Abstract
A comprehensive gene collection for S. oneidensis was constructed using the lambda recombinase (Gateway) cloning system. A total of 3584 individual ORFs (85%) have been successfully cloned into the entry plasmids. To validate the use of the clone set, three sets of ORFs were examined within three different destination vectors constructed in this study. Success rates for heterologous protein expression of S. oneidensis His- or His/GST- tagged proteins in E. coli were approximately 70%. The ArcA and NarP transcription factor proteins were tested in an in vitro binding assay to demonstrate that functional proteins can be successfully produced using the clone set. Further functional validation of the clone set was obtained from phage display experiments in which a phage encoding thioredoxin was successfully isolated from a pool of 80 different clones after three rounds of biopanning using immobilized anti-thioredoxin antibody as a target. This clone set complements existing genomic (e.g., whole-genome microarray) and other proteomic tools (e.g., mass spectrometry-based proteomic analysis), and facilitates a wide variety of integrated studies, including protein expression, purification, and functional analyses of proteins both in vivo and in vitro.
Introduction
Shewanella oneidensis MR-1, a facultatively anaerobic γ-proteobacterium, is capable of utilizing a diversity of organic compounds and metals to obtain energy needed for growth and survival. Since the genome sequence of S. oneidensis MR-1 was released in 2002 [1], many biological processes in S. oneidensis MR-1 have been investigated in a high-throughput manner by utilizing methods such as whole-genome microarrays, liquid chromatography-mass spectrometry (LC/MS), gas-chromatography-mass spectrometry (GC/MS), and nuclear magnetic resonance (NMR). These studies have greatly improved our understanding of lipopolysaccharides [2], small proteins [3], protein complexes [4] and metabolic flux [5]. In addition, new information has been obtained on regulatory responses to environmental stresses [6]–[12], heavy metals [13]–[16], and to an array of electron acceptors [17]. However, the resources necessary to support systematic in vivo and in vitro approaches for the analysis of protein function as well as protein-protein and DNA-protein interactions are still limited for S. oneidensis.
A comprehensive collection of individual open reading frames (ORFs) would be a powerful resource for S. oneidensis given its importance as a model organism for genetic and genomic studies of metal-reducing bacteria. A collection of such clones should be flexible and amenable for many in vivo and in vitro experimental systems as discussed in recent reports and reviews [18]–[22]. This resource would facilitate research in various aspects of functional genomics and proteomics in S. oneidensis, including but not limited to deciphering the functional activities of proteins, especially those involved in metal reduction [23]–[25], and building regulatory networks and interaction maps [26]–[27].
We describe here the generation and validation of a S. oneidensis clone set constructed using lambda recombinase (Gateway) technology [28]. The method circumvents restriction enzyme digestion and ligation of nucleic acid used in conventional cloning. By exploiting site-specific recombination, it provides a high-throughput means of cloning and subcloning ORF candidates of interest. The system involves cloning ORFs as PCR products into an entry plasmid that serves as the base for a set of clones. The entry vectors do not provide a means for protein expression or functional assays; they are a place-holder for the ORF. The ORF can then be transferred by site-specific recombination to a destination vector of choice. The conversion of the ORF to a destination vector allows for protein expression or functional studies in an organism of interest. With a wide variety of destination vectors, ORFs cloned into the entry vectors can be rapidly transferred for different applications.
Results
Updates in annotation of S. oneidensis genome
To create a comprehensive set of cloned genes for S. oneidensis the most important prerequisite is genome sequence information and the annotation for the complete set of predicted ORFs. The annotation of protein-coding genes in the S. oneidensis genome has changed over time. The genome was sequenced by TIGR (www.tigr.org) and the results were deposited in NCBI GenBank (AE014299 and AE014300) in 2002. S. oneidensis possesses a circular chromosome of 4,969,803 base pairs (bp) and a plasmid of 161,613 bp [1]. According to Heidelberg et al., there are a total of 4,758 and 173 predicted protein-encoding open reading frames on the chromosome and plasmid, respectively [1]. The final released version of annotation at www.tigr.org by TIGR, however, contains a total of 4,938 protein coding genes. After deposition, the genome was reannotated by NCBI and this version (chromosome & plasmid) contains only 4,467 protein coding genes (www.ncbi.nlm.nih.gov) [29]. The genome was annotated again by the Shewanella Federation [30], which further reduced the number of protein coding genes to 4,192. Moreover, the latest annotation also revealed frameshift sequencing errors in 105 ORFs. These errors resulted in a number of changes including truncated, merged, and split genes as well as genes with altered start codons.
Primer design, ORF amplification, and Cloning
The lambda recombination cloning method (Gateway) utilizes a DNA fragment flanked by att recombination sites that can be combined in vitro with an entry vector that also contains recombination sites and incubated with integration proteins to transfer the DNA fragment into the vector [28]. Consequently, the attB1 and attB2 sequences must be added during the PCR amplification at the 5′ end of each ORF-specific forward and reverse primer, respectively. The forward ORF-specific primers begin with the start codon except for genes containing a signal peptide sequence, with which the primers begin at the first codon after the signal peptide. This design prevents removal of any N-terminal fusion peptides in the destination vectors by host-cell signal peptidases. The reverse ORF-specific primers begin with the nucleotide before the stop codon in order to allow C-terminal fusion proteins to be expressed from destination vectors. Because of this design, the amino acids encoded in the final flanking att sequences are fused to each ORF-encoded sequence at the N- and C-termini of the proteins.
For the generation of the S. oneidensis clone set, 4,526 unique pairs of primers were initially designed based on the TIGR annotation. The changes in genome annotation, however, made a number of these primer pairs irrelevant with the end result and 4,030 primer pairs were valid for PCR amplification. All PCR amplification was performed in 96-well plates. From the first round of attempted amplification, a band corresponding to the specific ORF was obtained from 3504 ORFs. The second and third rounds produced 154 and 105 qualified PCR products, respectively. In total, 3,763 ORFs were generated with valid primer pairs by amplification, indicating a 93.3% success rate.
The recombination reactions used to insert the amplified PCR products into pDONR221 were performed in 96-well plates and subsequently were used to transform chemically competent DH5α E. coli cells. Initially, three colonies on LB plates containing kanamycin were picked and examined by PCR with M13 forward and reverse primers. Colonies were regarded as positive clones and stored unless a discrepancy was observed among three PCR products. For failed ORFs, more colonies were examined until three colonies exhibiting a correct size of PCR products were obtained. In total, 3,584 out of 3,763 amplified products were successfully captured in the pDONR221 entry vectors indicating a success rate of 95.4%. This represents approximately 85% of the 4,192 ORFs in S. oneidensis MR-1 genome according to the latest annotation. It is worth noting that the average size of ORFs failed in the recombination reaction was 2315 bp, nearly three times as large as the average size (845 bp) of all ORFs in the genome, implicating that the recombination reaction is less efficient for large genes.
