR cDNA as probes. (B) SUN-CoR mRNA and protein levels increase during differentiation of C2C12 cells into myocytes. Northern and Western analysis of C2C12 cells during the 4-day differentiation protocol using the mouse SUN-CoR coding region as a probe for Northern analysis and polyclonal anti-SUN-CoR antibody for Western blotting.
The Role of SUN-CoR in NHR Repression of Transcription.
If SUN-CoR plays a role in transcriptional repression by NHRs and its concentration is limiting under certain circumstances, addition of exogenous SUN-CoR would be expected to potentiate repression by NHRs. As shown in Fig. 3A, cotransfection of SUN-CoR with RevErb or TRβ1 resulted in a dose-dependent potentiation of transcriptional repression by the NHR. SUN-CoR increased repression 2–3-fold, which is similar to potentiation by other corepressors such as N-CoR or SMRT under similar conditions (18, 19). In contrast to its potentiation of repression by NHRs, SUN-CoR had no significant effect on repression mediated by the unrelated KRAB repression domain (28), which uses a corepressor distinct from those of NHRs (29). Thus, potentiation of repression by SUN-CoR is restricted to some but not all transcriptional repression domains. Addition of SUN-CoR in the absence of a repressor protein had no effect on transcription (data not shown).
If SUN-CoR is truly a corepressor, it would be expected to repress transcription when fused to a heterlogous DBD. In fact, when fused to the DBD of the yeast transcription factor Gal4 and transfected in to 293T cells, SUN-CoR repressed transcription from a minimal promoter downstream of five copies of the Gal4 DNA binding site (Fig. 3B). The Gal4 DBD had no effect on transcription (data not shown and Fig. 3C), and repression by SUN-CoR was dose dependent.
To map the repression domain within SUN-CoR, N- and C- terminal deletion mutants were fused to the Gal4 DBD and tested for transcriptional repression function. The N terminus of SUN-CoR seems to be required for transcriptional repression. The repression domain must include amino acids both N- and C-terminal to amino acid 50, as both 1–49 and 50–100 were insufficient to mediate dose-dependent repression, whereas a polypeptide composed of amino acids 1–100 is an active repressor (Fig. 3C). All Gal4 fusion proteins were of appropriate size and were expressed at approximately equal levels as determined by Western analysis of transfected cell extracts with anti-Gal4 antibody (data not shown).
Interaction with Nuclear Hormone Receptors.
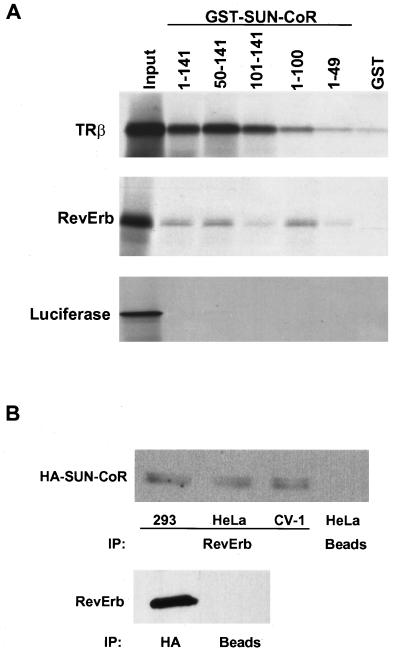
Next, we tested whether SUN-CoR could interact with RevErb as well as TR in vitro. For this purpose, and to map the receptor interaction domains of SUN-CoR, a series of GST fusion proteins was generated containing full-length SUN-CoR as well as several N- and C-terminal truncation mutants. These were tested for their ability to interact with 35S-labeled TR, RevErb, or luciferase proteins in vitro (Fig. 4A). SUN-CoR interacted with RevErb in vitro, and amino acids 50–100 of SUN-CoR were required for this interaction. SUN-CoR also interacted with TRβ (Fig. 4A) and TRα (data not shown), but binding to TR seemed to require a more C-terminal segment of the protein as GST constructs lacking amino acids 100–141 bound only slightly above background and more weakly than polypeptides containing the C terminus of SUN-CoR. Unlike N-CoR and SMRT, direct interactions between SUN-CoR and TR were not inhibited by thyroid hormone (data not shown). Fig. 4A also shows that luciferase did not bind to any of the SUN-CoR peptides, indicating that interactions between SUN-CoR and NHRs were specific. GST alone bound weakly or not at all to the 35S-labeled proteins.
Figure 4.
