Abstract
Myostatin (GDF-8) is a member of the transforming growth factor β superfamily of secreted growth and differentiation factors that is essential for proper regulation of skeletal muscle mass in mice. Here we report the myostatin sequences of nine other vertebrate species and the identification of mutations in the coding sequence of bovine myostatin in two breeds of double-muscled cattle, Belgian Blue and Piedmontese, which are known to have an increase in muscle mass relative to conventional cattle. The Belgian Blue myostatin sequence contains an 11-nucleotide deletion in the third exon which causes a frameshift that eliminates virtually all of the mature, active region of the molecule. The Piedmontese myostatin sequence contains a missense mutation in exon 3, resulting in a substitution of tyrosine for an invariant cysteine in the mature region of the protein. The similarity in phenotypes of double-muscled cattle and myostatin null mice suggests that myostatin performs the same biological function in these two species and is a potentially useful target for genetic manipulation in other farm animals.
The transforming growth factor β superfamily encompasses a large group of secreted growth and differentiation factors that play important roles in regulating development and tissue homeostasis (1). We have recently described a member of this family, myostatin, that is expressed specifically in developing and adult skeletal muscle and functions as a negative regulator of skeletal muscle mass in mice (2). Myostatin null mice generated by gene targeting show a dramatic and widespread increase in skeletal muscle mass. Individual muscles in myostatin null mice weigh 2- to 3-fold more than those of wild-type mice, primarily due to an increased number of muscle fibers without a corresponding increase in the amount of fat. To pursue potential therapeutic and agricultural applications of increasing muscle mass by inhibition of myostatin activity, we have been characterizing myostatin in animals other than mice. Here we report that the myostatin gene is highly conserved among vertebrate species and that two breeds of cattle that are characterized by increased muscle mass (double muscling), Belgian Blue (3) and Piedmontese (4), have mutations in the myostatin coding sequence. These results demonstrate that the function of myostatin has been highly conserved among vertebrates.
METHODS
Cloning of Myostatin.
Poly(A)-containing RNA was isolated from human (obtained from the International Institute for the Advancement of Medicine, Exton, PA), Holstein cow, sheep (Ruppersberger and Sons, Baltimore), pig (Bullock’s Country Meats, Westminster, MD), White Leghorn chicken (Truslow Farms, Chestertown, MD), turkey (kindly provided by D. Boyer and D. Miller, Wampler Foods, Oxford, PA) and zebrafish (kindly provided by S. Fisher and M. Halpern, Carnegie Institution of Washington) skeletal muscle tissue as described (5). cDNA libraries were constructed in the λZAP II vector (Stratagene) according to the instructions provided by the manufacturer and screened without amplification. Rat and baboon skeletal muscle cDNA libraries and a bovine (Holstein) genomic library were purchased from Stratagene. Library screening and analysis of clones were carried out as described (5), except that the final washes were carried out in 25 mM sodium phosphate (pH 8.5), 0.5 M NaCl, 2 mM EDTA, and 0.5% SDS at 65°C.
Mapping.
Fluorescence in situ hybridization was performed on human metaphase spreads (Bios, New Haven, CT) using a digoxigenin-labeled human genomic myostatin probe.
Sequencing of Bovine Genomic DNA.
Blood from cattle was spun at 3,400 × g for 15 min, resuspended in 150 mM NaCl and 100 mM EDTA, and digested with 200 μg⋅ml−1 proteinase K and 1% SDS at 44°C. Semen (Select Sires, Rocky Mount, VA) was digested in 50 mM Tris (pH 8.0), 20 mM EDTA, 1% sarcosyl, 0.2 M 2-mercaptoethanol, and 200 μg⋅ml−1 proteinase K. DNAs were purified on a CsCl gradient. Exons were amplified by PCR from 1 μg genomic DNA using primer pairs 133ACM 5′-CGCGGATCCTTTGGCTTGGCGTTGCTCAAAAGC-3′ and 134ACM 5′-CGCGGATCCTTCTCATGAACACTAGAACAGCAG-3′ (exon 1), 135ACM 5′-CGCGGATCCGATTGATATGGAGGTGTTCGTTCG-3′ and 136ACM 5′-CGCGGATCCGGAAACTGGTAGTTATTTTTCACT-3′ (exon 2), and 137ACM 5′-CGCGGATCCGAGGTAGGAGAGTGTTTTGGGATC-3′ and 138ACM 5′-CGCGGATCCCACAGTTTCAAAATTGTTGAGGGG-3′ (exon 3) at 94°C for 1 min, 52°C for 2 min, and 72°C for 2 min for 40 cycles. PCR products were digested with BamHI, subcloned into pBluescript, and sequenced.