DNA sequence validation of cloned genes
DNA sequence analysis was performed on a collection of the clones to examine the levels of mutations associated with PCR and the cloning procedure. DNA sequencing was attempted for each entry clone from both the 5′ and 3′ directions and at least one additional attempt was made if the quality of the first reads for a particular clone was poor. DNA sequence data were obtained for at least one direction for 85.7% (3,070 of 3,584) of the genes and junction regions in the S. oneidensis pDONR221 entry clone collection. A subset of sequences were analyzed to determine the frequency and source of errors among the cloned genes. A total of 107,071 of 107,083 (99.99%) of the insert bases sequenced were correct in the 159 clones examined. It was not determined whether the incorrect bases were errors that exist in the original genome sequence or introduced during the cloning process. This error frequency is of the same magnitude observed for bacterial whole genome sequencing efforts (typically approximated at 1 error in 10,000 bases). The 12 errors affect nine of the 159 clones examined. The clone for SO3896 contained two frameshifts of one and three nucleotides in length (accounting for four of the errors). The remaining eight errors, two frameshifts and six substitutions, were found in eight genes. While five of the six substitutions led to nonconservative amino acid changes (Met to Ala, Gly to Arg (twice), Ala to Val, and Ser to Pro), one resulted in a conservative substitution (Thr to Ser). Five clones, including SO3896, contained errors in the oligo-encoded 5′ or 3′ adapter att sequences that could affect the frequency of subsequently recombination into destination plasmids. In clones examined where the entire insert gene was sequenced, 31 of 32 clones were found to have no discrepancies with the published gene sequences. Therefore, the clone set contains relatively few mutations due to the PCR and cloning procedures.
Expression and characterization of S. oneidensis two-component response regulatory proteins
The advantage of the lambda recombinational cloning system is that a single set of clones can be converted to many different functional destination vectors. The efficiency of protein expression from the S. oneidensis clones in destination vectors was examined with a set of two-component transcriptional regulatory proteins. In order to respond to changing environments, bacteria such as S. oneidensis utilize global regulatory mechanisms such as the two-component regulatory system to simultaneously mediate the transcription of numerous operons. Two-component systems are composed of a sensor protein that is usually in the membrane and binds to an environmental signaling molecule. In response to the signal, the sensor protein autophosphorylates at a histidine residue and then transfers this phosphoryl group to a response regulatory protein that binds DNA at operons that make up the regulon to activate or repress transcription [31].
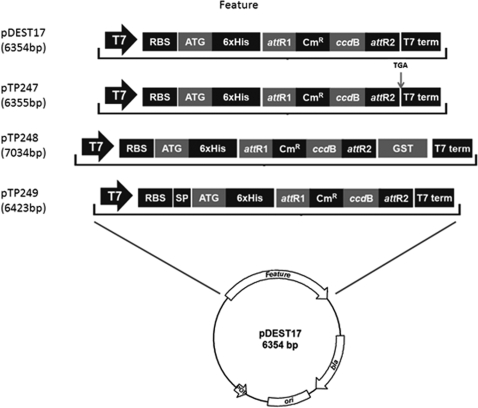
Based on the genome sequence annotation, thirty response regulatory proteins were selected and transferred from the pDONR221 plasmid to the pTP247 destination plasmid which is a modified version of the commercially available pDEST17 His-tag expression vector (Fig. 1). The stop codon of the destination vector pDEST17 is not in-frame with the upstream gene sequence when conversions are made using the pDONR221 vector resulting in the addition of 22 extra amino acids to the end of the gene product. A new, in-frame, stop codon (TGA) was introduced by site directed mutagenesis to create the pTP247 plasmid which avoids the addition of the extra 22 amino acids. LR Clonase (Invitrogen) was used to catalyze homologous recombination between the donor and destination vectors thus creating an expression plasmid for the ORF of interest. The pTP247-ORF plasmids were introduced into E. coli cells and protein expression was induced with IPTG. Twenty-one out of 30 of the proteins tested were expressed in E. coli cells as evaluated by the presence of a band of the expected size after resolving the soluble and insoluble fractions of whole cell protein lysates by SDS-PAGE and Coomassie Blue staining (Table 1). Expression of four of the proteins tested led to poor cell growth possibly due to a toxic effect of the introduced protein. Five of the proteins were not expressed under the conditions tested. The response regulatory proteins that were expressed were found predominantly in insoluble inclusion bodies. Formation of inclusion bodies is common for heterologous protein expression in E. coli and recent studies suggest optimization of growth conditions can lead to increased soluble protein expression [32].
Figure 1. Destination vectors constructed in this study. A variety of destination vectors derived from pDEST17 are shown.
Table 1. Expression data for S. oneidensis response regulator proteins in E. coli .
| ORF | Predicted protein length (a.a) | Protein expression | Soluble or Inclusion body | Annotation |
| SO2538 | 370 | Yes | Inc. body | Response regulator |
| SO4718 | 460 | Noa | Sigma-54 dependent Response regulator | |
| SO2426 | 237 | Noa | DNA-binding Response regulator | |
| SO0622 | 224 | Noa | DNA-binding Response regulator | |
| SO0351 | 198 | Yes | Inc. body | DNA-binding Response regulator |
| SO2193 | 230 | Yes | Inc. body | DNA-binding Response regulator |
| SO2104 | 219 | Yes | Inc. body | DNA-binding Response regulator |
| SO2540 | 192 | Noa | Response regulator | |
| SO2125 | 208 | Yes | Inc. body | chemotaxis protein CheD |
| SO1860 | 214 | Yes | Inc. body | DNA-binding Response regulator |
| SO1946 | 224 | Yes | Inc. body | Transcriptional regulatory protein, PhoP |
| SO4647 | 231 | Yes | Inc. body | DNA-binding Response regulator |
| SO0860 | 339 | Yes | Inc. body | Response regulator |
| SO2539 | 658 | Yes | Inc. body | Response regulator |
| SO2366 | 525 | Yes | Inc. body | Response regulator |
| SO1416 | 199 | Yes | Inc. body | DNA-binding Response regulator |
| SO3594 | 237 | Yes | Inc. body | Transcriptional regulatory protein, RstA, putative |
| SO0545 | 575 | No | Response regulator | |
| SO2541 | 441 | Yes | Inc. body | Response regulator |
| SO2547 | 121 | No | Response regulator | |
| SO0059 | 229 | Yes | Inc. body | Transcriptional regulatory protein, KdpE |
| SO1558 | 239 | Yes | Inc. body | Phosphate regulon Response regulator, PhoB |
| SO3982 | 209 | Yes | Soluble | DNA-biding nitrate/nitrite Response regulator |
| SO4003 | 357 | Yes | Soluble | Response regulator |
| SO4172 | 183 | No | DNA-binding Response regulator | |
| SO4388 | 227 | Yes | Inc. body | DNA-binding Response regulator |
| SO4477 | 228 | Yes | Inc. body | Transcriptional regulatory protein, CpxR |
| SO4633 | 241 | No | Transcriptional regulatory protein, OmpR | |
| SO4428 | 225 | No | DNA-binding Response regulator | |
| SO4487 | 220 | Yes | Inc. body | DNA-binding Response regulator |
expression of the protein has a strong negative effect on growth of E. coli host cells
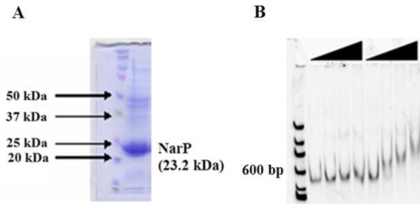
Protein expression and purification studies were pursued further for two of the response regulatory proteins, ArcA and NarP. As shown previously by Gao et al. [33], the purified ArcA protein can bind specifically to promoter DNA when phosphorylated/activated using carbamoyl phosphate according to electrophoretic gel mobility shift assays [33]. The non-phosphorylated protein did not show any specific DNA binding activity [33]. SO3982, showing sequence homology to E. coli NarP (72% similarity), was named as NarP in the latest annotation. NarP is a response regulator belonging to the LuxR family. Evidence from work in E. coli suggests that NarP may regulate its own expression [34], as well as expression of NrfA, a formate-dependent nitrite reductase enzyme [35]. In S. oneidensis, NarP and NrfA share the same promoter region, although the genes are transcribed in opposite directions. This promoter region was amplified via PCR and used in electrophoretic mobility shift assays. The cloned narP gene in the pDONR221 vector was transferred to the pTP247 His-tag destination vector and the NarP protein was expressed and purified in soluble form using metal affinity chromatography (Fig. 2A). The purified protein was phosphorylated with carbamoyl phosphate and electrophoretic mobility shift (EMSA) DNA binding studies indicated the phosphorylated SO3982 protein bound to a PCR product containing narP-nrfA promoter DNA while nonphosphorylated SO3982 protein did not (Fig. 2B). These results indicate that NarP phosphorylation is required for DNA binding. These studies demonstrate that the clone set and the pTP247 destination plasmid can be used to successfully express and purify proteins in E. coli.
