Abstract
We have asked whether comparative genome analysis and rat transgenesis can be used to identify functional regulatory domains in the gene locus encoding the hypothalamic neuropeptides oxytocin (OT) and vasopressin. Isotocin (IT) and vasotocin (VT) are the teleost homologues of these genes. A contiguous stretch of 46 kb spanning the Fugu IT-VT locus has been sequenced, and nine putative genes were found. Unlike the OT and vasopressin genes, which are closely linked in the mammalian genome in a tail-to-tail orientation, Fugu IT and VT genes are linked head to tail and are separated by five genes. When a cosmid containing the Fugu IT-VT locus was introduced into the rat genome, we found that the Fugu IT gene was specifically expressed in rat hypothalamic oxytocinergic neurons and mimicked the response of the endogenous OT gene to an osmotic stimulus. These data show that cis-acting elements and trans-acting factors mediating the cell-specific and physiological regulation of the OT and IT genes are conserved between mammals and fish. The combination of Fugu genome analysis and transgenesis in a mammal is a powerful tool for identifying and analyzing conserved vertebrate regulatory elements.
The pufferfish, Fugu rubripes, has a 400-Mb genome that is about 7.5 times smaller than the human genome (1). We have been exploring the utility of this genome as a “reference” vertebrate genome, and we are interested in the extent to which gene structure, linkage, and regulatory function have been conserved over the 400 million years since fish and tetrapods diverged (2–4). To explore this concept further, we have studied the evolutionarily ancient vertebrate genes that encode the neuropeptide hormones oxytocin (OT) and vasopressin (VP). Mammalian OT primarily is involved in parturition and lactation, and VP is involved in the maintenance of salt and water balance. The genes encoding the OT and VP preprohormones are highly homologous at both the structural and sequence levels and are linked in the mammalian genome in a tail-to-tail manner with intergenic regions ranging from 3 kb (mouse, ref. 5) to 12 kb (human, ref. 6; rat, ref. 7). The two genes are expressed in distinct magnocellular neurons of the supraoptic nuclei (SON) and paraventricular nuclei (PVN) of the hypothalamus. In addition, VP is expressed in the parvocellular neurons of the PVN and suprachiasmatic nuclei. Because no neuronal cell lines express OT and VP, transgenesis that uses mice and rats as hosts has become the method of choice for the functional analysis of the OT (8–10) and VP (11–14) genes.
In cyclostomes, the most primitive vertebrates, only the VP homologue called vasotocin (VT), is synthesized. However, teleosts contain two genes, VT and isotocin (IT), which probably have arisen by a duplication of the single parental gene. Teleost VT and IT are synthesized in separate neurons in the preoptic nuclei of the hypothalamus (15, 16). Although VT has been implicated in the maintenance of salt-water balance, the biological roles of VT and IT are ambiguous (17). We have cloned and sequenced the Fugu IT-VT locus and shown that it contains at least nine genes. To identify conserved regulatory elements, we have generated transgenic rats bearing a Fugu cosmid from the IT-VT locus and analyzed the expression of the IT and VT genes in transgenic rats.
MATERIALS AND METHODS
Cloning and Sequence Analysis.
A Fugu genomic cosmid library (constructed in lawrist 4 by Greg Elgar, UK HGMP Resource Centre, Cambridge) was screened with a full-length rat VP cDNA (11), and five overlapping cosmids were isolated. Of these five, two cosmids 55F6 (43-kb insert) and 370B2 (39-kb insert) were selected for detailed restriction mapping, subcloning, and sequencing. The sequences were obtained by a combination of “shotgun” sequencing and primer walking on an Applied Biosystems 373 automatic DNA sequencer. Sequences were searched for homology by using the nonredundant protein database of the National Center for Biotechnology Information. Hypothetical gene sequences were predicted by using xgrail (version 1.3c).
Generation of Transgenic Rats.