SUN-CoR associates with nuclear receptors in vitro and in cells. (A) The C terminus of SUN-CoR interacts with TR and RevErb in vitro. RevErb, TRβ1, and luciferase were translated in vitro in the presence of [35S]methionine and analyzed by affinity pull-down with the indicated SUN-CoR peptides fused to GST and bound to glutathione-Sepharose. (B) RevErb and SUN-CoR associate in cells. 293, HeLa, and CV-1 cells were transfected with HA-SUN-CoR and RevErb expression constructs. Cell extracts were immunoprecipitated with anti-RevErb antibody (Upper), anti-HA antibody (Lower), or protein A/G agarose beads alone (“beads”). The immunoprecipitated proteins were visualized by Western analysis with anti-HA antibody (Upper) or anti-RevErb antibody (Lower).
We also investigated whether SUN-CoR and RevErb associate in cells. As SUN-CoR does not seem to be expressed in the cultured cell lines (data not shown), we cotransfected RevErb and HA-tagged SUN-CoR into 293, HeLa, and CV1 cells. Lysates of these cells were immunoprecipitated with anti-RevErb antibody, and the RevErb-associated proteins were analyzed by Western blotting with anti-HA antibody (Fig. 4B). HA-SUN-CoR coimmunoprecipitated with RevErb in all three transfected cell lines, whereas beads alone did not pull down appreciable amounts of SUN-CoR (Fig. 4B Upper). Conversely, immunoprecipitation with HA antibody followed by Western with anti-RevErb antibody also revealed the SUN-CoR interaction with RevErb (Fig. 4B Lower). Thus, RevErb associates with SUN-CoR in vitro and in cells.
Interaction with Corepressors.
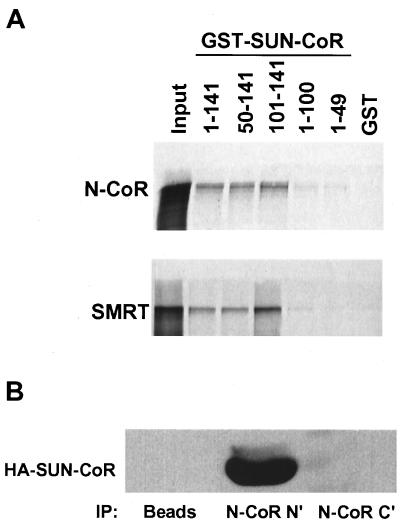
As N-CoR and SMRT are known to function as corepressors for TR (6, 7), and N-CoR is a corepressor for RevErb (18, 19), we considered whether SUN-CoR might be a part of the nuclear receptor repression complex. To test this, we first looked at the ability of SUN-CoR to interact with N-CoR and SMRT in vitro. The same series of GST-SUN-CoR fusion proteins were used to test binding of SUN-CoR peptides to 35S-labeled full-length N-CoR and SMRT. Both corepressors interacted with SUN-CoR and required the extreme C terminus of SUN-CoR (amino acids 100–141) for binding (Fig. 5A). The C-terminal half of N-CoR (amino acids 1510–2453) bound to SUN-CoR with the same pattern as full-length N-CoR (data not shown), indicating that SUN-CoR does not interact with the N-CoR repression domains but rather with the part of N-CoR that contains the nuclear receptor interaction domain.
Figure 5.
SUN-CoR interacts with nuclear receptor corepressors in vitro and in cells. (A) The C terminus of SUN-CoR interacts with N-CoR and SMRT in vitro. Full-length N-CoR and SMRT were translated in vitro in the presence of [35S]methionine and analyzed by affinity pull-down with the indicated GST fusion proteins bound to glutathione-Sepharose. (B) SUN-CoR associates with endogenous N-CoR. HeLa cells were transfected with HA-SUN-CoR, and cell extracts were immunoprecipitated with anti-N-CoR N- and C-terminal antibodies or protein A/G agarose beads alone (“beads”). The immunoprecipitated proteins were visualized by Western analysis with anti-HA antibody.
To show that SUN-CoR associates with endogenous N-CoR in vivo, we transfected HA-SUN-CoR into HeLa cells and immunoprecipitated extracts with anti-N-CoR antibody followed by visualization of coimmunoprecipitated SUN-CoR by Western analysis with anti-HA antibody (Fig. 5B). The N-terminal N-CoR antibody immunoprecipitated HA-SUN-CoR. The amount of transfected SUN-CoR associated with endogenous N-CoR seemed to be considerably more than was observed with transfected RevErb as shown above. Interestingly, the C-terminal N-CoR antibody, which was raised to the region of N-CoR that is involved in SUN-CoR (and nuclear receptor) interaction, did not coimmunoprecipitate SUN-CoR (Fig. 5B), suggesting that this antibody disrupted the N-CoR–SUN-CoR interaction.
DISCUSSION
SUN-CoR is a protein involved in transcriptional repression by nuclear hormone receptors. The C terminus of SUN-CoR interacts with TR and RevErb in vitro and associates with RevErb in cells, SUN-CoR potentiates repression by both receptors in cells, and the N terminus of SUN-CoR contains an intrinsic repression domain. Thus, SUN-CoR seems to be a nuclear receptor corepressor.
