Abstract
SUMMARY
Macrophages from certain inbred mouse strains are rapidly killed (<90 min) by anthrax lethal toxin (LT). LT cleaves cytoplasmic MEK proteins at 20 min and induces caspase-1 activation in sensitive macrophages at 50-60 min, but the mechanism of LT-induced death is unknown. Proteasome inhibitors block LT-mediated caspase-1 activation and can protect against cell death, indicating that the degradation of at least one cellular protein is required for LT-mediated cell death. Proteins can be degraded by the proteasome via the N-end rule, in which a protein's stability is determined by its N-terminal residue. Using amino acid derivatives that act as inhibitors of this pathway, we show that the N-end rule is required for LT-mediated caspase-1 activation and cell death. We also found that bestatin methyl ester, an aminopeptidase inhibitor protects against LT in vitro and in vivo and that the different inhibitors of the protein degradation pathway act synergistically in protecting against LT. We identify c-IAP1, a mammalian member of the inhibitor of apoptosis protein (IAP) family, as a novel N-end rule substrate degraded in macrophages treated with LT. We also show that LT-induced c-IAP1 degradation is independent of the IAP-antagonizing proteins Smac/DIABLO and Omi/HtrA2, but dependent on caspases.
INTRODUCTION
Bacillus anthracis lethal toxin (LT) is composed of the receptor binding component protective antigen (PA) and the enzymatic component lethal factor (LF) (Leppla, 2006). LF is a zinc metalloprotease that cleaves members of the mitogen-activated protein kinase kinase family (MEKs) (Duesbery et al., 1998;Pellizzari et al., 1999;Vitale et al., 2000). Macrophages from certain inbred mouse strains are rapidly and uniquely killed by LT, while macrophages from others strains are resistant (Friedlander et al., 1993;Singh et al., 1989). While a number of cellular responses to LT have been characterized, the mechanism for LT-induced macrophage death is unknown. Caspase-1 activation by LT is required for LT-induced macrophage death (Boyden and Dietrich, 2006) and is a very late event relative to MEK cleavage by the toxin (Wickliffe et al., 2007). The caspase-1 substrates IL-1β and IL-18 are released extracellularly following LT treatment (Cordoba-Rodriguez et al., 2004;Wickliffe et al., 2007) but are not required for LT-mediated cytotoxicity (Wickliffe et al., 2007). The specific role of caspase-1 and MEK cleavage in LT-induced macrophage death remains unknown.
The 26S proteasome, a multi-subunit complex responsible for the regulated degradation of intracellular proteins, was previously shown to be required for LT-induced macrophage death (Tang and Leppla, 1999). We recently showed that LT-mediated caspase-1 activation requires proteasome activity (Wickliffe et al., 2007). Inhibitors of the proteasome, including lactacystin and MG-132, block the cytotoxic effects of LT on murine macrophages without affecting MEK cleavage (Tang and Leppla, 1999), showing that the proteasome-mediated degradation of an unknown substrate or substrates is necessary for LT toxicity. Identification of the substrates targeted for breakdown by toxin in macrophages and the pathway of this degradation will provide insight into the mechanism of LT-induced cell death.
Before a protein is degraded by the proteasome, it must be marked for degradation, and the most significant modification that tags a protein for proteasome-mediated degradation is the formation of a polyubiquitin chain on the target substrate. Although the mechanism of ubiquitination and the required enzymes have been well characterized (Glickman and Ciechanover, 2002), the degradation signals that initially mark a protein for ubiquitination are less well understood. Proteins may be targeted for ubiquitination by phosphorylation or by certain amino acid motifs present in the protein's native structure, such as PEST elements (regions rich in Pro, Glu, Ser, and Thr) (Hershko and Ciechanover, 1998). One of the best studied degradation signals is a destabilizing N-terminal residue. Along with an internal lysine residue that serves as the site of polyubiquitin chain formation, this signal forms the basis for the N-end rule, in which a protein's half-life is determined by its N-terminal residue (Bachmair et al., 1986;Gonda et al., 1989;Varshavsky, 1997). The N-end rule is organized hierarchically, with destabilizing residues falling into three categories. Primary destabilizing residues are divided into Type 1 (basic residues His, Lys, and Arg) and Type 2 (bulky, hydrophobic residues Ile, Tyr, Trp, Leu, and Phe) groups (Varshavsky, 1997). Few N-end substrates have been verified in biological systems, and they include Drosophila IAP1 (DIAP1), an inhibitor of apoptosis (Ditzel et al., 2003); the Saccharomyces cerevisiae proteins Gpa1 (the α subunit of a G protein) (Madura and Varshavsky, 1994), SSC1 (a subunit of cohesion) (Rao et al., 2001), and CUP9 (a protein involved in peptide import) (Byrd et al., 1998); the mammalian proteins RGS4 and RGS16 (GTPase-activating proteins) (Davydov and Varshavsky, 2000;Lee et al., 2005) and the γ2 subunit of G proteins (Hamilton et al., 2003); the viral proteins HIV-1 integrase (Mulder and Muesing, 2000) and Sindbis virus RNA polymerase (De Groot et al., 1991); and the Listeria protein p60 following its secretion into the host cytoplasm (Sijts et al., 1997).