Figure 2. SDS-PAGE of S. oneidensis NarP expressed from the destination vector pTP247.
A. Lane 1: protein marker; Lane 2: Soluble protein after lysis of E. coli expressing NarP protein. B. Electrophoretic gel shift assays of NarP and phosphorylated NarP-P. Increasing concentrations (0, 0.65, 1.3, 2.6, and 5 µM) of unphosphorylated NarP and phosphorylated NarP are shown. DNA is stained with CyberGreen. The 100bp ladder was used in the experiments.
Construction of additional destination vectors for protein expression
Two additional destination vectors that are useful for protein expression were constructed for this study (Fig. 1). The pTP249 plasmid is a destination vector for the expression of proteins in E. coli that are secreted to the periplasmic space. It was constructed by inserting a DNA fragment encoding the E. coli pelB secretion signal sequence into the pDEST17 plasmid. Upon conversion of genes from the pDONR221 plasmid to pTP249, the proteins are secreted to the periplasm of E. coli. This plasmid was constructed in order to express periplasmic proteins in their normal environment. This is particularly important for the expression of proteins containing disulfide bonds that will be oxidized in the periplasm but not the cytoplasm.
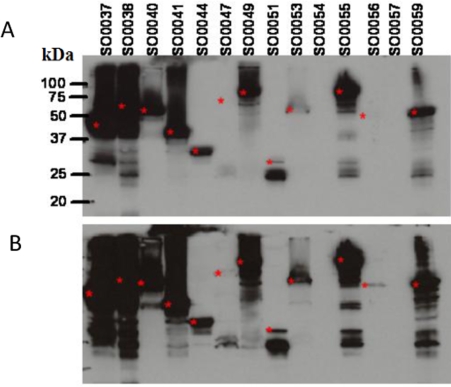
The pTP248 destination vector was constructed in order to express proteins with purification tags at both the N and C-termini of the protein. After conversion of genes from the clone set in pDONR221 to pTP248 and expression in E. coli, the proteins contain a 6x His-tag at their N-terminus and the GST protein fused to the C-terminus. A set of S. oneidensis genes in pDONR221 were converted to the pTP248 vector and protein expression was evaluated upon IPTG induction in E. coli to estimate the percentage of gene products that can be expressed from this construct. These genes, from the SO0037 to SO0059 ORF region of the chromosome, encode proteins with a range of different functions including metabolism, transcription regulation, transporters and conserved hypotheticals. As seen in Fig. 3, approximately 60–70% of the S. oneidensis gene products were expressed in E. coli and detected in whole cell lysates by Western blotting using an anti-GST antibody.
Figure 3. Expression of S. oneidensis ORFs from the pTP248 double-tag destination plasmid.
A. The protein expression levels of ORFs were evaluated from whole cell protein lysates of E. coli by SDS-Page fractionation and immunodetection using anti-GST antibody. The S. oneidensis ORF number is shown above each lane. The red asterisks indicate the position of a band that is consistent with the predicted molecular weight of the fusion protein. Note that for ORFs SO0047 and SO0056, a band is not visible in panel A but is visible in the longer film exposure of panel B. The predicted molecular weights (kDa) of the fusion proteins are: SO0037, 40.7; SO0038, 65.1; SO0040, 61.2; SO0041, 41.4; SO0044, 39.7; SO0047, 75.1; SO0049, 86.9; SO0051, 34.3; SO0053, 66.8; SO0054, 74.2; SO0055, 94.7; SO0056, 59.3; SO0057, 79.9; SO0059, 57.1. B. Identical to A. but with a longer film exposure.
Phage display of Shewanella ORFs
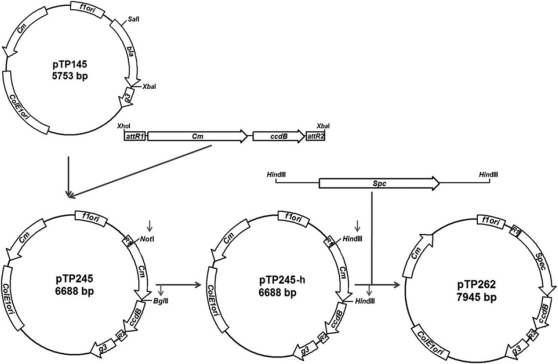
Phage display is an approach for the study of interactions between proteins, proteins and ligands, and proteins and DNAs. Several different types of bacteriophages have been used as platforms for phage display but the most commonly used is M13 bacteriophage [36]. The phage display is one of the techniques chosen by the Shewanella Federation for functional analyses thus a great effort was made to create and validate a phage display system for the bacterium. A destination vector was made by inserting a cassette containing attR sites flanking a chlorampenicol resistance gene and the ccdB gene for counter selection, into the pTP145 phage display vector [37]. The pTP145 phage display plasmid allows display of the TEM-1 β-lactamase enzyme as a fusion to the M13 gene III capsid. The attR cassette was inserted between the β-lactamase signal sequence and gene III which, upon recombination conversion with a pDONR221 vector would place a S. oneidensis ORF in position to be displayed on the surface of M13 bacteriophage (Fig. 4). This destination vector was named pTP262.
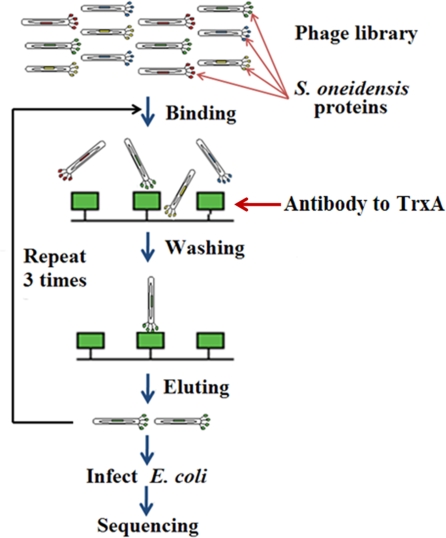
Figure 4. Phage display destination vector constructed in this study.
M13 filamentous phage display vector pTP262 is a destination plasmid that can be used for insertion of ORFs from the pDONR clone set to create phage display plasmids.
In order to test the usefulness of the bacteriophage M13 phage display system implemented with pTP262, a sub-library was constructed and tested for the ability to isolate a gene by biopanning. The phage display system was tested using a rabbit polyclonal antibody to the E. coli thioredoxin protein (Materials and Methods). The S. oneidensis thioredoxin protein is 76% identical to the E. coli protein and it was reasoned that the polyclonal antibody against the E. coli protein would also bind the S. oneidensis protein. The library to be used for phage display consisted of 80 clones from the SO0337 to SO0516 region of the genome which includes SO0406 encoding thioredoxin (trxA). Plasmid DNA from each pDONR221 clone was prepared and the set of 80 plasmids was pooled. The pooled plasmids were then converted from the pDONR221 vector to the pTP262 destination vector in batch using LR Clonase recombination enzyme [28]. The reaction products were introduced into E. coli by electroporation and the transformed cells were spread on agar plates containing chloramphenicol. Six colonies were picked and analyzed by restriction enzyme digestion to ensure that the conversion reaction was efficient and the library contained inserts into the pTP262 plasmid. Five of six clones contained inserts and the five inserts were of different size indicating the reaction of efficient and the library is diverse. Approximately 10,500 colonies on the agar plates were pooled to create the sub-library of S. oneidensis genes in pTP262.