All animals were cared for in accordance with National Institutes of Health guidelines. DNA of cosmid 55F6 was linearized by digesting with RsrII, which has a unique site in the vector arm and is used for microinjection. Transgenic rats were generated by microinjection of fertilized one-cell Sprague–Dawley rat eggs according to procedures previously described (18). Founders were identified by Southern hybridization analysis of the tail DNA by using Sau3A fragments of the cosmid 55F6 as probes. These probes do not hybridize to the DNA of wild-type rats. Independent transgenic lines were generated by crossing founders with wild-type Sprague–Dawley mates. Only Southern blot-positive transgenic rats were used for expression analysis.
Northern Blot Analyses.
Total cellular RNA was extracted from 8- to 10-week-old rats and analyzed by Northern blotting as described (8). The Northern filters were probed with gene-specific oligonucleotides end-labeled with [γ-32P]ATP. The rat VP probe is a 48-mer corresponding to the sequences encoding the last 16 amino acids of VP exon III (19). The rat OT probe is a 41-mer complementary to the seven bases of the 5′ untranslated region and 34 bases of the coding sequence (5′-CCAAGCAGGCAGCAAGCGAGACTGGGGCAGGCCATGGCGTT-3′). The Fugu IT probe is a 40-mer complementary to the last 11 bases of the last exon and 29 bases of the 3′ untranslated region (5′-ACACCTTGTTGCCGGGAGAAGCCCGTTAGTGAGTCTGATG-3′). The Fugu VT probe is a 40-mer complementary to the last 40 bases of the last exon (5′-TCAGTACTCGCTCTGTCCTCTGCTGCTCATGTGCAGCAGA-3′). The IT and VT antisense oligos were complementary to the sequences of the IT and VT probes, respectively. The levels of RNA were determined by densitometric scanning of autoradiograms by using the Visage 110 (BioImage, Ann Arbor, MI) with the sunview program, normalized by the measured level of α-tubulin RNA, and expressed as a percentage of the mean of the control group ± SE.
In Situ Hybridization.
Tissues were frozen in OCT, and serial sections of 10 μm were cut in a cryostat. The sections were hybridized with 35S(dATP)-labeled probes (20) or simultaneously with 35S(dATP) and digoxigenin(dUTP)-labeled probes (21). The hybridized digoxigenin-labeled probes were detected with a peroxidase-conjugated antibody recognizing digoxigenin (Boehringer Mannheim). Oligonucleotide probes used in these experiments were the same as those used in the Northern analysis. Antisense oligos to IT and VT probes were used as controls.
RESULTS
Structure of the Fugu IT-VT Locus.
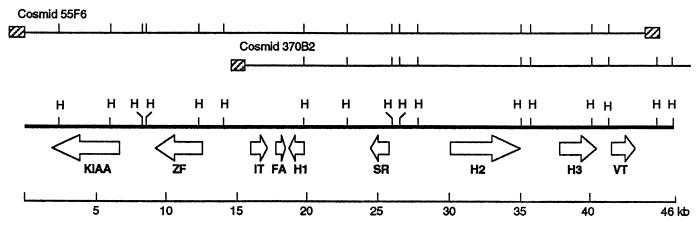
By using a rat VP cDNA as a probe we isolated overlapping Fugu cosmid genomic clones and sequenced a contiguous stretch of 46 kb (Fig. 1). The Fugu IT and VT genes, in contrast to their mammalian counterparts, are found in tandem, in a head-to-tail array, separated by 24.4 kb. In addition to the IT and VT genes, the locus contained seven more genes: a zinc finger (ZF) protein gene, a fatty acid binding protein gene, a sepiapterin reductase gene, a gene with homology to a human cDNA of unknown function isolated from a myeloblast cell line (accession no. 1665795), and three hypothetical genes. The ZF gene is a novel ZF gene, which comprises five exons that code for a putative protein of 490 residues containing 11 ZF domains of C2H2 type. Seven GT-AC16–46 repeats and a single CTAT27 repeat are the only repetitive elements found in this locus.
Figure 1.
Schematic representation of the Fugu IT-VT locus. A 46-kb sequence (thick line) was obtained from cosmids 55F6 and 370B2. Only HindIII sites (H) are shown. Arrows represent complete genes and indicate the direction of transcription. Hatched rectangles represent the cosmid arms (not to scale). KIAA, gene with homology to a human cDNA (KIAA0264) whose function is not known; FA, fatty acid binding protein gene; SR, sepiapterin reductase gene; H1, H2, and H3, hypothetical genes.