The role of the proteasome in LT toxicity having been established, results from previous studies suggested a possible role for the N-end rule in LT-mediated death. Various amino acid derivatives as well as the aminopeptidase inhibitor bestatin were shown to protect against LT (Klimpel et al., 1994). By removing N-terminal amino acids, aminopeptidases may be involved in LT-induced death by exposing destabilizing residues. In this report, we further investigate the role of the N-end rule in LT toxicity. We tested additional amino acid derivatives that inhibit the N-end rule and show that all tested derivatives with Type II destabilizing residues protect macrophages against LT mediated death, suggesting that this pathway of protein degradation is required for the cytotoxic effect of LT. We also show that the N-end rule acts upstream of caspase-1 activation. Finally, we show that cellular inhibitor of apoptosis protein 1 (c-IAP1), a mammalian inhibitor of apoptosis protein and a homolog of the Drosophila N-end rule substrate DIAP1, is a novel target of the N-end pathway. This protein is degraded in response to LT in a manner in bone marrow-derived macrophages (BMDM). To our knowledge, this is the first report of a bacterial toxin initiating N-end rule pathway-based degradation of a cellular protein.
RESULTS
Amino acid derivatives protect against LT-mediated macrophage death
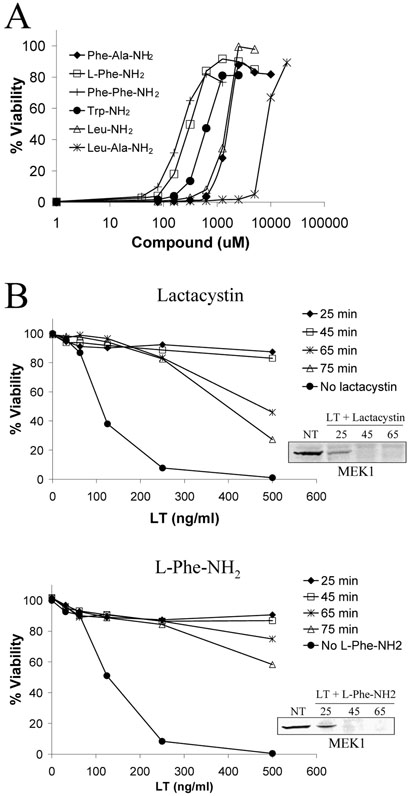
Previous studies showed that cytosolic protein degradation mediated by the proteasome is a required event in LT-mediated macrophage death (Tang and Leppla, 1999). We previously observed that amino acid derivatives containing Phe and Leu residues protect macrophages from LT-induced death (Klimpel et al., 1994). Similar amino acid derivatives and dipeptides have been shown to compete with N-end rule substrates for Type 1 or Type 2 binding sites on E3 ubiquitin ligases, thereby inhibiting ubiquitination and degradation (Baker and Varshavsky, 1991). To determine if the N-end rule is required for LT-mediated cytotoxicity, we tested a number of amino acid derivatives with both Type I and Type II primary destabilizing N-terminal residues for protection against LT. All tested amino acid derivatives with Type II destabilizing residues (L-Phe-NH2, L-Trp-NH2, L-Leu-NH2, Phe-Ala-NH2, Phe-Phe-NH2, Leu-Ala-NH2) showed dose-dependent protection against LT (Fig. 1A). In contrast, compounds with Type I destabilizing residues (Arg-Ala-NH2 and His-NH2) did not provide any protection against LT (data not shown). These results suggest that the N-end rule may be required for LT toxicity and that the substrate(s) whose degradation is required for LT-mediated death contains a Type II destabilizing N-terminal residue.
Figure 1. Protection against LT-induced macrophage death with amino acid derivatives.
(A) RAW264.7 cells were treated with amino acid derivatives for 45 min prior to LT addition (1 μg/ml), and cell viability was assessed at 3 h by MTT staining. (B) RAW264.7 cells were treated with LT (1 μg/ml), and lactacystin (15 μM) or L-Phe-NH2 (1 mM) were added back to cells at the indicated times. Viability was determined by MTT after a 3-h toxin treatment. In the insets, RAW264.7 cells were treated with LT (1 μg/ml) in the presence and absence of lactacystin or L-Phe-NH2 for the indicated times (min), and MEK1 cleavage was following in cell lysates by Western blot using an antibody whose epitope spans the LF cleavage site of MEK1. NT indicates no-treatment control cells.
L-Phe-NH2 and lactacystin protect against LT without inhibiting MEK cleavage (Fig. 1B). To determine approximately when the N-end rule and the proteasome are required for LT-induced death, add-back experiments were performed in which L-Phe-NH2 and lactacystin were added 25, 45, 65, or 75 min post-LT addition. With both compounds, addition by 45 min provided full protection from LT at all tested toxin concentrations (Fig. 1B). At 65 and 75 min drug add-back times, protection began to fall with higher LT concentrations (Fig. 1B). However, within a given cell population, events are not completely synchronized, and it is likely that the degradation of substrates by the proteasome does not occur concurrently, providing a possible explanation for why partial protection was observed with the later add-back times.