Southern Blot Analysis of Mutant Sequences.
One-fifth of exon 3 amplification products were electrophoresed on 2% agarose gels, blotted to nylon membranes, hybridized with 32P-labeled 13-mers as described (6), and washed in 30 mM sodium citrate, 300 mM NaCl, and 0.1% SDS. Primers used were 146 ACM 5′-ATGAACACTCCAC-3′ (Holstein wild-type sequence, nucleotides 936–948), 145ACM 5′-TTGTGACAGAATC-3′ (Belgian Blue mutation, nucleotides 931–936 with 948–954), 673SJL 5′-GAGAATGTGAATT-3′ (Holstein wild-type sequence, nucleotides 1050–1062), and 674SJL 5′-GAGAATATGAATT-3′ (Piedmontese mutation, G1056A).
RESULTS AND DISCUSSION
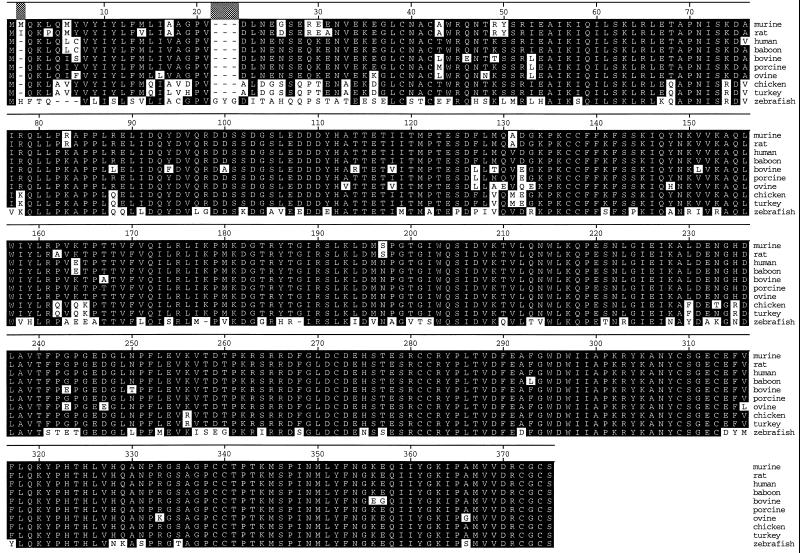
To clone the myostatin gene from other species, cDNA libraries were constructed from RNA isolated from skeletal muscle tissue and screened with a mouse myostatin probe corresponding to the conserved C-terminal region, which is mature, active portion of the molecule. An alignment of the predicted amino acid sequences of murine, rat, human, baboon, bovine, porcine, ovine, chicken, turkey, and zebrafish myostatin, deduced from nucleotide sequence analysis of full-length cDNA clones, is shown in Fig. 1. All of these sequences contain a putative signal sequence for secretion and a putative RXXR proteolytic processing site (amino acids 263–266) followed by a region containing the conserved C-terminal cysteine residues found in all transforming growth factor β family members (1). As seen from this alignment, myostatin is highly conserved across species. In fact, the sequences of murine, rat, human, porcine, chicken, and turkey myostatin are 100% identical in the C-terminal region following the putative proteolytic processing site, and baboon, bovine, and ovine myostatin contain only one to three amino acid differences in the mature protein. Zebrafish myostatin is considerably more diverged and is only 88% identical to the others in this region.