Southern analysis revealed that the IT and VT genes are present as single copies in the Fugu genome (data not shown). The two genes have a three-exon structure similar to the VT genes in cyclostomes (22), other teleosts (23), and chicken (24), and the VP and OT genes in mammals (5, 6, 25, 26). The genomic sequence of the teleost IT gene has been characterized only in the white sucker (27), but, unlike the Fugu IT, both copies of IT genes in this tetraploid teleost are devoid of introns. The Fugu IT and VT code for preprohormones of similar length (155 and 153 amino acid residues, respectively) and show considerable homology in their nucleotide and amino acid sequences. The homology is highest in the second exon (85% of nucleotides and 87% of amino acid residues are identical), which codes for the neurophysin molecule.
Transgenic Rats Bearing Fugu Genomic Cosmid.
Transgenic rats were generated by microinjecting linearized Fugu cosmid 55F6 into fertilized one-cell stage rat eggs. Altogether about 900 eggs were injected with the linearized cosmid. Sixty percent of them survived microinjection and were transferred to 20 surrogate mothers. Of the 50 pups born, six were transgenic. One of the founders (designated 46.5) was sterile, and two (46.3 and 46.4) did not show germ-line transmission. The other three, 46.1, 46.2 and 46.6, which showed germ-line transmission, were used for expression studies. Integration of the entire cosmid insert into the genome of these lines was confirmed by probing Southern blots of RsrII-digested genomic DNA with HindIII fragments of the cosmid 55F6.
Expression of the Fugu IT and VT Transgenes in Rats.
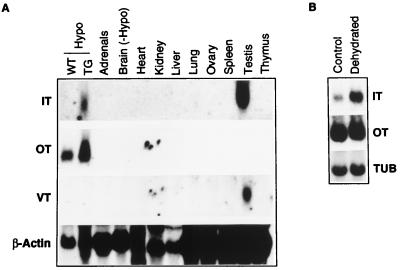
Northern analysis of various transgenic rat tissues revealed IT RNA in the hypothalamus and testis of rats from transgenic lines 46.1 (Fig. 2A), whereas in the other two lines, 46.2 and 46.6, IT transcripts were detected only in the testis. VT gene expression was detected in the testis from all three lines of transgenic rats. The sizes of IT and VT transcripts in the hypothalamus and testis of transgenic rats were about 1,000 bases, similar to the sizes of IT and VT mRNA expressed in the Fugu brain (data not shown).
Figure 2.
(A) Expression of Fugu genes in transgenic rats. WT-Hypo, hypothalamus from wild-type rats; TG-Hypo, hypothalamus from transgenic rats; Brain (−Hypo), brain without hypothalamus. Adrenal glands and ovary from three rats were pooled to obtain adequate RNA for Northern analysis. RNA was fractionated on a 1.2% gel, transferred to nylon membrane (Amersham), and probed with various probes. Each lane contains approximately 20 μg (hypothalamus) or 50 μg (other tissues) of total RNA. IT and VT probes did not hybridize to RNA from the testis of wild-type rats (data not shown). (B) Effect of dehydration on levels of transgene expression. Each lane contains approximately 20 μg of total RNA from the hypothalamus of transgenic rat that was given tap water (control) or deprived of fluid for 3 days (dehydrated). A oligonucleotide probe recognizing rat α-tubulin (TUB) was used as internal standard for quantitation.
Expression of the Fugu IT Transgene in Rat Hypothalamus.