Bestatin protects macrophages from LT-induced death and mice from lethal doses of LT
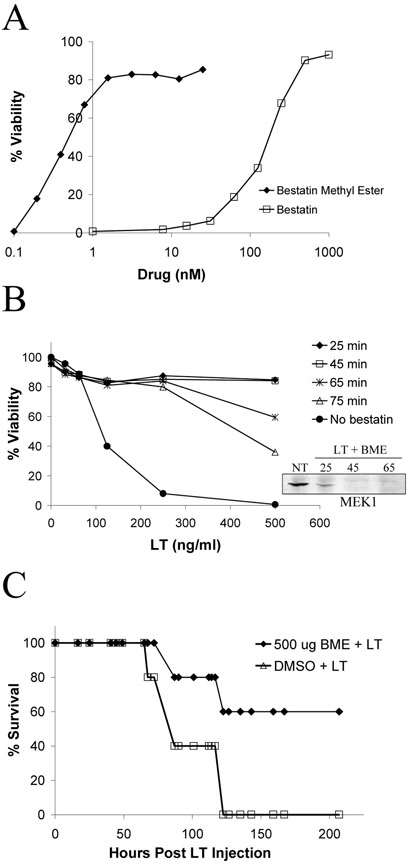
Protein degradation by the N-end rule is dependent on a destabilizing N-terminal residue, but all eukaryotic proteins are synthesized with a stabilizing methionine residue at the N-terminus (Varshavsky, 1997). Therefore, processing is required to expose a destabilizing residue prior to a substrate's degradation by the proteasome. This event may be initiated by aminopeptidases. The Streptomyces olivoreti peptide bestatin, which inhibits leucine, alanine, and arginine aminopeptidases (Umezawa et al., 1976), protects macrophages against LT (Klimpel et al., 1994). We tested the cell-permeable methyl ester derivative of bestatin and found it protected against LT at 1000-fold lower concentrations than bestatin with no effect on MEK cleavage (Fig. 2A). The aminopeptidase inhibitors actinonin (Gordon et al., 1962) and amastatin (Tieku and Hooper, 1992), which inhibit a variety of aminopeptidases, including leucine aminopeptidase and aminopeptidases A, N, and W, showed no protection against LT-induced death at concentrations up to 1 mM (data not shown), indicating that an aminopeptidase specifically sensitive to bestatin is involved in macrophage death. Bestatin methyl ester add-back experiments similar to those with L-Phe-NH2 and lactacystin produced full protection against LT with a 45 min add back time and LT-dependent decreasing levels of protection with later times (Fig. 2B).
Figure 2. Bestatin methyl ester protects against LT-induced macrophage death and lethality in mice.
(A) RAW264.7 cells were treated with bestatin or bestatin methyl ester for 45 min prior to LT addition (1 μg/ml) for 3 h. In the inset, RAW264.7 cells were treated with LT (1 μg/ml) in the presence and absence of bestatin methyl ester for the indicated times (min), and MEK1 cleavage was following in cell lysates by Western blot. NT indicates no-treatment control cells. (B) RAW264.7 cells were treated with LT (1 μg/ml), and bestatin methyl ester (10 μM) was added back to cells at the indicated times. After a 3-h toxin treatment, viability was assessed by MTT staining. (C) BALB/cJ mice were injected (100 μl; i.v.) with 500 μg bestatin methyl ester (BME) followed 5 min later by injection (1 ml; i.p.) of 100 μg LT. Control animals were i.v. injected with 100 μl of a 10% DMSO solution prior to LT challenge. Animals were monitored for survival. n = 5 for each group.
We further tested the protective effects of bestatin methyl ester against lethal LT treatment in mice. BALB/cJ mice were injected with various doses of bestatin methyl ester 5 min prior to challenge with a lethal dose of LT. A dose of 500 μg bestatin methyl ester injected i.v. partially protected mice from LT (Fig.2C).
Synergy between inhibitors of the protein degradation pathway
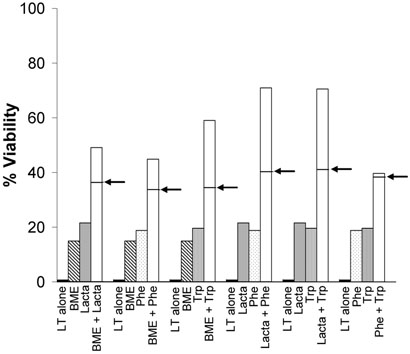
We next asked whether protective amino acid derivatives would act synergistically with other inhibitors of the protein degradation pathway to protect cells against LT-induced death. RAW264.7 cells were treated with sub-protective concentrations of lactacystin, bestatin methyl ester, L-Phe-NH2, and L-Trp-NH2 alone or in all combinations, and we determined whether the resultant protection against LT was greater than the additive effect expected if the drugs acted on the same target. The combination of L-Phe-NH2 and L-Trp-NH2 produced an additive protective effect as would be expected if both compounds are targeting the same binding sites of E3 ligases (Fig. 3). The greatest effects were seen with the combinations of lactacystin and either L-Phe-NH2 or L-Trp-NH2 (Fig. 3). With these treatments, individual compound concentrations that alone produced 20% viability together yielded 70% viability, 30% higher than an additive effect. The other combinations of drugs produced only minimal synergistic protective effects against LT (Fig. 3).