SUN-CoR has no homology to the known nuclear receptor corepressors N-CoR and SMRT and is considerably smaller in size than these proteins. It is interesting to note that several other small proteins have been shown to be involved in transcriptional repression. SAP18 is a small protein that copurifies with the Sin3A repression complex and enhances Sin3-mediated repression (23). DR1 is a 20-kDa protein that has been shown to repress PolII transcription by interacting with TATA-binding protein and precluding the entry of TFIIA and TFIIB into the preinitiation complex (30). DR1 seems to act as a universal repressor of PolII transcription, although it is possible that, like SUN-CoR, DR1 is recruited to specific promoters by a subset of sequence-specific DNA binding proteins to repress transcription of particular genes. SUN-CoR has no homology to DR1 or SAP18.
The detailed mechanisms by which SUN-CoR potentiates NHR repression remain to be determined; however, three potential roles for SUN-CoR are suggested by our data. First, we have shown that SUN-CoR is up-regulated during cellular differentiation and is expressed in many fully differentiated tissues. Many genes, particularly those involved in cell growth and division, must be permanently suppressed in differentiated cells. It is possible that SUN-CoR is up-regulated in differentiated cells to assist in this function. Interestingly, RevErb is induced during adipocyte differentiation (31) but down-regulated during myogenesis (ref. 16 and data not shown), suggesting that SUN-CoR may be a physiological corepressor for RevErb in some but not all situations.
In addition, we have shown that SUN-CoR interacts with NHRs as well as other corepressors both in vitro and in vivo. Through these multiple interactions, SUN-CoR may stabilize the NHR repression complex, which also contains Sin3A, histone deacetylase, and other proteins that associate with these. Because N-CoR and SMRT are limiting cofactors that are also involved in repression by other proteins such as the Mad family of bHLH proteins, the presence of SUN-CoR in differentiated tissues may preferentially target N-CoR or SMRT to NHRs rather than other sequence-specific DNA binding proteins.
Finally, SUN-CoR contains an intrinsic repression domain. This domain is distinct from the region of SUN-CoR that interacts with N-CoR/SMRT, indicating that repression by SUN-CoR is not solely due to recruitment of N-CoR or SMRT. It is possible that SUN-CoR function is independent of the N-CoR–SMRT pathway. SUN-CoR may repress transcription by interacting with elements of the basal transcription apparatus, analogous to DR1 which interacts with TATA binding protein. Preliminary data suggest that SUN-CoR may interact with TATA binding protein (data not shown) although the significance of this interaction remains to be determined. Alternatively, the SUN-COR repression domain may contain or recruit enzymatic activities such as deacetylase or phosphatase activities, which may modify histones or the carboxyl-terminal domain of PolII, resulting in transcriptional repression.
Thus, SUN-CoR is a nuclear receptor corepressor that may function as an alternative to the N-CoR–SMRT pathway or as part of a larger repression complex. These complexes seem to integrate signals from many families of transcription factors as well as signal transduction pathways. Just as the coactivator CBP has been shown to integrate signals from a variety of activators, the N-CoR complex is involved in repression by NHRs as well as the Mad family of bHLH proteins (20, 21). However, these complexes are not universal, as N-CoR is not involved in repression by the ets repressor factor ERF (20), and SUN-CoR does not effect repression by the KRAB repression domain. This suggests that several independent mechanisms of transcriptional activation and repression exist that may be mediated by different protein complexes.
Acknowledgments
We thank Caleb Kallen for help with Northern analysis and Thorsten Heinzel for help in preparation of anti-N-CoR antibodies. This work was supported by National Institutes of Health Grant DK45586 (M.A.L.). I.Z. was supported by the Medical Scientist Training Program (5P32GM07170).
ABBREVIATIONS
- NHR
nuclear hormone receptor
- TR
thyroid hormone receptor
- DBD
DNA binding domain
- HA
hemagglutinin
- GST
glutathione S-transferase
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF031426).