Phage were prepared from the library using helper phage as previously described [37]. Three rounds of biopanning were performed whereby a polyclonal antibody against the highly similar E. coli thioredoxin was immobilized and the phage library was allowed to bind, unbound phage were washed away and bound phage were eluted and amplified for the next round of panning (Fig. 5). After the third round of biopanning, the eluted phages were used to infect E. coli. Six clones were chosen from the round 3 population and the identity of the gene for each of the six clones was determined by DNA sequencing to be trxA, which encodes thioredoxin. This result indicates the trxA phage clone was enriched during the panning process and dominated the population after three rounds. Therefore, it was concluded that the thioredoxin was efficiently displayed on the surface of the phage after expression from the pTP262 destination vector. These results suggest that pooling sets of ORF clones is an efficient means of converting the clone set to a phage display vector and that antibody-antigen interactions can be enriched by phage display.
Figure 5. In vitro biopanning protocol.
Briefly, phages were incubated with immobilized anti-E. coli thioredoxin antibody, non-binders were removed by washing, bound phage were eluted, amplified, and the process was repeated. Eluted phage clones from round 3 were DNA sequenced.
Discussion
While molecular cloning with restriction enzymes has been a well-established procedure for more than three decades, the emergence of the Gateway (Invitrogen), the Creator (Clontech), the Univector [38], and the MAGIC [39] cloning systems makes the creation of an ORF repositories feasible. Such site-specific recombination based cloning systems have been successfully employed for building a number of clone sets [18], [20]–[22], [40]–[42]. In recent years, the lambda recombinase (Gateway) system has been become popular because of its high-fidelity and cost efficient properties [18], [20], [22], [41]–[42]. In this study, an ORF clone set for S. oneidensis was created using the lambda recombinase system [28]. ORFs within entry vectors in this system can be readily transferred into multiple destination vectors, making the clone set a useful resource for research groups studying this microorganism, especially those expressing proteins in vivo or in vitro and using a variety of experimental analyses [26], [43]–[46].
Due to the acquisition of new experimental data and advanced sequence analysis methods, annotation of protein-coding genes in the S. oneidensis genome has become increasingly accurate. The clone set was constructed using primer pairs based on the most recent annotation. As outlined above, 3,584 ORFs were captured in the pDONR221entry vector, with a success rate of 93.3 % for PCR amplification and 95.4% for cloning. These rates are in agreement with those observed from the work on Brucella melitensis [41]. Despite multiple attempts at PCR amplification and cloning, a number of ORFs were not amplified and/or captured in the donor vector. It appears that the ORF size affects the BP cloning efficiency negatively. In average, the ORFs failed to be captured in the BP reaction were nearly three times larger than the ORFs in the genome. The S. oneidensis clone set was validated by sequencing a total of 107,083 insert bases. 12 mutations were identified, corresponding to one error per 8924 bp or a mutation rate of 0.01%. This is significantly lower than those observed in the other clone sets [18], [40]. There are multiple factors that could contribute to mutations. The mutation rate is 1 error every 2673 bp in the Treponema pallidum clone set, which employed AdvanTaq DNA polymerase (Clontech) for PCR amplification [40]. The higher fidelity of Pfu turbo DNA polymerase used in this study likely accounts for the difference in mutation rates. On the other hand, a mutation rate of 1 error every 450 bp obtained in the Pseudomonas aeruginosa clone set was suggested by authors to be largely due to the high-GC-content ORFs [18]. Strikingly, PCR amplification during construction of this P. aeruginosa clone set yielded a success rate of 99%. Taq polymerase is reported to have 4-10 times higher processitivity than Pfu turbo polymerase although the enzymes differ little in amplification efficiency [47]. Nonetheless, it is important to amplify ORFs for a clone set with an enzyme of high amplification efficiency that also has a low error rate.
Although the Gateway system offers multiple destination vectors for a variety of analyses, we developed a series of destination plasmids to better suit the needs of this study. The backbone of these plasmids is pDEST17, one of the commercial destination vectors specifically designed for the Gateway system (Invitrogen). With introduction of a new, in-frame, stop codon, the resulting His-tag expression vector pTP247 functions the same as pDEST17 without the addition of the extra 22 amino acids to the end of the gene product. Other plasmids were designed for expression of proteins with a His-GST double-tag (pTP248) and for secreted or periplasmic proteins (pTP249) as well as for M13 phage display (pTP262). Except for pTP249, all these plasmids were used to validate selected clones from the collection. The newly constructed destination plasmids are suitable for use with other clone sets or with individual genes.
To establish that the S. oneidensis clone set could be used for protein expression and functional studies, three sets of ORFs were examined for expression of His-tag proteins, expression of His/GST-tag proteins, or for effective display on phage. A total of 21 out of 30 (70%) predicted two-component transcriptional regulators from S. oneidensis were successfully expressed in the His-tag format. While the reason for failure in expression of five proteins is unknown, expression of four genes was toxic to E. coli growth. In-depth studies with both ArcA and NarP revealed that purified proteins were suitable for in vitro binding assays, further validating the use the clone set for protein expression [33]. A similar success rate was obtained for expression of 14 His-GST double tagged proteins examined by Western blotting using an anti-GST antibody.
The use of the S. oneidensis clone set for functional studies was tested using a phage display system. Phage display is a useful method for studying protein-ligand interactions [36]. The method involves the fusion of peptides or proteins to a coat protein of a bacteriophage. This results in display of the fused protein on the exterior of the phage, while the DNA encoding the fusion resides within the virion. The physical linkage between the displayed protein and the DNA encoding it allows screening of vast numbers of proteins for ligand-binding properties [36]. With this technology, a phage clone encoding thioredoxin TrxA was isolated from a sub-library consisting of 80 clones.
It is evident that the S. oneidensis clone set can be used for expression of functional S. oneidensis proteins in E. coli using the appropriate destination vectors. Destination vectors are available for a number of protein expression formats as well as for functional studies such as phage display and yeast two-hybrid assays. Note that it should be possible to construct a destination vector that can be used to expression proteins directly in S. oneidensis. In combination with whole-genome microarrays and proteomics databases [3], [6], the clone set is a useful resource for integrated studies at the whole genome level. The functionality of many genes cannot be determined from sequence similarity alone. Studies on S. oneidensis have revealed many surprising findings about proteins that have been well-studied in other model organisms such as E. coli. These include the observation that reduction pathways for all electron acceptors but TMAO share a pivot electron transfer protein CymA [48], that an Arc system without ArcB mediates aerobic growth [33], [49], that the E. coli Fnr analog has no significant role in anaerobiosis [50], and that Crp functions under anaerobic but not aerobic conditions [51], to name a few. In this respect, the clone set will be particularly useful because it facilitates protein expression, purification, and a variety of functional analyses
Methods
Bacterial strains, plasmids, primers, and culture conditions
A list of Escherichia coli strains, plasmids, and primers used in this study is given in Table 2. The cloning to obtain master clones, protein expression, and the Gateway destination cloning were performed using E. coli strains DH5α, BL21(DE3)*, and DB3.1, respectively. Other recombinant DNA experiments were performed using E. coli XL1-Blue (Stratagen). Unless otherwise noted, E. coli strains and S. oneidensis MR-1 were grown in Luria-Bertani (LB, Difco) medium at 37°C and room temperature for genetic manipulation, respectively. When needed, the growth medium was supplemented with antibiotics at the following concentrations: ampicillin at 50 µg/ml, kanamycin at 50 µg/ml, chloramphenicol at 12.5 µg/ml, and spectinomycin at 50 µg/ml.