Figure 3. Synergy between protective treatments.
RAW264.7 cells were pretreated with lactacystin (“Lacta”, 950 nM), bestatin methyl ester (BME, 75 nM), L-Phe-NH2 (Phe, 85 μM), and L-Trp-NH2 (Trp, 225 μM) alone or in all treatment combinations 30 min prior to LT addition (1 μg/ml) for 3 h. Cell viability was determined by MTT staining. In the bar corresponding to the treatment combinations, the arrow points to the calculated additive effects of each inhibitor individually.
LT-induced caspase-1 activation requires proteasome activity and the N-end rule
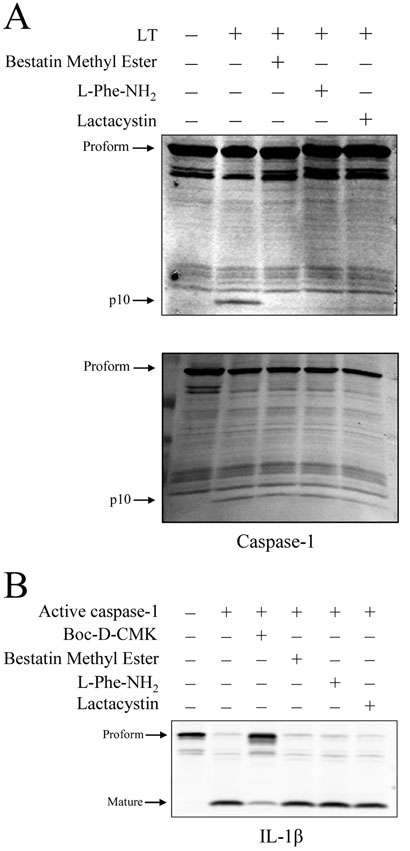
Following the early event of MEK cleavage, caspase-1 activation in LT-sensitive macrophages begins at 50-60 min (Wickliffe et al., 2007) and is required for LT-induced death (Boyden and Dietrich, 2006). We had previously shown that LT-induced caspase-1 activation is prevented in the presence of lactacystin (Wickliffe et al., 2007), and we now asked whether the N-end rule participates in LT-induced death upstream or downstream of caspase-1 activation. LT-induced caspase-1 activation, as evidenced by the appearance of the p10 fragment of the active enzyme, was prevented by bestatin methyl ester, L-Phe-NH2, and lactacystin in BALB/cJ BMDM (Fig. 4A, top panel). In similar experiments, the three compounds were added to cells 65 min after LT addition, an add-back time that did not protect cells against LT. As expected, caspase-1 activation was not inhibited by the compounds in these lysates (Fig. 4A, bottom panel), suggesting that the protective ability of these compounds is related to their ability to prevent caspase-1 activation. To determine whether bestatin methyl ester, L-Phe-NH2, or lactacystin directly inhibit caspase-1 proteolytic activity, cell extracts of LPS-primed RAW264.7 cells were prepared as a source of pro-IL-1β and treated with active, recombinant caspase-1. Enzyme activity was measured by the levels of mature IL-1β. At the concentrations used in treating cells, bestatin methyl ester, L-Phe-NH2, and lactacystin had no effect on caspase-1 cleavage of pro-IL-1β (Fig. 4B), indicating that none of the compounds inhibit caspase-1 proteolytic activity directly.
Figure 4. Inhibition of LT-induced caspase-1 activation.
(A) BALBc/J BMDM were treated with LT (1 μg/ml; 85 min) in the presence and absence of lactacystin (15 μM), L-Phe-NH2 (1 mM), or bestatin methyl ester (10 μM). In the top panel, cells were pretreated with the compounds for 40 min prior to LT addition. In the bottom panel, compounds were added to cells 65 min following LT addition. Cell lysates were prepared, and the p10 fragment of active caspase-1 detected by Western blot analysis. (B) LPS-primed RAW264.7 cell lysates prepared using sucrose lysis buffer served as a source of pro-IL-1β and were treated with active recombinant caspase-1 (2 h; 37°C) in the presence and absence of Boc-D-CMK (400 μM), lactacystin (20 μM), bestatin methyl ester (10 μM), or L-Phe-NH2 (1 mM). Reaction mixtures were subject to SDS-PAGE and Western blot using an IL-1β antibody that recognizes both the proform and the mature form of the cytokine.
c-IAP1 is degraded in macrophages treated with LT in a proteasome-dependent manner
The N-end rule was initially discovered and characterized using test substrates, and despite the presence of the pathway in all eukaryotic cells, the identification of physiological substrates has proved challenging. We attempted to identify possible N-end rule substrates that may be involved in cell death. Of the less than twenty presently identified substrates, the only protein linked to cell death is DIAP1, a Drosophila inhibitor of apoptosis protein (IAP), which was shown to be degraded by the N-end rule following treatment with diap1 dsRNA (Ditzel et al., 2003). IAP family proteins suppress apoptosis primarily by binding and inhibiting caspases (Deveraux and Reed, 1999;Salvesen and Duckett, 2002). In addition to inhibiting caspase activity directly, IAP proteins possess E3 ubiquitin ligase activity (Yang et al., 2000) and can inhibit cell death by promoting the ubiquitination and degradation of death inducer proteins such as Smac/DIABLO (Hu and Yang, 2003) or caspases (Huang et al., 2000) themselves.