Figure 1.
Amino acid sequence alignment of murine, rat, human, baboon, bovine, porcine, ovine, chicken, turkey, and zebrafish myostatin. Shaded residues indicate amino acids matching the consensus. Amino acids are numbered relative to the human sequence. Dashed lines indicate gaps.
The high degree of sequence conservation of myostatin across species suggests that the function of myostatin has also been conserved. To determine whether myostatin plays a role in regulating muscle mass in animals other than mice, we investigated the possibility that mutations in the myostatin gene might account for the increased muscle mass observed in double-muscled livestock breeds. Double muscling, which has been observed in many breeds of cattle for the past 190 years, appears to be inherited as a single major autosomal locus with several modifiers of phenotypic expression, resulting in incomplete penetrance (7). In the most extensively studied double-muscled breed of cattle, Belgian Blue, the double muscling phenotype (Fig. 2) segregates as a single genetic locus designated muscular hypertrophy (mh) (8). The mh mutation, which is partially recessive, causes an average increase in muscle mass of 20–25%, a decrease in mass of most other organs (9, 10), and a decrease in intramuscular fat and connective tissue (11). The mh locus is tightly linked to markers on a region of bovine chromosome 2 (12) that is syntenic to a region of human chromosome 2 (2q32) (13) to which we had mapped the human myostatin gene by fluorescence in situ hybridization (data not shown).
Figure 2.
A fullblood Belgian Blue bull showing the double muscling phenotype.
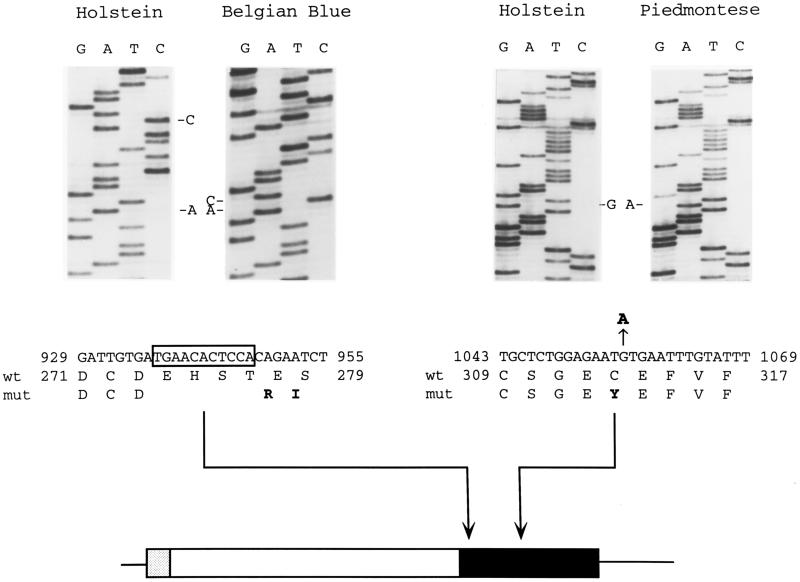
The similarities in phenotype between the myostatin null mice and the Belgian Blue cattle breed and the similar map positions of the myostatin gene and the mh locus suggested the bovine homolog of myostatin as a candidate gene for the mh locus. To determine whether the bovine myostatin gene is mutated in the Belgian Blue breed, all three exons of the gene from the full-blood Belgian Blue bull shown in Fig. 2 were amplified by PCR, subcloned, and sequenced. The Belgian Blue myostatin coding sequence was identical to the Holstein sequence except for a deletion of nucleotides 937–947 in the third exon (Fig. 3). This 11-nucleotide deletion causes a frame-shift which is predicted to result in a truncated protein that terminates 14 codons downstream of the site of the mutation. The deletion is expected to be a null mutation because it occurs after only the first 7 amino acids of the C-terminal region, resulting in a loss of 102 amino acids (amino acids 274–375). This mutation is similar to the targeted mutation in myostatin null mice in which the entire region encoding the mature protein was deleted (2). By Southern blot analysis, using oligonucleotides corresponding to the wild-type or mutant sequence, this mutation was found in both alleles in 14/14 fullblood Belgian Blue cattle examined (data not shown).