Table 2. Strains, plasmids, and primers used in this study.
| Strain or plasmid | Characteristic or relevant feature | Reference or source |
| E. coli | ||
| DH5α | F- φ80lacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17 (rk−, mk+) phoA supE44 λ- thi-1 gyrA96 relA1 | Invitrogen |
| BL21 | F- ompT hsdSB (r B −m B −) gal dcm rne131 (DE3) | Invitrogen |
| DB3.1 | F- gyrA462 endA1 glnV44 Δ(sr1-recA) mcrB mrr hsdS20(rB −, mB −) ara14 galK2 lacY1 proA2 rpsL20(Smr) xyl5 | Invitrogen |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 (rk −, mk −) supE44 relA1 lac (F− proAB lacI q Z ΔM15 Tn10 (tetr)) | Stratagene |
| Plasmids | ||
| pDONR221 | Entry vector, Kmr | Invitrogen |
| pDEST17 | Destination expression vector of the Gateway system (His-tag), Apr | Invitrogen |
| pDEST24 | Destination expression vector of the Gateway system (GST), GST donor, Apr | Invitrogen |
| pTP145 | Phage display vector, Cmr | 37 |
| pKRP13 | Spectinomycin resistance gene donor, Spr | 53 |
| pTP245 | Phage display vector derived from pTP145, containing the attR1-Cmr-ccdB-attR2 sequence, Apr | This study |
| pTP247 | Expression vector derived from pDEST17, containing a stop codon at the position 1846, Apr | This study |
| pTP248 | Expression vector derived from pDEST17, containing both His- and GST tags, Apr | This study |
| pTP249 | Expression vector derived from pDEST17, containing a DNA fragment encoding a pelB signal sequence, Apr | This study |
| pTP262 | Phage display vector derived from pTP145, containing the attR1-Sper-ccdB-attR2 sequence, Cmr, Spr | This study |
| Primers | ||
| attB1 forward | GGGGACAAGTTTGTACAAAAAAGCAGGCTTC-ORF specific bps1 | Invitrogen |
| attB2 reverse | GGGGACCACTTTGTACAAGAAAGCTGGGTN-ORF specific bps2 | Invitrogen |
| DONRF-2 | GTTCCCTACTCTCGCGTTAACG | This study |
| DEST17-STOP | CAGCTTTCTTGTACAAAGTGGTTTGATTCGAGG | This study |
| PELB-F | TATGAAATACCTGCTGCCGACCGCTGCTGCTGGTCTGCTGCTCCTCGCTGCCCAGCCGGCGATGGCCCA | This study |
| PELB-R | ACTTTATGGACGACGGCTGGCGACGACGACCAGACGACGAGGAGCGACGGGTCGGCCGCTACCGGGTAT | This study |
| DEST-XHOI | GGGCTCGAGCTCTCGAATCAACAAGTTTGTACAAAAAAGCTGAACGAGAAACGTAAAATG | This study |
| DEST-XBAI | GGCGTCTAGACAACCACTTTGTACAAGAAAGCTGAACGAGAAACGTAAAATGATATAAATATC | This study |
| NPAP-F | TTGCGAGGCTCAGTTTTATCAC | This study |
| NPAP-R | CCAGAATTGGCGTAAAACATCA | This study |
The 25 bp attB1site is underlined.
The 25 bp attB2 site is underlined, N represents A, G, C, or T.
Primer Design
The predicted S. oneidensis MR-1 open reading frames (ORFs) based on the annotation by TIGR (www.tigr.org) were used for primer design. Signal peptides were determined using Signal-P prediction (http://www.cbs.dtu.dk/services/SignalP/) and the corresponding encoding DNA sequences were removed prior to primer design. Further analysis using PRIMEGENS was performed to identify repetitive ORFs, for which one pair of primers was designed for a group [52]. In total, 4526 unique pairs of primers were designed for S. oneidensis. The general guidelines for design were from manufacturer's recommendations (Invitrogen). The attB1 and attB2 sequences, listed in Table 2, were included at the 5′ ends of each of the ORF specific primer pairs. To maintain the proper reading frame between the attB1 and attB2 regions and the ORF, additional nucleotides (TC) and N (A, G, C, or T) were added to the 3′ ends of the attB1 and attB2 sequences, respectively, before the ORF-specific nucleotides (Table 2). As a result, one of four amino acids (Tyr, His, Asn, or Asp) will be inserted as the first amino acid C-terminal to the ORF as part of the att fusion region when the protein is translated from a destination vector. The ORF-specific region of the forward primers began with the start codon and the reverse primers began with the final nucleotide before the stop codon. The natural stop codon for each ORF was not included in the reverse primers. The annealing temperature of these primers was kept in the range of from 54 to 65°C by adjusting the length of primers (18–28 bp). The primers, given in Table S1, were synthesized by MWG (mwg-biotech.com) in 96-well format.
PCR amplification of the ORFs
Amplification of the ORFs was performed in a 96-well format using in a GeneAmp PCR System 9700 thermal cycler (Applied Biosystems). The first attempt was made in a volume of 25 µl, with 3.125 U of Pfu Turbo polymerase, 1.5 X final concentration of a 10X cloned Pfu DNA polymerase buffer, 500 µM of each dNTP, 0.5 µM of each of Forward and Reverse primers, and 100 ng of genomic DNA based on the manufacturer's recommendation (Stratagene). Thermocycling was carried out using an initial denaturation step at 94°C for 2 min, followed by 30 cycles of denaturation at 94°C for 30 seconds, annealing at 55°C for 30 seconds, and elongation at 72°C for 2 min. After a final incubation at 72°C for 7 minutes, the reaction was held at 4°C. For longer ORFs (>2 kb), elongation time at 72°C was proportionally extended. PCR products were examined on a 1% agarose gel to confirm the size of the amplicons. For ORFs that failed to display the correct size of PCR product, at least three attempts were made with modified amplification conditions based on the annealing temperature of the corresponding primers. For ORFs that exhibited multiple bands including one of the correct size, gel purification of the correct size products was performed using QIAquick Gel Extraction Kit (Qiagen).
BP recombinational cloning
PCR products of correct size were cloned into pDONR221 using the BP reaction kit (Invitrogen) following the protocol recommended by the manufacturer. Chemically competent E.coli DH5α cells (Invitrogen) were transformed with 2 µl of the BP reaction, spread onto LB-kanamycin plates, and incubated overnight. Kanamycin resistant E.coli DH5α transformants were screened using colony PCR with universal vector primers M13 Forward and Reverse. Visualization of a PCR product corresponding to the size of the ORF indicated the insertion of the ORF into the vector.
Plasmid DNA isolation and DNA sequencing
Entry plasmid DNA was isolated for each clone using the QIAprep Spin Miniprep or QIAprep 96 Turbo Miniprep kits (Qiagen). Plasmid DNA was sequenced using Big Dye chemistry (Applied Biosystems) with the M13 Forward and M13 Reverse primers available from Invitrogen and/or with an additional forward primer DONRF-2 (Table 2). Sequence reads were compared to the S. oneidensis whole genome sequence or the pDEST17 plasmid sequence using BLASTn and BLASTx (NCBI, NLM). Discrepancies were verified by manual examination of the sequence traces. Custom oligonucleotides were purchased from Integrated DNA Technologies (idtdna.com).