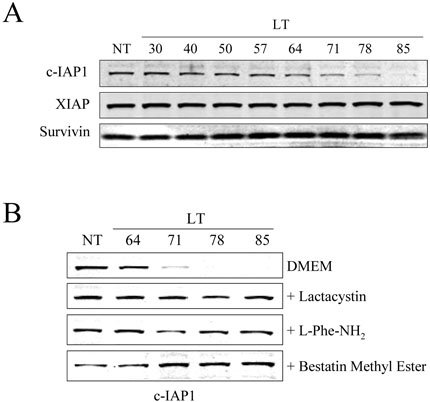
To determine whether IAP protein levels change in response to LT, whole cell lysates of BALB/cJ BMDM and RAW264.7 cells treated with LT for various times were analyzed by Western blot using antibodies to XIAP, c-IAP1, and survivin, three representative mammalian IAP proteins. We observed no change in the levels of survivin or XIAP with LT treatment times up to 85 min (Fig. 5A). However, levels of c-IAP1 began to decrease in BMDM lysates beginning approximately 65-70 min after toxin addition, and the protein was almost completely degraded by 85 min (Fig. 5A). LT-induced c-IAP1 degradation did not occur in the LT-resistant BMDM isolated from C57BL/6J mice (data not shown). Interestingly, c-IAP1 was also not degraded following LT treatment of the immortalized RAW264.7 macrophages (data not shown), which are sensitive to toxin, indicating that any link that c -IAP1 degradation has to cell death may be limited to primary cells. To investigate if LT-induced c-IAP1 degradation was dependent on the N-end rule or the proteasome, BALB/cJ BMDM were treated with LT in the presence of lactacystin, L-Phe-NH2, or bestatin methyl ester. c-IAP1 degradation was prevented with these three drugs (Fig. 5B), suggesting that LT-induced c-IAP1 breakdown requires the N-end rule, a bestatin-sensitive enzyme, and the proteasome.
Figure 5. c-IAP1 is degraded in LT-treated BMDM.
(A) BALB/cJ BMDM were treated with LT (1 μg/ml) for various times (min). Cell lysates were prepared and levels of c-IAP1, XIAP, and survivin were analyzed by Western blot. (B) BALBc/J BMDM were treated with LT for the indicated times (min) in the presence or absence of lactacystin (20 μM), L-Phe-NH2 (1 mM), or bestatin methyl ester (10 μM). c-IAP1 levels in cell lysates were then determined by Western blotting.
Smac/DIABLO and Omi/HtrA2 are not translocated to the cytosol with LT treatment
Aminopeptidases may play a general role in exposing destabilizing N-terminal residues and thereby targeting proteins for degradation. Having shown that c-IAP1 is degraded in response to LT treatment, we wanted to identify other proteins known to interact with IAPs that may be involved in this specific degradation event. The anti-apoptotic activity of IAPs is antagonized by the pro-apoptotic mitochondrial proteins Smac/DIABLO (Du et al., 2000;Verhagen et al., 2000) and Omi/HtrA2 (Hegde et al., 2002). Following treatment with an apoptotic stimulus, these proteins are translocated from mitochondria to the cytosol where they bind IAP proteins, effectively preventing the IAPs from binding caspases. Furthermore, Smac/DIABLO has been reported to induce the autoubiquitination and degradation of c-IAP1 and c-IAP2 (Yang and Du, 2004), and Omi/HtrA2 is a serine protease that cleaves c-IAP proteins (Yang et al., 2003).
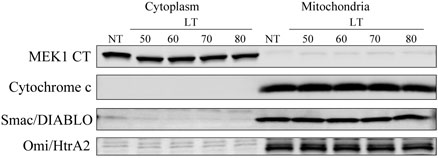
Because IAPs also induce the ubiquitination and degradation of Smac/DIABLO (Hu and Yang, 2003), we first examined levels of Smac/DIABLO and Omi/HtrA2 with LT treatment in BALB/cJ BMDM and RAW264.7 cells and observed no change, showing that neither protein is a target of the E3 ligase activity of the IAP proteins following LT treatment (data not shown). To determine if Smac/DIABLO and Omi/HtrA2 are translocated to the cytoplasm with LT treatment, BALB/cJ BMDM and RAW264.7 cells were treated with LT for various times, and cytosolic and mitochondrial fractions were prepared. Purity of the lysates was confirmed by analyzing the fractions by Western blotting using antibodies to cytochrome c and the C-terminus of MEK1 as mitochondrial and cytoplasmic markers, respectively (Fig. 6). The distribution of Smac/DIABLO and Omi/HtrA2 was then determined by Western blot. With LT treatment times up to 80 min, both Smac/DIABLO and Omi/HtrA2 were observed only in the mitochondrial fractions and never in the cytoplasmic fractions (Fig. 6). These results indicate that Smac/DIABLO and Omi/HtrA2 are not involved in LT-induced c-IAP1 degradation. Additionally, the absence of cytochrome c translocation from mitochondria to cytoplasm (Fig. 6) confirms that the classic apoptosis pathways are not involved in LT-induced macrophage death. These results also confirm that the MEK proteins are cleaved by LF but not degraded in sensitive macrophages through time points immediately preceding cell death. A similar cleavage of MEK proteins by LF without concomitant degradation is observed in resistant macrophages over longer times (data not shown).