Figure 3.
Myostatin mutations in Belgian Blue (Left) and Piedmontese (Right) cattle compared with wild-type Holstein cattle. The nucleotides immediately preceding (A936) and following (C948) the Belgian Blue 11-nucleotide deletion are marked. Nucleotide and amino acid sequences are given below and numbered relative to wild type. The Belgian Blue 11-nucleotide deletion (Δ937–947) is boxed, and the Piedmontese G1056A transition is marked. Bold letters indicate nucleotide and amino acid changes. Arrows identify the locations of the mutations in the myostatin coding sequence. Shading indicates the signal sequence (gray), pro region (white) and mature C-terminal region (black).
We also sequenced the myostatin gene in another cattle breed, Piedmontese, in which double muscling occurs at an extremely high frequency (4). The Piedmontese sequence contained 2 nucleotide changes relative to the Holstein sequence. One was a C to A transversion in exon 1, resulting in a conservative substitution of leucine for phenylalanine (amino acid 94). The second was a G to A transition in exon 3, resulting in a cysteine to tyrosine substitution in the mature region of the protein (amino acid 313) (Fig. 3). By Southern blot analysis, this mutation was found in both alleles in 10/10 double-muscled Piedmontese cattle examined. This mutation is likely to result in a complete or almost complete loss of function, as this cysteine residue is invariant not only among all myostatin sequences but also among all known members of the transforming growth factor β superfamily (1). This cysteine residue is known to be one of the amino acids involved in forming the intramolecular cystine knot structure in members of this superfamily for which the three-dimensional structure is known (14–17). Furthermore, when the corresponding cysteine in activin A (cysteine-44) was mutated to alanine, the mutant protein had only 2% of wild-type receptor binding and biological activity (18).
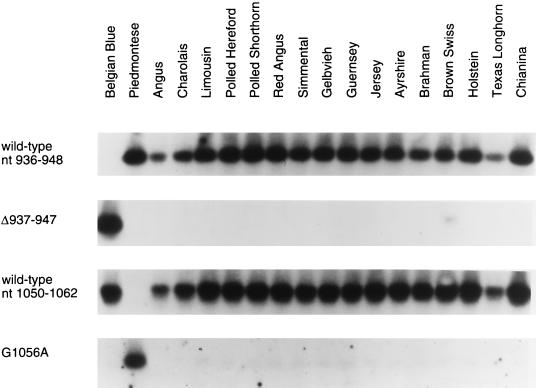
The similar map positions of the myostatin gene and the mh locus and the identification of relatively severe mutations in the myostatin gene of two different double-muscled cattle breeds suggest that these mutations are responsible for the double muscling phenotype. To further support this hypothesis, we analyzed DNA isolated from 120 individual fullblood or purebred cattle in 16 other breeds that are not classified as double-muscled (11 Angus, 11 Charolais, 10 Holstein, 10 Brown Swiss, 10 Polled Hereford, 10 Gelbvieh, 9 Simmental, 9 Jersey, 9 Guernsey, 9 Ayrshire, 7 Limousin, 4 Brahman, 4 Polled Shorthorn, 4 Red Angus, 2 Chianina, and 1 Texas Longhorn) for the presence of each of these mutations (Fig. 4). By Southern blot analysis, the cysteine to tyrosine substitution present in the Piedmontese breed was not detected in any of the 120 individuals. The 11-nucleotide deletion present in the Belgian Blue breed was detected in one allele of a single Red Angus non-double-muscled full-blood bull. In this regard, it has been suggested that the double muscling phenotype that is occasionally seen in many breeds may be due to a single mutation or very few mutations that migrated into many of the European breeds of cattle during the development of the modern breeds (7). Our results demonstrate that myostatin mutations which cause double muscling have occurred at least twice in cattle.