Plasmid constructions
a) Construction of pTP247
The stop codon of the destination vector pDEST17 (Invitrogen) is not in-frame with the upstream gene sequence when conversions are made using the pDONR221 vector. This results in the addition of 22 extra amino acids to the converted gene product. The position of the stop codon in pDEST17 was changed by site-directed mutagenesis using primer DEST17-STOP (Table 2). Compared with pDEST17, the new plasmid, named pTP247, contains a stop codon (TGA) at position 1846.
b) Construction of pTP248
To create a His/GST double tagged destination vector, a SalI–NheI fragment containing the GST gene was released from pDEST24 by restriction enzyme digestion, and then inserted into pDEST17 between the SalI and NheI sites. The resulting plasmid, named pTP248, contains a His tag and a GST tag flanking the target gene at the N-terminal and C-terminal sites, respectively.
c) Construction of pTP249
To create a destination plasmid that can be used to express secreted or periplasmic proteins, a DNA fragment encoding a pelB signal sequence was created by annealing synthetic oligonucleotides (Table 2) and inserting the resulting DNA fragment into the NdeI restriction site of the pDEST17 plasmid. The resulting plasmid was named as pTP249.
d) Construction of pTP262
In order to construct a destination vector for M13 phage display, the attR1-Cmr-ccdB-attR2 region of the pDEST17 destination vector was amplified by PCR using primers DEST-XHOI and DEST-XBAI (Table 2).
The resulting product was digested with the XhoI and XbaI restriction enzymes and inserted into the pTP145 phage display vector digested with SalI and XbaI [37]. The resulting plasmid, named pTP245, has the attR1 site fused adjacent to the blaTEM-1 signal sequence for protein secretion and the attR2 site fused adjacent to M13 gene III for phage display [37]. The pTP145 plasmid, however, contains a chloramphenicol resistance gene and therefore the Cmr gene present in the attR cassette is not useful for selection. To insert a useful selectable marker, the Cmr gene in the attR cassette was replaced with a spectinomycin resistance gene from the plasmid pKRP13 [53]. This was accomplished by creating HindIII restriction sites at the NotI and BglII sites of pTP245 which flank the Cmr resistance gene in the attR1-attR2 cassette by oligonucleotide directed mutagenesis. The Cmr resistance gene was then replaced by inserting the spectinomycin gene on 2.0 kbp HindIII fragment of pKRP13 [53]. The resulting plasmid, named pTP262, contains an attR1-Spr-ccdB-attR2 cassette between the signal sequence of β-lactamase and M13 phage gene III (Fig. 3).
Expression of two-component regulatory proteins
The experiments to determine protein expression levels of two-component response regulatory proteins in E. coli were initiated by converting the relevant pDONR-ORF plasmids to the pTP247 His-tag destination plasmid using a Clonase reaction according to manufacturer instructions (Invitrogen). The resulting pTP247-ORF plasmids were introduced into E. coli BL21(DE3)* for protein expression experiments. E. coli BL21(DE3)* strains containing the expression plasmids were inoculated into 15 ml of LB with ampicillin and grown to an OD600≈1.0, at which point protein expression was induced by the addition of IPTG to a concentration of 1 mM and the cultures were grown overnight. After overnight growth, 1 ml of each induced culture was centrifuged at 10K rpm for 1 min. to pellet cells. The supernatant was discarded and the cell pellet was resuspended in 300 µl of bacterial protein extraction reagent (B-PER, Pierce) and 6 µl of 10 mg/ml lysozyme was added. The cell suspension was frozen at −80°C and then thawed at room temperature and centrifuged at 13K rpm for 10 min. The precipitated proteins in the pellet were resuspended in 30 µl SDS gel loading buffer and 20 µl of soluble supernatant was mixed with 30 µl of SDS gel loading buffer. The protein preparations were then heated to 95°C and fractionated by SDS-PAGE. The SDS-PAGE gels were stained with Coomassie blue and the level of protein expression was judged by the presence or absence of a band of the expected size on the gel.
Purification of NarP protein and use in gel shift assays
For purification of NarP, the pTP247-narP was introduced into E. coli BL21(DE3)*. This strain was grown in 1 liter of LB with ampicillin to an OD600 of 1.0 and induced with 1 mM IPTG and grown overnight. The cells were centrifuged and the pellet was resuspended in 50 ml of 1X Equilibration buffer (50 mM sodium phosphate buffer, pH 7.0, 300 mM NaCl). A protein lysate was made from the resuspended cells using a microfluidizer instrument. The lysate was centrifuged at 14K rpm for 20 min. to remove insoluble material. NarP was purified from the protein lysate by metal affinity chromatography using a TALON resin (Clontech) that had been equilibrated in 1X Equilibration buffer and protein was eluted with 50 mM sodium phosphate, 300 mM NaCl, and 150 mM immidazole. The purity of the eluted fractions was evaluated by SDS-PAGE coomassie-stained gels. Fractions containing >90% pure NarP were pooled and dialyzed against 50 mM HEPES buffer. The NarP protein was concentrated by ultrafiltration using an Amicon Ultra-15 column and the protein concentration was determined using a Bradford assay [54]. Electrophoretic mobility shift assays were performed for NarP with minor modifications as described for ArcA by Gao et al. [33]. The modification for the NarP experiments is that the shifted and unshifted DNA was visualized using SYBR Green staining rather than by labeling the DNA with 33P. The target narP-nrfA promoter DNA for the EMSA experiments is a 527 bp PCR product obtained using the primers NPAP-F and NPAP-R (Table 2). This region corresponds to nucleotide coordinates 4117504 to 4118031 in the Shewanella oneidensis MR1 genome sequence [1].
Western blotting of Shewanella proteins expressed from pTP248
To examine expression levels of His/GST double tagged proteins, the so0037 to so0059 ORFs were converted from the pDONR221 plasmid to the pTP248 plasmid using LR clonase according to manufacturer instructions. The SO-orf-pTP248 clones were introduced into E. coli BL21(DE3)* by electroporation and individual cultures were grown in 10 ml of LB supplemented with ampicillin to an OD600 of approximately 0.6 and protein expression was induced by adding IPTG to a final concentration of 0.3 mM. The cultures were then grown overnight. The samples were centrifuged to pellet cells and the cells were resuspended in SDS-PAGE loading buffer. The amount of loading buffer used was adjusted to the OD600 of the cultures to maintain a constant OD600 to loading buffer ratio. A total of 15 µl of each sample was fractionated by SDS-PAGE and the proteins were transferred to a nitrocellulose filter. Western blotting was then performed as previously described [55] using a 1:5000 dilution of rabbit anti-GST polyclonal antibody purchased from Amersham.
Phage display biopanning
Plasmid DNA from pDONR clones within the so0337 to so0516 region was prepared and the set of 80 plasmids was pooled. The pooled plasmids were then converted from the pDONR vector to the pTP262 destination vector in batch using Clonase recombination enzyme in 10 µl reaction volume. The reaction products were introduced into E. coli XL1-Blue by electroporation and the transformed cells were spread on agar plates containing chloramphenicol. Six colonies were picked and analyzed by restriction enzyme digests to ensure the conversion occurred efficiently and the library consisted of inserts into the pTP262 plasmid. Five of the six colonies analyzed contained an insert and the inserts were of different size. The colonies on the agar plates were then pooled to create a sub-library of S. oneidensis genes in pTP262.