Figure 6. Distribution of Smac/DIABLO and Omi/HtrA2 in cells treated with LT.
BALB/cJ BMDM cells were treated with LT (1 μg/ml) for the indicated times (min). Mitochondrial and cytoplasmic fractions were prepared using a digitonin-based method, and the purity of the resulting fractions was confirmed by examining the distribution of MEK1 and cytochrome c. Levels of Smac/DIABLO and Omi/HtrA2 in the fractions over the LT treatment time course was followed by Western blot. NT refers to no-treatment control.
LT-induced degradation of c-IAP1 is a caspase-dependent event
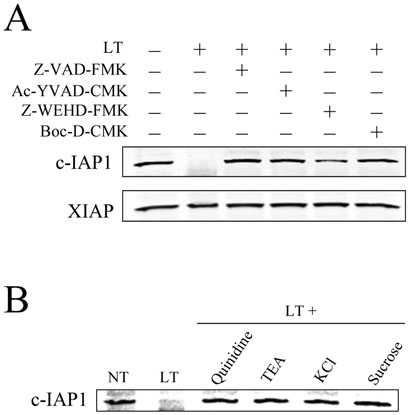
In response to various apoptotic stimuli, c-IAP1 is cleaved and/or degraded in a caspase-dependent manner (Clem et al., 2001;Herrera et al., 2002). Importantly, cleavage of the N-end rule substrate DIAP1 by caspases produces the destabilizing residue that leads to the protein's degradation via the proteasome (Ditzel et al., 2003). The late timing of LT-induced caspase-1 activation (50-60 min) roughly correlates with the observed decrease in levels of c-IAP1. To determine if caspases are involved in LT-induced c-IAP1 degradation, BALB/cJ BMDM were treated with LT in the presence of the pan-caspase inhibitors Z-VAD-FMK and Boc-D-CMK and the caspase-1-specific inhibitors Ac-YVAD-CMK and Z-WEHD-FMK. All tested inhibitors blocked LT-induced c-IAP1 degradation (Fig. 7A), suggesting that this degradation event is caspase-dependent, occurring downstream of caspase-1 activation. Our previous work indicates that the signaling event for LT-induced caspase-1 activation is ion fluxes, specifically potassium efflux (Wickliffe et al., 2007). We have shown that inhibiting ion fluxes by treating cells with LT in the presence of elevated levels of KCl and sucrose or the potassium channel blockers quinidine and TEA prevents caspase-1 activation (Wickliffe et al., 2007). Here, we also found that c-IAP1 degradation was prevented with the above treatments (Fig. 7B), further supporting the result that c-IAP1 degradation occurs downstream of increases in plasma membrane permeability and caspase-1 activation.
Figure 7. Inhibition of LT-induced c-IAP1 degradation.
(A) BALBc/J BMDM were treated with LT (85 min; 1 μg/ml) following a 1 h pretreatment with caspase inhibitors (all at 100 μM). (B) To inhibit potassium efflux, BALBc/J BMDM were treated with LT (1 μg/ml; 85 min) in DMEM containing 130 mM KCl or 300 mM sucrose or after a 25 min pretreatment with 250 μM quinidine or 100 mM TEA. Levels of c-IAP1 were determined by Western blot.
DISCUSSION
LT induces rapid (80-90 min) cell death in macrophages from certain mouse strains. Few LT-induced cellular events have been identified after LF entry into the cytosol. The MEK proteins are cleaved beginning 15-20 min after LT addition to cells, followed by ion fluxes at 45 min (Hanna et al., 1992). Caspase-1 activation which requires a functional Nalp1b protein (Boyden and Dietrich, 2006) as well as potassium fluxes and an active proteasome occurs at 60 min (Wickliffe et al., 2007). The proteasome is required for LT-toxicity (Tang and Leppla, 1999), indicating that the degradation of an intracellular protein is required for caspase-1 activation and macrophage lysis. Expanding upon the role of the proteasome, we now show that the N-end rule of protein degradation is specifically required for LT-induced death.
Protein degradation via the N-end rule is based on the presence of a destabilizing N-terminal residue that marks a protein for ubiquitination and subsequent breakdown by the proteasome. The original N-terminal residue of all eukaryotic proteins is methionine, a stabilizing residue in the N-end rule pathway (Varshavsky, 1996). Therefore, a processing event, possibly mediated by aminopeptidases, is required to expose a destabilizing N-terminal residue. Destabilizing N-terminal residues are classified as Type I or Type II, based on binding to distinct sites on the N-end rule E3 ligase (Varshavsky, 1996). In this work we show that the aminopeptidase inhibitor bestatin and amino acid derivatives with Type II destabilizing residues that can act as competitive inhibitors of the N-end rule show dose-dependent protection against LT. We therefore hypothesized that the breakdown of a protein via the N-end rule is required for LT-mediated macrophage death.