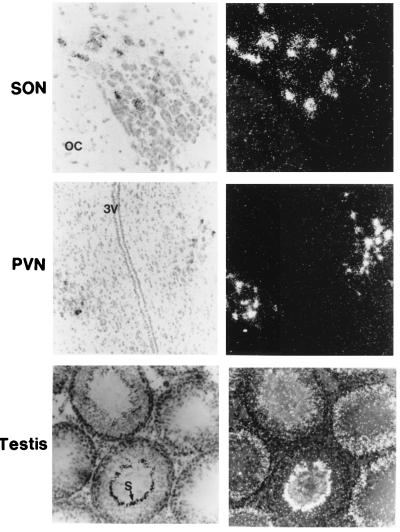
Because Northern analysis showed that IT gene expresses in the hypothalamus of rats from the transgenic line 46.1, we performed in situ hybridization of brain sections from this line to identify the cell type in which IT gene is expressed. The in situ analysis clearly showed that IT RNA is expressed specifically in the magnocellular neurons of the SON and PVN in the hypothalamus (Fig. 3). In situ hybridization revealed a similar pattern of IT gene expression in the hypothalamus from the other two transgenic lines, 46.2 and 46.6, in which expression of IT gene was not detected by Northern analysis. Probing of brain sections from the three transgenic lines with VT probe did not detect VT transcripts in any part of the brain (data not shown).
Figure 3.
In situ hybridization of brain and testis sections from transgenic rats. Tissue sections were hybridized with 35S-labeled isotocin probe. OC, optic chiasma; 3V, third ventricle; S, spermatozoa. (Left) Sections were photographed under bright-field. The same sections were photographed in the dark-field (Right) to show the distribution of silver grains. IT transcripts were found only in the SON and PVN of the hypothalamus. No VT transcripts were found in any part of the brain. The IT probe did not hybridize to brain sections from the wild-type rats. The antisense oligonucleotide for IT probe did not bind to any cells in the brain or testis from transgenic rats (not shown).
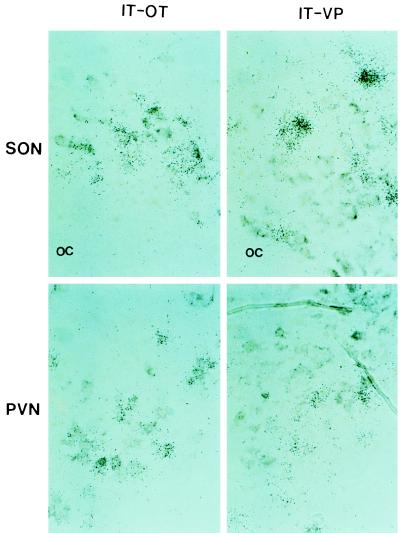
To determine if IT gene expresses in the oxytocinergic and/or vasopressinergic neurons in the SON and PVN, we hybridized adjacent brain sections simultaneously with two probes, either IT probe labeled with 35S and OT probe labeled with digoxigenin, or IT probe labeled with 35S and VP probe labeled with digoxigenin. Probing with IT and VP showed that the IT gene expresses in magnocellular neurons distinct from those expressing VP transcripts in the SON and PVN. In the adjacent sections that were probed with IT and OT probes, IT and OT transcripts were found to express in the same magnocellular neurons in both SON and PVN (Fig. 4).
Figure 4.
Double in situ hybridization analysis. Adjacent sections of brain were hybridized simultaneously with 35S-labeled IT probe and digoxigenin-labeled OT probe (IT-OT) or with 35S-labeled IT probe and digoxigenin-labeled VP probe (IT-VP). Silver grains represent the hybridized radioactive probe, and the gray-brown patches are the result of color reaction of the hybridized digoxigenin probe. The IT transcripts colocalize with endogenous OT transcripts in the SON and PVN, but not with the VP transcripts. OC, optic chiasma.
Expression of the Fugu IT and VT Transgenes in the Rat Testis.
In situ hybridization of testis sections showed that the Fugu IT and VT genes express in the seminiferous tubules of testis from all three expressing transgenic lines. A representative section of testis probed with 35S-labeled IT is shown in Fig. 3. The distribution of IT and VT signals within the seminiferous tubules suggests that these genes are expressed in the Sertoli cells of the seminiferous tubules. Differences were seen in the levels of IT and VT signals in seminiferous tubules containing germ cells at different stages of spermatogenesis. The IT and VT transcripts were abundant in seminiferous tubules with early-stage germ cells, whereas their levels dropped in tubules with fully developed spermatids (Fig. 3), suggesting a developmental-stage-specific regulation of their expression.
Regulation of the Fugu IT Transgene Expression by Physiological Stimuli.