Biopanning was performed by coating a microtiter ELISA well with 10 µg/ml of rabbit polyclonal anti-E. coli thioredoxin antibody (Sigma). After absorption, the wells were blocked with 3% dry milk, 0.05% Tween 20 in 1x TBS. Approximately 1×1011 phage were added to the well in 1x TBS and incubated to allow binding. After 10 cycles of washing with 1x TBS, bound phages were eluted using 0.2 M glycine, pH 2.2. The elution was neutralized with 1 M Tris pH 8.0 and the eluate was used to infect E. coli TG1. VCS M13 helper phage were added and the culture was grown overnight and phages were prepared as previously described [37]. This process was repeated three times to enrich for phage that displayed a protein that bound the anti-thioredoxin antibody. After the third round, the eluted phage were used to infect E. coli TG1 and the transfections were spread on LB agar plates containing chloramphenicol to isolate single colonies. Six colonies were picked and the phagemid DNA was isolated and the DNA sequence of the ORF region was determined by automated DNA sequence using Big Dye Terminator and an Applied Biosystems 3100 capillary DNA sequencer.
Supporting Information
S. oneidensis clone set ORF-specific primers.
(0.69 MB XLS)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was supported by The U.S. Department of Energy under the Genomics: GTL Program through Shewanella Federation, Office of Biological and Environmental Research, Office of Science. Oak Ridge National Laboratory is managed by University of Tennessee-Battelle LLC for the Department of Energy under contract DOE-AC05-00OR22725.
References
- 1.Heidelberg JF, Paulsen IT, Nelson KE, Gaidos EJ, Nelson WC, et al. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat Biotech. 2002;20:1118–1123. doi: 10.1038/nbt749. [DOI] [PubMed] [Google Scholar]
- 2.Korenevsky AA, Vinogradov E, Gorby Y, Beveridge TJ. Characterization of the lipopolysaccharides and capsules of Shewanella spp. Appl Environ Microbiol. 2002;68:4653–4657. doi: 10.1128/AEM.68.9.4653-4657.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Romine MF, Elias DA, Monroe ME, Auberry K, Fang R, et al. Validation of Shewanella oneidensis MR-1 small proteins by amt tag-based proteome analysis. OMICS. 2004;8:239–254. doi: 10.1089/omi.2004.8.239. [DOI] [PubMed] [Google Scholar]
- 4.Tang X, Yi W, Munske GR, Adhikari DP, Zakharova NL, et al. Profiling the membrane proteome of Shewanella oneidensis MR-1 with new affinity labeling probes. J Proteome Res. 2007;6:724–734. doi: 10.1021/pr060480e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang YJ, Hwang JS, Wemmer DE, Keasling JD. Shewanella oneidensis MR-1 fluxome under various oxygen conditions. Appl Environ Microbiol. 2007;73:718–729. doi: 10.1128/AEM.01532-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao H, Wang Y, Liu X, Yan T, Wu L, et al. Global transcriptome analysis of the heat shock response of Shewanella oneidensis. J Bacteriol. 2004;186:7796–7803. doi: 10.1128/JB.186.22.7796-7803.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao H, Yang ZK, Wu L, Thompson DK, Zhou J. Global transcriptome analysis of the cold shock response of Shewanella oneidensis MR-1 and mutational analysis of its classical cold shock proteins. J Bacteriol. 2006;188:4560–4569. doi: 10.1128/JB.01908-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y, Gao W, Wang Y, Wu L, Liu X, et al. Transcriptome analysis of Shewanella oneidensis MR-1 in response to elevated salt conditions. J Bacteriol. 2005;187:2501–2507. doi: 10.1128/JB.187.7.2501-2507.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qiu X, Daly MJ, Vasilenko A, Omelchenko MV, Gaidamakova EK, et al. Transcriptome analysis applied to survival of Shewanella oneidensis MR-1 exposed to ionizing radiation. J Bacteriol. 2006;188:1199–1204. doi: 10.1128/JB.188.3.1199-1204.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qiu X, Sundin GW, Wu L, Zhou J, Tiedje JM. Comparative analysis of differentially expressed genes in Shewanella oneidensis MR-1 following exposure to UVC, UVB, and UVA radiation. J Bacteriol. 2005;187:3556–3564. doi: 10.1128/JB.187.10.3556-3564.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kolker E, Picone AF, Galperin MY, Romine MF, Higdon R, et al. Global profiling of Shewanella oneidensis MR-1: Expression of hypothetical genes and improved functional annotations. Proc Natl Acad Sci. 2005;102:2099–2104. doi: 10.1073/pnas.0409111102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leaphart AB, Thompson DK, Huang K, Alm E, Wan X-F, et al. Transcriptome profiling of Shewanella oneidensis gene expression following exposure to acidic and alkaline pH. J Bacteriol. 2006;188:1633–1642. doi: 10.1128/JB.188.4.1633-1642.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bencheikh-Latmani R, Williams SM, Haucke L, Criddle CS, Wu L, et al. Global transcriptional profiling of Shewanella oneidensis MR-1 during Cr(VI) and U(VI) reduction. Appl Environ Microbiol. 2005;71:7453–7460. doi: 10.1128/AEM.71.11.7453-7460.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown SD, Martin M, Deshpande S, Seal S, Huang K, et al. Cellular response of Shewanella oneidensis to strontium stress. Appl Environ Microbiol. 2006;72:890–900. doi: 10.1128/AEM.72.1.890-900.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown SD, Thompson MR, VerBerkmoes NC, Chourey K, Shah M, et al. Molecular dynamics of the Shewanella oneidensis response to chromate stress. Mol Cell Proteomics. 2006;5:1054–1071. doi: 10.1074/mcp.M500394-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Chourey K, Thompson MR, Morrell-Falvey J, VerBerkmoes NC, Brown SD, et al. Global molecular and morphological effects of 24-hour chromium(VI) exposure on Shewanella oneidensis MR-1. Appl Environ Microbiol. 2006;72:6331–6344. doi: 10.1128/AEM.00813-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beliaev AS, Klingeman DM, Klappenbach JA, Wu L, Romine MF, et al. Global transcriptome analysis of Shewanella oneidensis MR-1 exposed to different terminal electron acceptors. J Bacteriol. 2005;187:7138–7145. doi: 10.1128/JB.187.20.7138-7145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LaBaer J, Qiu Q, Anumanthan A, Mar W, Zuo D, et al. The Pseudomonas aeruginosa PA01 gene collection. Genome Res. 2004;14:2190–2200. doi: 10.1101/gr.2482804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marsischky G, LaBaer J. Many paths to many clones: a comparative look at high-throughput cloning methods. Genome Res. 2004;14:2020–2028. doi: 10.1101/gr.2528804. [DOI] [PubMed] [Google Scholar]
- 20.Aguiar JC, LaBaer J, Blair PL, Shamailova VY, Koundinya M, et al. High-throughput generation of P. falciparum functional molecules by recombinational cloning. Genome Res. 2004;14:2076–2082. doi: 10.1101/gr.2416604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park J, Hu Y, Murthy TVS, Vannberg F, Shen B, et al. Building a human kinase gene repository: Bioinformatics, molecular cloning, and functional validation. Proc Natl Acad Sci. 2005;102:8114–8119. doi: 10.1073/pnas.0503141102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu Y, Rolfs A, Bhullar B, Murthy TVS, Zhu C, et al. Approaching a complete repository of sequence-verified protein-encoding clones for Saccharomyces cerevisiae. Genome Res. 2007;17:536–543. doi: 10.1101/gr.6037607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu H, Bilgin M, Bangham R, Hall D, Casamayor A, et al. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- 24.Phizicky EM, Grayhack EJ. Proteome-scale analysis of biochemical activity. Crit Rev Biochem Mol Biol. 2006;41:315–327. doi: 10.1080/10409230600872872. [DOI] [PubMed] [Google Scholar]
