Abstract
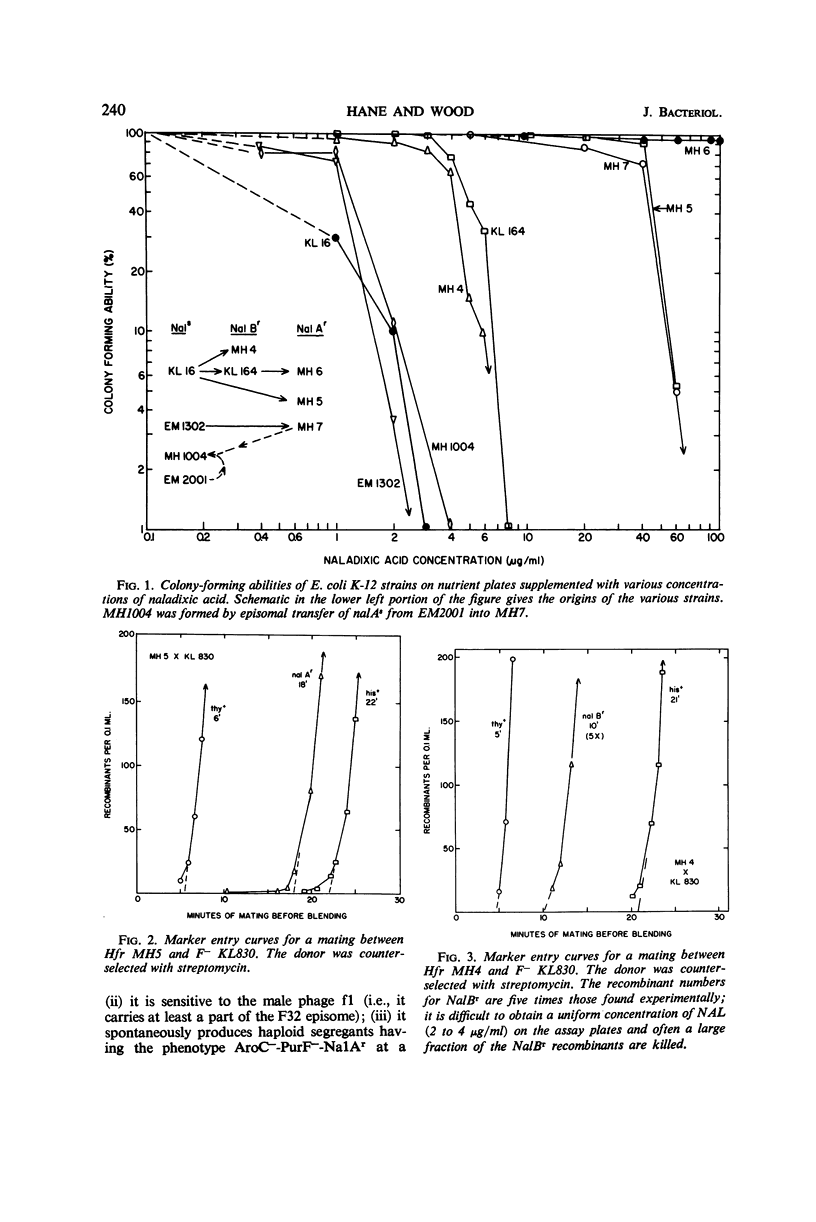
Escherichia coli K-12 strains tested so far (approximately 20) can be separated into three groups on the basis of their abilities to form colonies on nutrient agar supplemented with nalidixic acid (NAL): (i) Nals or wild type (no growth at 1 to 2 μg/ml); (ii) NalAr (growth at 40 μg/ml or higher); and (iii) NalBr (growth at 4 μg/ml, but no growth at 10 μg/ml). The NalAr group has a spectrum of sensitivity ranging from 60 to over 100 μg/ml. All Hfr strains of the NalAr and NalBr groups transfer NAL resistance to recipient cells at genetic loci which are at 42.5 ± 0.5 and 51 ± 1 min, respectively, on the Taylor-Trotter map. Some members of the NalAr group also have the genetic locus for NalBr. The nalAs allele is completely dominant to nalAr in a partial diploid configuration. In haploids, nalAr-nalBr is phenotypically NalAr; nalAr-nalBs is NalAr; and nalAs-nalBr is NalBr. The map location of nalA and the easy differentiation between NalAr and NalAs allow this marker to be used as a counterselector in bacterial conjugation experiments.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barbour S. D. Effect of nalidixic acid on conjugational transfer and expression of episomal lac genes in Escherichia coli K12. J Mol Biol. 1967 Sep 14;28(2):373–376. doi: 10.1016/s0022-2836(67)80016-0. [DOI] [PubMed] [Google Scholar]
- Bastarrachea F., Willetts N. S. The elimination by acridine orange of F30 from recombination-deficient strains of Escherichia coli K12. Genetics. 1968 Jun;59(2):153–166. doi: 10.1093/genetics/59.2.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresler S. E., Lanzov V. A., Lukjaniec-Blinkova A. A. On the mechanism of conjugation in Escherichia coli K 12. Mol Gen Genet. 1968;102(4):269–274. doi: 10.1007/BF00433718. [DOI] [PubMed] [Google Scholar]
- Cuzin F., Buttin G., Jacob F. On the mechanism of genetic transfer during conjugation of Escherichia coli. J Cell Physiol. 1967 Oct;70(2 Suppl):77–88. doi: 10.1002/jcp.1040700407. [DOI] [PubMed] [Google Scholar]
- GOSS W. A., DEITZ W. H., COOK T. M. MECHANISM OF ACTION OF NALIDIXIC ACID ON ESCHERICHIA COLI.II. INHIBITION OF DEOXYRIBONUCLEIC ACID SYNTHESIS. J Bacteriol. 1965 Apr;89:1068–1074. doi: 10.1128/jb.89.4.1068-1074.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollom S., Pritchard R. H. Effect of inhibition of DNA synthesis on mating in Escherichia coli K12. Genet Res. 1965 Nov;6(3):479–483. doi: 10.1017/s0016672300004365. [DOI] [PubMed] [Google Scholar]
- LOW B., WOOD T. H. A QUICK AND EFFICIENT METHOD FOR INTERRUPTION OF BACTERIAL CONJUGATION. Genet Res. 1965 Jul;6:300–303. doi: 10.1017/s001667230000416x. [DOI] [PubMed] [Google Scholar]
- Low B. Formation of merodiploids in matings with a class of Rec- recipient strains of Escherichia coli K12. Proc Natl Acad Sci U S A. 1968 May;60(1):160–167. doi: 10.1073/pnas.60.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall E. Dominance studies with stable merodiploids in the D-serine deaminase system of Escherichia coli K-12. J Bacteriol. 1967 Dec;94(6):1982–1988. doi: 10.1128/jb.94.6.1982-1988.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor A. L., Trotter C. D. Revised linkage map of Escherichia coli. Bacteriol Rev. 1967 Dec;31(4):332–353. doi: 10.1128/br.31.4.332-353.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]