Rats from transgenic line 46.1 were subjected to osmotic challenge by withdrawing dietary water for 3 days. Their littermates fed with tap water served as the control group. Total RNA extracted from the hypothalamus and testis from dehydrated and control groups of transgenic rats were analyzed by Northern blotting to determine the effect of osmotic challenge on the expression of the IT transgene by using α-tubulin as an internal control. The osmotic stimulus induced a 6-fold increase in the abundance of the IT transcripts in the transgenic rat hypothalamus (control: 100 ± 20; dehydrated 583 ± 128: P < 0.05; n = 5 in each group) (Fig. 2B). This was in parallel with the response of the endogenous OT transcripts, which showed a 2-fold increase (control, 100 ± 15; dehydrated 192 ± 13; P < 0.05). However, unlike the endogenous OT transcripts, which showed an increase in their length in dehydrated rats, no apparent increase was seen in the size of the Fugu IT mRNA (Fig. 2B). No significant difference was found in the levels of IT transcripts in the testes from normal and dehydrated rats (data not shown). Neither Northern analysis nor in situ hybridization detected VT expression in the hypothalamus of dehydrated transgenic rats.
DISCUSSION
Teleosts have a body plan similar to the higher vertebrates and possess all the complex physiological functions found in higher vertebrates. Thus, it might be expected that teleosts and mammals contain a similar repertoire of genes. Because the genome of the Fugu is 7.5 times smaller than that of mammals, we predicted a higher gene density in the Fugu than in higher vertebrates (1). The 46 kb from the Fugu IT and VT locus contains at least nine genes. This corresponds to a gene density of one gene per 5 kb and is similar to that reported for the Fugu homologue of the human AD3 locus, which contains seven protein-coding genes in a span of 40 kb (28).
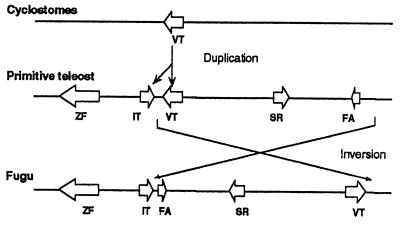
The head-to-tail arrangement of the Fugu IT and VT genes, as compared with the tail-to-tail orientation of the mammalian OT and VP genes, suggests a localized rearrangement in this locus. One possible model of rearrangement is shown in Fig. 5. Inverted duplication is a common feature of gene amplification events in mammalian cells and has been proposed to be the primary event in the amplification of DNA (29). Tail-to-tail arrangement of genes that are believed to have arisen by duplication of an ancestral gene, such as the α- and β-globin genes from the Atlantic salmon (30), supports this hypothesis. It therefore is possible that duplication of the ancestral VT gene of cyclostomes resulted in a tail-to-tail orientation of the two copies of genes. Subsequently, in the lineage that led to the Fugu, a fragment of DNA containing the VT gene has undergone inversion, resulting in a head-to-tail orientation of the IT and VT genes. We have not found any unusual sequences such as transposons in the Fugu IT-VT locus that might have been involved in such an inversion. Our model predicts a tail-to-tail orientation of IT and VT genes in some primitive fish that gave rise to the tetrapod lineage.
Figure 5.
A simple model to show the rearrangement in the IT-VT locus. The open arrows represent genes (VT; IT; FA, fatty acid binding protein; SR, sepiapterin reductase; and ZF) and indicate the direction of their transcription.
We used these genes to study the conservation of regulatory elements by generating six independently derived lines of transgenic rats bearing a Fugu cosmid (55F6) from the IT-VT locus. In this case, the rat is a favored host over mouse for VP and OT transgenes, because the physiological and central nervous systems of rats can be studied easily.
Northern analysis and in situ hybridization showed that the Fugu IT gene specifically is expressed in the rat hypothalamus and testis, whereas the VT gene is expressed only in the testis. Although hypothalamic expression of IT gene was detected by Northern blotting in only one of the rat lines (46.1), in situ hybridization showed specific expression of IT in the SON and PVN in all three expressing lines of rats. Failure to detect IT transcripts by Northern blotting in the two lines, 46.2 and 46.6, appears to be caused by overall lower level of expression.