- 25.Salamat-Miller N, Fang J, Seidel CW, Smalter AM, Assenov Y, et al. A network-based analysis of polyanion-binding proteins utilizing yeast protein arrays. Mol Cell Proteomics. 2006;5:2263–2278. doi: 10.1074/mcp.M600240-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Li S, Armstrong CM, Bertin N, Ge H, Milstein S, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu X, Gerstein M, Snyder M. Getting connected: analysis and principles of biological networks. Genes Dev. 2007;21:1010–1024. doi: 10.1101/gad.1528707. [DOI] [PubMed] [Google Scholar]
- 28.Hartley JL, Temple GF, Brasch MA. DNA cloning using in vitro site specific recombination. Genome Res. 2000;10:1788–1795. doi: 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daraselia N, Dernovoy D, Tian Y, Borodovsky M, Tatusov R, et al. Reannotation of Shewanella oneidensis genome. OMICS. 2003;7:171–175. doi: 10.1089/153623103322246566. [DOI] [PubMed] [Google Scholar]
- 30.Romine MF, Carlson TS, Norbeck AD, McCue LA, Lipton MS. Identification of Mobile Elements and Pseudogenes in the Shewanella oneidensis MR-1 Genome. Appl Environ Microbiol. 2008;74:3257–3265. doi: 10.1128/AEM.02720-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoch JA. Two-component and phosphorelay signal transduction. Curr Opin Microbiol. 2000;3:165–170. doi: 10.1016/s1369-5274(00)00070-9. [DOI] [PubMed] [Google Scholar]
- 32.Forstner M, Leder L, Mayr LM. Optimization of protein expression systems for modern drug discovery. Expert Rev Proteomics. 2007;4:67–78. doi: 10.1586/14789450.4.1.67. [DOI] [PubMed] [Google Scholar]
- 33.Gao H, Wang X, Yang ZK, Palzkill T, Zhou J. Probing regulon of ArcA in Shewanella oneidensis MR-1 by integrated genomic analyses. BMC Genomics. 2008;9:42. doi: 10.1186/1471-2164-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Darwin AJ, Stewart V. Expression of the narX, narL, narP, and narQ genes of Escherichia coli K-12: regulation of the regulators. J Bacteriol. 1995;177:3865–3869. doi: 10.1128/jb.177.13.3865-3869.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tyson KL, Cole JA, Busby SJW. Nitrite and nitrate regulation at the promoters of two Escherichia coli operons encoding nitrite reductase: identification of common target heptamers for both NarP- and NarL-dependent regulation. Mol Microbiol. 1994;13:1045–1055. doi: 10.1111/j.1365-2958.1994.tb00495.x. [DOI] [PubMed] [Google Scholar]
- 36.Kehoe JW, Kay BK. Filamentous phage display in the new millennium. Chem Rev. 2005;105:4056–4072. doi: 10.1021/cr000261r. [DOI] [PubMed] [Google Scholar]
- 37.Huang W, McKevitt M, Palzkill T. Use of the arabinose pbad promoter for tightly regulated display of proteins on bacteriophage. Gene. 2000;251:187–197. doi: 10.1016/s0378-1119(00)00210-9. [DOI] [PubMed] [Google Scholar]
- 38.Liu Q, Li MZ, Leibham D, Cortez D, Elledge SJ. The univector plasmid-fusion system, a method for rapid construction of recombinant DNA without restriction enzymes. Curr Biol. 1998;8:1300–1309. doi: 10.1016/s0960-9822(07)00560-x. [DOI] [PubMed] [Google Scholar]
- 39.Li MZ, Elledge SJ. MAGIC, an in vivo genetic method for the rapid construction of recombinant DNA molecules. Nat Genet. 2005;37:311–319. doi: 10.1038/ng1505. [DOI] [PubMed] [Google Scholar]
- 40.McKevitt M, Patel K, Smajs D, Marsh M, McLoughlin M, et al. Systematic cloning of Treponema pallidum open reading frames for protein expression and antigen discovery. Genome Res. 2003;13:1665–1674. doi: 10.1101/gr.288103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dricot A, Rual JF, Lamesch P, Bertin N, Dupuy D, et al. Generation of the Brucella melitensis ORFeome version 1.1. Genome Res. 2004;14:2201–2206. doi: 10.1101/gr.2456204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gelperin DM, White MA, Wilkinson ML, Kon Y, Kung LA, et al. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 2005;19:2816–2826. doi: 10.1101/gad.1362105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 44.Gavin A-C, Bosche M, Krause R, Grandi P, Marzioch M, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 45.Reboul J, Vaglio P, Rual J-F, Lamesch P, Martinez M, et al. C. elegans ORFeome version 1.1: experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat Genet. 2003;34:35–41. doi: 10.1038/ng1140. [DOI] [PubMed] [Google Scholar]
- 46.McKevitt M, Brinkman MB, McLoughlin M, Perez C, Howell JK, et al. Genome scale identification of Treponema pallidum antigens. Infect Immun. 2005;73:4445–4450. doi: 10.1128/IAI.73.7.4445-4450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arezi B, Xing W, Sorge JA, Hogrefe HH. Amplification efficiency of thermostable DNA polymerases. Anal Biochem. 2003;321:226–235. doi: 10.1016/s0003-2697(03)00465-2. [DOI] [PubMed] [Google Scholar]
- 48.Schwalb C, Chapman SK, Reid GA. The tetraheme cytochrome cyma is required for anaerobic respiration with dimethyl sulfoxide and nitrite in Shewanella oneidensis. Biochem. 2003;42:9491–9497. doi: 10.1021/bi034456f. [DOI] [PubMed] [Google Scholar]
- 49.Gralnick JA, Brown CT, Newman DK. Anaerobic regulation by an atypical Arc system in Shewanella oneidensis. Mol Microbiol. 2005;56:1347–1357. doi: 10.1111/j.1365-2958.2005.04628.x. [DOI] [PubMed] [Google Scholar]
- 50.Maier TM, Myers CR. Isolation and characterization of a Shewanella putrefaciens MR-1 electron transport regulator etrA mutant: reassessment of the role of EtrA. J Bacteriol. 2001;183:4918–4926. doi: 10.1128/JB.183.16.4918-4926.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saffarini DA, Schultz R, Beliaev A. Involvement of cyclic AMP (cAMP) and cAMP receptor protein in anaerobic respiration of Shewanella oneidensis. J Bacteriol. 2003;185:3668–3671. doi: 10.1128/JB.185.12.3668-3671.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu D, Li G, Wu L, Zhou J, Xu Y. PRIMEGENS: robust and efficient design of gene-specific probes for microarray analysis. Bioinformatics. 2002;18:1432–1437. doi: 10.1093/bioinformatics/18.11.1432. [DOI] [PubMed] [Google Scholar]
- 53.Reece KS, Phillips GJ. New plasmids carrying antibiotic-resistance cassettes. Gene. 1995;165:141–142. doi: 10.1016/0378-1119(95)00529-f. [DOI] [PubMed] [Google Scholar]
- 54.Bradford MM. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 55.Palzkill T, Le Q-Q, Venkatachalam KV, LaRocco M, Ocera H. Evolution of antibiotic resistance: several different amino acid substitutions in an active site loop alter the substrate profile of β-lactamase. Mol Microbiol. 1994;12:217–229. doi: 10.1111/j.1365-2958.1994.tb01011.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S. oneidensis clone set ORF-specific primers.
(0.69 MB XLS)