Figure 4.
Representative Southern blot hybridization showing the presence of the Belgian Blue and Piedmontese mutant sequences only in double-muscled breeds of cattle. Exon 3 PCR products were hybridized to oligonucleotide probes spanning the wild-type sequence of the region of the Belgian Blue mutation (top row), the Belgian Blue mutation Δ937–947 (second row), the wild-type sequence at nucleotide 1,056 (third row), and the Piedmontese mutant sequence at nucleotide 1,056 (bottom row). Differences in band intensity reflect differences in amounts of PCR products loaded, as judged by ethidium bromide staining (data not shown). Homozygosity for the mutations was seen only in double-muscled cattle and not in any conventional cattle as described in the text (P < 0.001 by χ2).
Finally, to rule out the presence of other myostatin mutations in non-double-muscled breeds, we determined the complete sequence of the myostatin coding region of 11 of these breeds (Angus, Charolais, Brown Swiss, Polled Hereford, Gelbvieh, Guernsey, Ayrshire, Limousin, Brahman, Polled Shorthorn, and Texas Longhorn). This analysis revealed only polymorphisms that were either silent changes in the coding sequences or were present in the introns and untranslated regions.
Unlike in mice, a myostatin null mutation in cattle causes a reduction in sizes of internal organs and only a modest increase in muscle mass (20–25% in the Belgian Blue breed as compared with 200–300% in myostatin-deficient mice). It is possible that cattle may be nearer to a maximal limit of muscle size after generations of selective breeding for large muscle mass, unlike mice, which have not been similarly selected. In this regard, even in cattle breeds that are not heavily muscled, the myostatin sequence contains two adjacent nonconservative amino acid differences (EG vs. KE) in the C-terminal region, compared with all other species examined. Although the functional significance of these differences is unknown, it is possible that these two changes represent a partial loss-of-function allele that became fixed in the population during many years of cattle breeding.
For agricultural applications, there are some disadvantages to double-muscled cattle, namely the reduction in female fertility, lower viability of offspring, and delay in sexual maturation (19). However, in the Belgian Blue breed, the increased muscle mass and increased feed efficiency largely offset these drawbacks (20). The fact that a null mutation in the myostatin gene in cattle results in animals that are still viable and fertile and produce high-quality meat demonstrates the potential value of producing an increase in muscle mass in other meat animals such as sheep, pig, chicken, turkey, and fish by disrupting myostatin function. Indeed, the high degree of sequence conservation in animals ranging from mammals to birds to fish suggests that the biological function of myostatin has been conserved widely throughout the animal kingdom.
Acknowledgments
We thank Dee Garrels and Chet Pennington (Lakeview Belgian Blue Ranch, Stockton, MO) for providing blood and photographs of Belgian Blue cattle. This work was supported by research grants from the Edward Mallinckrodt, Jr., Foundation and MetaMorphix, Inc. (to S.-J.L). Under an agreement between MetaMorphix, Inc. and the Johns Hopkins University, the authors are entitled to a share of sales royalty received by the University from MetaMorphix, Inc. The University, A.C.M., and S.-J.L. also own MetaMorphix stock, which is subject to certain restrictions under University policy. S.-J.L. is a consultant to MetaMorphix, Inc. The terms of this arrangement are being managed by the University in accordance with its conflict of interest policies.
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [baboon (accession no. AF019619), bovine (accession no. AF019620), chicken (accession no. AF019621), ovine (accession no. AF019622), porcine (accession no. AF019623), rat (accession no. AF019624), turkey (accession no. AF019625), zebrafish (accession no. AF019626), and human (accession no. AF019627)].
A commentary on this article begins on page 12249.