In mammals, the OT and VP genes are expressed in different magnocellular neurons of the SON and PVN of the hypothalamus. Similarly, VT and IT transcripts are synthesized in distinct neurons in the preoptic nuclei of teleost hypothalamus (15, 16). The double in situ analysis of brain sections from transgenic rats clearly showed that the Fugu IT gene is expressed in oxytocinergic, but not in vasopressinergic, magnocellular neurons in the SON and PVN. These results demonstrate that the cis-acting sequences directing the cell-specific expression of the OT gene are conserved in the Fugu IT-VT locus, and that these elements are recognized by the trans-acting factors of the rat neuronal cells.
Mammalian VP and OT gene expression is modulated by physiological stimuli and the stages of gestation and lactation (31). In the rat, osmotic challenge results in an increase in the amounts of both the VP and OT mRNAs, as well as a lengthening of their poly(A) tails (32, 33). To analyze the physiological response of the IT gene, we subjected transgenic rats to an osmotic challenge. This stimulus induced a 6-fold increase in the level of IT expression in the hypothalamus of the transgenic rats. These results demonstrate that the cis-acting elements mediating the osmotic-stimulus regulated expression of the OT gene in the hypothalamus are conserved in the Fugu IT gene and are recognized by the rat trans-acting factors.
The physiological significance of the expression of the Fugu IT and VT genes in the rat testis is not clear. In rats and mice, neither VP nor OT transcripts could be detected in the testis by Northern blotting. However, OT transcripts have been detected in the rat testis by PCR (8, 34). In cattle, high levels of OT RNA, but not VP RNA, have been detected in the testis by Northern analysis (8). The expression of IT and VT genes in teleost testis has not been studied, and therefore, we do not known whether the expression of the Fugu IT and VT genes in the rat testis is caused by uncontrolled transcription or the reflection of their expression pattern in the fish.
The lack of expression of the VT gene in the rat hypothalamus suggests that the regulatory elements mediating the hypothalamic expression of the VT gene all may not be present on cosmid 55F6. Transgenic studies in rats and mice have indicated that the critical elements required for the hypothalamic expression of VP reside in the 3′ region (13, 14), and some of these sequences may be present outside of the 800 bp of the 3′ region of the VT gene on cosmid 55F6. We now are testing this hypothesis by generating transgenic rats bearing another Fugu cosmid (370B2, see Fig. 1), which contains about 10 kb of sequence downstream of the VT gene.
The promoter region of the Fugu IT gene contains a potential cAMP-responsive element, TGACGTCA, and an estrogen, retinoic acid, and thyroxine-responsive element, TTGACC, (2,518 bp and 446 bp upstream of the first codon, respectively) that are conserved in the mammalian OT promoter region (31). We have found other conserved sequences by comparing homologous regions from the Fugu IT and rat OT genes (Fig. 6), but the functional significance of these sequences is unknown. Complementary sequence (GAAAAATCACAA) of one of these sequences, TTGTGATTTTTC, has a high homology to the osmotic response element characterized in the aldose reductase genes from rabbit (CGGAAAATCAC, ref. 35) and humans (TGGAAAATCAC, ref. 36). It is possible that this conserved sequence mediates the osmotic-stimulus regulated expression of the OT and IT genes in the hypothalamus.
Figure 6.
Conserved sequences in the promoter and 3′ regions of the Fugu IT and the rat OT genes. Homologous regions from the Fugu IT-VT locus and the rat OT-VP locus (7) were compared by using Martinez/Needleman–Wunsch algorithm of the lasergene software (DNAstar, Madison, WI). Only sequences that show at least seven conserved bases at a stretch are shown. The numbers in A and B refer to the position of the bases in relation to the first base of the translation initiation codon and the termination codon, respectively. ∗ denote conserved bases.
It previously has been shown in transgenic mice that a conserved noncoding sequence in the Fugu Hoxb-4 gene linked to a lacZ reporter construct was able to direct expression to neural tube similar to the endogenous mouse sequence (2). We have extended this study to show that the Fugu IT gene is specifically transcribed in rats and contains conserved elements of sequence that direct expression of the IT gene in the specific oxytocinergic neurons in the rat hypothalamus. The locus also must contain sequences involved in the response to an osmotic stimulus. Fish and rats are separated by 400 million yr, and irrelevant sequences will have had sufficient time to randomize, allowing significance to be assigned to any sequence that is totally conserved. Because the Fugu genome is compact, transgenic rats can be generated with cosmids containing several genes at the same time. It should be relatively easy to identify and characterize conserved regulatory elements by using an approach that combines comparative Fugu and mammalian genome analysis, and transgenesis in the rat.