References
- 1.Hanna-Rose W, Hansen U. Trends Genet. 1996;12:229–234. doi: 10.1016/0168-9525(96)10022-6. [DOI] [PubMed] [Google Scholar]
- 2.Han M, Grunstein M. Cell. 1988;55:1137–1145. doi: 10.1016/0092-8674(88)90258-9. [DOI] [PubMed] [Google Scholar]
- 3.Durrin L K, Mann R K, Kayne P S, Grunstein M. Cell. 1991;65:1023–1031. doi: 10.1016/0092-8674(91)90554-c. [DOI] [PubMed] [Google Scholar]
- 4.Sternglanz R. Trends Biochem Sci. 1996;21:357–358. [PubMed] [Google Scholar]
- 5.Baniahmad A, Kohne A C, Renkawitz R. EMBO J. 1992;11:1015–1023. doi: 10.1002/j.1460-2075.1992.tb05140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horlein A J, Naar A M, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass C K, Rosenfeld M G. Nature (London) 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 7.Chen J D, Evans R M. Nature (London) 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 8.Chen J D, Umesono K, Evans R M. Proc Natl Acad Sci USA. 1996;93:7567–7571. doi: 10.1073/pnas.93.15.7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sande S, Privalsky M L. Mol Endocrinol. 1996;10:813–825. doi: 10.1210/mend.10.7.8813722. [DOI] [PubMed] [Google Scholar]
- 10.Danielian P S, White R, Lees J A, Parker M G. EMBO J. 1992;11:1025–1033. doi: 10.1002/j.1460-2075.1992.tb05141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barettino D, Vivanco-Ruiz M d M, Stunnenberg H G. EMBO J. 1994;13:3039–3049. doi: 10.1002/j.1460-2075.1994.tb06603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durand B, Saunders M, Gaudon C, Roy B, Losson R, Chambon P. EMBO J. 1994;13:5370–5382. doi: 10.1002/j.1460-2075.1994.tb06872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyajima N, Horiuchi R, Shibuya Y, Fukushige S, Matsubara K, Toyoshima K, Yamamoto T. Cell. 1989;57:31–39. doi: 10.1016/0092-8674(89)90169-4. [DOI] [PubMed] [Google Scholar]
- 14.Lazar M A, Hodin R A, Darling D S, Chin W W. Mol Cell Biol. 1989;9:1128–1136. doi: 10.1128/mcb.9.3.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chawla A, Lazar M A. J Biol Chem. 1993;268:16265–16269. [PubMed] [Google Scholar]
- 16.Downes M, Carozzi A J, Muscat G E O. Mol Endocrinol. 1995;9:1666–1678. doi: 10.1210/mend.9.12.8614403. [DOI] [PubMed] [Google Scholar]
- 17.Harding H P, Lazar M A. Mol Cell Biol. 1995;15:4791–4802. doi: 10.1128/mcb.15.9.4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zamir I, Harding H P, Atkins G B, Horlein A, Glass C K, Rosenfeld M G, Lazar M A. Mol Cell Biol. 1996;16:5458–5465. doi: 10.1128/mcb.16.10.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zamir I, Zhang J, Lazar M A. Genes Dev. 1997;11:835–846. doi: 10.1101/gad.11.7.835. [DOI] [PubMed] [Google Scholar]
- 20.Heinzel T, Lavinsky R M, Mullen T-M, Soderstrom M, Laherty C D, Torchia J, Yuang W-M, Brard G, Ngo S D, Davie J R, Seto E, Eisenman R N, Rose D W, Glass C K, Rosenfeld M G. Nature (London) 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 21.Alland L, Muhle R, Hou H, Potes J, Chin L, Schreiber-Agus N, DePinho R A. Nature (London) 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- 22.Nagy L, Kao H-Y, Chakvarkti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- 24.Kallen C B, Lazar M A. Proc Natl Acad Sci USA. 1996;93:5793–5796. doi: 10.1073/pnas.93.12.5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dumas B, Harding H, Choi H-S, Lehmann K, Chung M, Lazar M A, Moore D D. Mol Endocrinol. 1994;8:996–1005. doi: 10.1210/mend.8.8.7997240. [DOI] [PubMed] [Google Scholar]
- 26.Harding H P, Lazar M A. Mol Cell Biol. 1993;13:3113–3121. doi: 10.1128/mcb.13.5.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Juodka B, Spiess E, Angiolillo A, Joswig G, Rothbarth K, Werner D. Nucleic Acids Res. 1995;23:1359–1366. doi: 10.1093/nar/23.8.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Margolin J F, Friedman J R, Meyer W K, Vissing H, Thiesen H J, Rauscher F J. Proc Natl Acad Sci USA. 1994;91:4509–4513. doi: 10.1073/pnas.91.10.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friedman J R, Fredericks W J, Jensen D E, Speicher D W, Huang X P, Neilson E G, Rauscher F J. Genes Dev. 1996;10:2067–2078. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- 30.Yeung K C, Inostroza J A, Mermelstein F H, Kannabiran C, Reinberg D. Genes Dev. 1994;8:2097–2109. doi: 10.1101/gad.8.17.2097. [DOI] [PubMed] [Google Scholar]
- 31.Chawla A, Schwarz E J, Dimaculangan D D, Lazar M A. Endocrinology. 1994;135:798–800. doi: 10.1210/endo.135.2.8033830. [DOI] [PubMed] [Google Scholar]