Although it is most likely that the amino acid derivatives tested in this study are preventing the breakdown of a protein required for LT toxicity, much in the manner that proteasome inhibitors protect, we must also consider the possibility that these compounds can act in accelerating degradation of a different protein, as reported in the case of the yeast protein Cup9 (Turner et al., 2000). Because amino acid derivatives are often utilized in cell-free systems to assess degradation of proteins and because the more cell-permeable amide derivatives tested in our studies have never been previously used, it is difficult to draw comparisons between the concentrations required for the protection against LT toxicity in this study (1 mM), those required for inhibition of degradation in other systems (10 mM range) and that required for the more rare acceleration of degradation pathways (1 μM range).
To date, very few physiological N-end rule substrates have been identified, and among those, only one, an apoptosis inhibiting Drosophila IAP protein, plays a role in cell death (Ditzel et al., 2003). We therefore investigated the effects of LT on mammalian IAP proteins. We found that c-IAP1 (but not x-IAP and survivin) is degraded in response to LT, with levels beginning to decrease after a 60 min LT treatment, and that this LT-initiated late degradation event is dependent on the N-end rule and proteasome activity. Since c-IAP degradation in response to apoptotic stimuli has been shown to depend on caspases (Herrera et al., 2002;Messmer et al., 2001) and caspase-1 is required for LT-induced death (Boyden and Dietrich, 2006), we tested the role of caspases in LT-mediated c-IAP1 breakdown and found degradation was prevented with caspase inhibitors. As c-IAP1 and XIAP are directly cleaved by caspases (Clem et al., 2001;Deveraux et al., 1999), it will be important to determine whether c-IAP1 is specifically a target of caspase-1.
Although c-IAP1 represents a novel mammalian N-end rule substrate and the first protein shown to be degraded by the proteasome in response to LT treatment, it is likely not the essential N-end rule substrate required for macrophage lysis. The absence of c-IAP1 degradation in LT-treated RAW264.7 cells indicates that this event is not absolutely required for LT-mediated cell death and may be specific to BMDM. As precursor bone marrow cells differentiate to become macrophages, levels of IAPs increase over time, and the resistance of these cells to apoptosis during differentiation has been attributed to the increasing levels of IAPs (Lin et al., 2001). Therefore, it is possible that IAP proteins are regulated differently and play a different role in cell survival in BMDM than in immortalized macrophage lines. Additionally, c-IAP1 may simply be a member of a group of proteins broken down prior to cell death in these cells.
c-IAP1 cleavage and degradation occurs following treatment with a variety of apoptotic stimuli (Clem et al., 2001;Herrera et al., 2002;Messmer et al., 2001;Yang et al., 2000), but we found that the pro-apoptotic, IAP-antagonizing proteins Smac/DIABLO and Omi/HtrA2 are not translocated to the cytoplasm following LT treatment, indicating that neither are involved in c-IAP1 breakdown or LT-mediated death. Furthermore, IAP proteins primarily function through inhibition of the classic pro-apoptotic caspases 3, 7, and 9 (Roy et al., 1997;Salvesen and Duckett, 2002), but these caspases do not play a role in LT-mediated macrophage lysis (data not shown). Among the IAP proteins, c-IAP1 is uniquely involved in TNF-α signaling, playing a pro-apoptotic role by inducing the ubiquitination and proteasome-mediated degradation of the adaptor protein TRAF-2 (Li et al., 2002). We observed no change in TRAF2 levels following LT treatment (data not shown), and this activity of c-IAP1 is also unlikely to play a role in LT-induced death as macrophages do not release TNF-α in response to LT (Erwin et al., 2001;Moayeri et al., 2003). Therefore, although the protective effects of proteasome inhibitors against apoptosis have been linked to the ability of the inhibitors to stabilize IAP levels (Sohn et al., 2006;Yang et al., 2000), we find it unlikely that c-IAP1 breakdown is required for macrophage death.
Further supporting the existence of other N-end rule candidates and proteasome targets required for LT-mediated lysis, we found that the proteasome and the N-end rule are required for caspase-1 activation but that c-IAP1 degradation occurs downstream of caspase-1 activation. Therefore, the proteasome appears to be involved in the breakdown of more than one protein by the N-end rule after LT treatment. The fact that LF acts as a protease cleaving at N-terminal sequences of MEK proteins offers the attractive hypothesis that the toxin itself is responsible for initiating N-end rule mediated breakdown of a crucial substrate or substrates. This substrate is not any of the LT-targeted MEK proteins, which are cleaved but not broken down hours after cleavage of their N-termini.