Acknowledgments
We thank Greg Elgar for the Fugu cosmid library, Ms. Nachiammal Kambadhasan and her staff for maintenance of the Institute of Molecular and Cell Biology transgenic rat colony, and Miss Boon Hui Tay for technical assistance.
ABBREVIATIONS
- OT
oxytocin
- VP
vasopressin
- SON
supraoptic nuclei
- PVN
paraventricular nuclei
- VT
vasotocin
- IT
isotocin
- ZF
zinc finger
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. U90880).
References
- 1.Brenner S, Elgar G, Sandford R, Macrae A, Venkatesh B, Aparicio S. Nature (London) 1993;366:265–268. doi: 10.1038/366265a0. [DOI] [PubMed] [Google Scholar]
- 2.Aparicio S, Morrison A, Gould A, Gilthorpe J, Chaudhuri C, Rigby P, Krumlauf R, Brenner S. Proc Natl Acad Sci USA. 1995;92:1684–1688. doi: 10.1073/pnas.92.5.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Venkatesh B, Tay B H, Elgar G, Brenner S. J Mol Biol. 1996;259:655–665. doi: 10.1006/jmbi.1996.0347. [DOI] [PubMed] [Google Scholar]
- 4.How G F, Venkatesh B, Brenner S. Genome Res. 1996;6:1185–1191. doi: 10.1101/gr.6.12.1185. [DOI] [PubMed] [Google Scholar]
- 5.Hara Y, Battey J, Gainer H. Mol Brain Res. 1990;8:319–324. doi: 10.1016/0169-328x(90)90045-f. [DOI] [PubMed] [Google Scholar]
- 6.Sausville E, Carney D, Battey J. J Biol Chem. 1985;260:10236–10241. [PubMed] [Google Scholar]
- 7.Schmitz E, Mohr E, Richter D. DNA Cell Biol. 1991;10:81–91. doi: 10.1089/dna.1991.10.81. [DOI] [PubMed] [Google Scholar]
- 8.Ang H-L, Ungefroren H, de Bree F, Foo N C, Carter D A, Burbach J P, Ivell R, Murphy D. Endocrinology. 1991;128:2110–2117. doi: 10.1210/endo-128-4-2110. [DOI] [PubMed] [Google Scholar]
- 9.Young W S, III, Reynolds K, Shepard E A, Gainer H, Castel M. J Neuroendocrinol. 1990;2:917–925. doi: 10.1111/j.1365-2826.1990.tb00660.x. [DOI] [PubMed] [Google Scholar]
- 10.Ho M-Y, Carter D A, Ang H L, Murphy D. J Biol Chem. 1995;270:27199–27205. doi: 10.1074/jbc.270.45.27199. [DOI] [PubMed] [Google Scholar]
- 11.Foo N-C, Funkhouser J M, Carter D A, Murphy D. J Biol Chem. 1994;269:659–667. [PubMed] [Google Scholar]
- 12.Ang H-L, Carter D A, Murphy D. EMBO J. 1993;12:2397–2409. doi: 10.1002/j.1460-2075.1993.tb05894.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grant F D, Reventos J, Gordon J W, Kawabata S, Miller M, Majzoub A. Mol Endocrinol. 1993;7:659–667. doi: 10.1210/mend.7.5.8100353. [DOI] [PubMed] [Google Scholar]
- 14.Zeng Q, Carter D A, Murphy D. J Neuroendocrinol. 1994;6:469–477. doi: 10.1111/j.1365-2826.1994.tb00610.x. [DOI] [PubMed] [Google Scholar]
- 15.Goossens N, Dierickx K, Vandesande F. Gen Comp Endocrinol. 1977;32:371–375. doi: 10.1016/0016-6480(77)90216-7. [DOI] [PubMed] [Google Scholar]
- 16.Hyodo S, Urano A. J Comp Physiol B. 1991;161:549–556. doi: 10.1007/BF00260744. [DOI] [PubMed] [Google Scholar]
- 17.Urano K, Kubokawa K, Hiraoka S. Fish Physiol. 1994;13:101–132. [Google Scholar]