References
- 1.McPherron A C, Lee S-J. In: Growth Factors and Cytokines in Health and Disease. LeRoith D, Bondy C, editors. 1B. Greenwich, CT: JAI; 1996. pp. 357–393. [Google Scholar]
- 2.McPherron A C, Lawler A M, Lee S-J. Nature (London) 1997;387:83–90. doi: 10.1038/387083a0. [DOI] [PubMed] [Google Scholar]
- 3.Hanset R. In: Muscle Hypertrophy of Genetic Origin and Its Use to Improve Beef Production. King J W B, Ménissier F, editors. The Hague, The Netherlands: Nijhoff; 1982. pp. 437–449. [Google Scholar]
- 4.Masoero G, Poujardieu B. In: Muscle Hypertrophy of Genetic Origin and Its Use to Improve Beef Production. King J W B, Ménissier F, editors. The Hague, The Netherlands: Nijhoff; 1982. pp. 450–459. [Google Scholar]
- 5.Lee S-J. Mol Endocrinol. 1990;4:1034–1040. doi: 10.1210/mend-4-7-1034. [DOI] [PubMed] [Google Scholar]
- 6.Gärtner J, Moser H, Valle D. Nat Genet. 1992;1:16–23. doi: 10.1038/ng0492-16. [DOI] [PubMed] [Google Scholar]
- 7.Ménissier F. In: Muscle Hypertrophy of Genetic Origin and Its Use to Improve Beef Production. King J W B, Ménissier F, editors. The Hague, The Netherlands: Nijhoff; 1982. pp. 387–428. [Google Scholar]
- 8.Hanset R, Michaux C. Genet Sel Evol. 1985;17:359–368. doi: 10.1186/1297-9686-17-3-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ansay M, Hanset R. Livest Prod Sci. 1979;6:5–13. [Google Scholar]
- 10.Hanset R. In: Breeding for Disease Resistance in Farm Animals. Owen J B, editor. Wallingford, U.K.: CAB International; 1991. pp. 467–478. [Google Scholar]
- 11.Hanset R, Michaux C, Dessy-Doize C, Burtonboy G. In: Muscle Hypertrophy of Genetic Origin and Its Use to Improve Beef Production. King J W B, Ménissier F, editors. The Hague, The Netherlands: Nijhoff; 1982. pp. 341–349. [Google Scholar]
- 12.Charlier C, Coppieters W, Farnir F, Grobet L, Leroy P L, Michaux C, Mni M, Schwers A, Vanmanshoven P, Hanset R, Georges M. Mamm Genome. 1995;6:788–792. doi: 10.1007/BF00539005. [DOI] [PubMed] [Google Scholar]
- 13.Solinas-Toldo S, Lengauer C, Fries R. Genomics. 1995;27:489–496. doi: 10.1006/geno.1995.1081. [DOI] [PubMed] [Google Scholar]
- 14.Daopin S, Piez K A, Ogawa Y, Davies D R. Science. 1992;257:369–373. doi: 10.1126/science.1631557. [DOI] [PubMed] [Google Scholar]
- 15.Schlunegger M P, Grütter M G. Nature (London) 1992;358:430–434. doi: 10.1038/358430a0. [DOI] [PubMed] [Google Scholar]
- 16.Griffith D L, Keck P C, Sampath T K, Rueger D C, Carlson W D. Proc Natl Acad Sci USA. 1996;93:878–883. doi: 10.1073/pnas.93.2.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mittl P R, Priestle J P, Cox D A, McMaster G, Cerletti N, Grütter M G. Protein Sci. 1996;5:1261–1271. doi: 10.1002/pro.5560050705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mason A J. Mol Endocrinol. 1994;8:325–332. doi: 10.1210/mend.8.3.8015550. [DOI] [PubMed] [Google Scholar]
- 19.Ménissier F. In: Muscle Hypertrophy of Genetic Origin and Its Use to Improve Beef Production. King J W B, Ménissier F, editors. The Hague, The Netherlands: Nijhoff; 1982. pp. 23–53. [Google Scholar]
- 20.Hanset R, Michaux C, Stasse A. Genet Sel Evol. 1987;19:225–248. doi: 10.1186/1297-9686-19-2-225. [DOI] [PMC free article] [PubMed] [Google Scholar]