- 18.Murphy D, Carter D A. Transgenesis Techniques, Principles and Protocols. Totowa, NJ: Humana; 1993. [Google Scholar]
- 19.Murphy D, Levy A, Lightman S L, Carter D A. Proc Natl Acad Sci USA. 1989;86:9002–9005. doi: 10.1073/pnas.86.22.9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Young W S, III, Mezey E, Siegel R E. Mol Brain Res. 1986;1:231–241. doi: 10.1016/0169-328x(86)90029-x. [DOI] [PubMed] [Google Scholar]
- 21.Young W S., III Neuropeptides. 1989;13:271–275. doi: 10.1016/0143-4179(89)90081-4. [DOI] [PubMed] [Google Scholar]
- 22.Heierhorst J, Lederis K, Richter D. Proc Natl Acad Sci USA. 1992;89:6798–6802. doi: 10.1073/pnas.89.15.6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morley S D, Schonrock C, Heierhorst J, Figueroa J, Lederis K, Richter D. Biochemistry. 1990;29:2506–2511. doi: 10.1021/bi00462a011. [DOI] [PubMed] [Google Scholar]
- 24.Hamann D, Hunt N, Ivell R. J Neuroendocrinol. 1992;4:505–513. doi: 10.1111/j.1365-2826.1992.tb00198.x. [DOI] [PubMed] [Google Scholar]
- 25.Ivell R, Richter D. Proc Natl Acad Sci USA. 1984;81:2006–2010. doi: 10.1073/pnas.81.7.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruppert S, Scherer G, Schutz G. Nature (London) 1984;308:554–557. doi: 10.1038/308554a0. [DOI] [PubMed] [Google Scholar]
- 27.Figueroa J, Morley S D, Heierhorst J, Krentler C, Lederis K, Richter D. EMBO J. 1989;8:2873–2877. doi: 10.1002/j.1460-2075.1989.tb08435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trower M K, Orton S M, Purvis I J, Sanseau P, Riley J, Christodoulou C, Burt D, See C G, Elgar G, Sherrington R, Rogaev E I, St. George-Hyslop P, Brenner S, Dykes C W. Proc Natl Acad Sci USA. 1996;93:1366–1369. doi: 10.1073/pnas.93.4.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fried M, Feo S, Heard E. Biochim Biophys Acta. 1991;1090:143–155. doi: 10.1016/0167-4781(91)90095-4. [DOI] [PubMed] [Google Scholar]
- 30.Wagner A, Deryckere F, McMorrow T, Gannon F. J Mol Evol. 1994;38:28–35. doi: 10.1007/BF00175492. [DOI] [PubMed] [Google Scholar]
- 31.Young W S., III J Neuroendocrinol. 1992;4:527–540. doi: 10.1111/j.1365-2826.1992.tb00201.x. [DOI] [PubMed] [Google Scholar]
- 32.Carter D A, Murphy D. J Biol Chem. 1989;264:6601–6603. [PubMed] [Google Scholar]
- 33.Murphy D, Carter D A. Mol Endocrinol. 1990;4:1051–1059. doi: 10.1210/mend-4-7-1051. [DOI] [PubMed] [Google Scholar]
- 34.Foo N C, Carter D, Murphy D, Ivell R. Endocrinology. 1991;128:2118–2128. doi: 10.1210/endo-128-4-2118. [DOI] [PubMed] [Google Scholar]
- 35.Ferraris J D, Williams C K, Jung K, Bedford J J, Burg M B, Garcia-Perez A. J Biol Chem. 1996;271:18318–18321. doi: 10.1074/jbc.271.31.18318. [DOI] [PubMed] [Google Scholar]
- 36.Ko B C B, Ruepp B, Bohren K M, Gabbay K H, Chung S S M. J Biol Chem. 1997;272:16431–16437. doi: 10.1074/jbc.272.26.16431. [DOI] [PubMed] [Google Scholar]