A link between the proteasome, caspase-1 activation, and cell death was previously reported for the bacterial pathogen Shigella flexneri (Hilbi et al., 2000). Similar to LT, Shigella induces cell death via a caspase-1-dependent mechanism that has been linked to a protein degradation event mediated by the serine protease tripeptidyl protease II (Hilbi et al., 2000) (TPPII). Inhibitors of this protease, including AAF-CMK, protect macrophages against Shigella-induced caspase-1 activation and death. We found that AAF-CMK also protected macrophages against LT (data not shown). However, this protection was likely due to the compound's inhibition of the chymotrypsin-like activity of the proteasome (Bury et al., 2001;Geier et al., 1999), as the TPPII-specific inhibitor butabindide (Rose et al., 1996) did not protect against LT (data not shown). It is currently unknown whether LT-mediated death requires the breakdown of a single protein essential to cell death by the proteasome or a group of proteins and whether protein degradation pathways in addition to the N-end rule are involved in LT toxicity.
In summary, we have shown that the N-end rule of protein degradation is required for LT-mediated caspase activation and toxicity. We further present c-IAP1 as a new mammalian N-end rule substrate to add to the very limited number of previously identified physiological substrates. The LT-mediated breakdown of c-IAP1 also represents the first example of a bacterial toxin initiating the N-end rule pathway-based degradation of a cellular protein. Although we believe c-IAP1 degradation is not essential for LT toxicity, it is one of the few known LT-induced events that occurs downstream of caspase-1 activation, and therefore provides a starting point from which to study caspase-1-dependent death mechanisms. Whether c-IAP1 degradation specifically participates in LT-induced death in BMDM by exerting a unique protective effect against LT in these cells remains under study. The crucial involvement of the proteasome and the N-end rule in LT-induced death occurs after the early event of MEK cleavage (20 min) but before caspase-1 activation (50-60 min), which is in turn required for c-IAP1 degradation. The substrate(s) whose degradation is essential for caspase-1 activation and thereby LT cytotoxicity in all sensitive cell types remain unknown, and future studies will aim to identify these substrates and their role in caspase-1 activation and cell death.
EXPERIMENTAL PROCEDURES
Materials
PA and LF were purified in our laboratory as described previously (Park and Leppla, 2000;Ramirez et al., 2002;Varughese et al., 1998). In all assays, working stocks of toxin were prepared in Dulbecco's modified Eagle medium (DMEM). Concentrations of LT refer to the amount of each component (i.e. 1 μg/ml LT is 1 μg LF plus 1 μg PA/ml). The amino acid derivatives L-Phe-NH2, L-Trp-NH2, and L-Leu-NH2, the aminopeptidase inhibitors amastatin and actinonin, the potassium channel blockers quinidine and tetraethylammonium chloride (TEA), and lipopolysaccharide (LPS) from E. coli serotype 011:B4 were from Sigma (St. Louis, MO). Phe-Phe-NH2, Phe-Ala-NH2, Leu-Ala-NH2, His-NH2, and Arg-Ala-NH2 were from Bachem (King of Prussia, PA). Anti-MEK1 NT antibody, lactacystin, bestatin, bestatin methyl ester, and the caspase inhibitors Ac-YVAD-CMK and Boc-D-CMK were purchased from Calbiochem (San Diego, CA). AAF-CMK (Ala-Ala-Phe-chloromethylketone·TFA) was from Biomol (Plymouth Meeting, PA) and butabindide from Tocris (Ellisville, MO). Anti-c-IAP1, anti-IL-1β, anti-Smac/DIABLO, and anti-Omi/HtrA2 antibodies and the caspase inhibitors Z-WEHD-FMK (Z-W-E(OMe)-H-D(OMe)–FMK) and Z-VAD-FMK (Z-V-A-D(OMe)-FMK) were from R&D Systems (Minneapolis, MN). Anti-XIAP antibody was from BD Transduction Labs (San Jose, CA). Survivin, cytochrome c, caspase-1 p10, MEK1 CT, and actin antibodies were from Santa Cruz (Santa Cruz, CA). Active recombinant caspase-1 was purchased from MBL International (Woburn, MA). Anti-rabbit, anti-goat, and anti-mouse infrared-dye-conjugated secondary antibodies were from Rockland Immunochemicals (Gilbertsville, PA).
Cell culture
RAW264.7 cells and L929 mouse fibroblast cells were grown in DMEM supplemented with 10% fetal bovine serum, 10 mM HEPES, and 50 μg/ml gentamicin (all from Invitrogen, Carlsbad, CA) at 37°C in 5% CO2. Bone marrow-derived macrophages (BMDM) were derived from BALB/cJ mice (Jackson Laboratories, Bar Harbor, ME). Bone marrow cells were plated in DMEM (supplemented as described above) containing 30% L929 cell culture supernatant, grown for 7-10 days, and used in assays at 90% confluence following removal of unattached cells.
Cytotoxicity assays
RAW264.7 cells were plated in 96-well plates and grown overnight to 80-90% confluence prior to assays. In protection assays, cells were pretreated with compounds at various concentrations for 30-45 min prior to the addition of 1 μg/ml LT. Cells were incubated with LT for 3 h prior to the addition of MTT [3-(4, 5-dimethylthiazo-2-yl)-2, 5-diphenyltetrazolium bromide] (Sigma, St. Louis, MO) at a final concentration of 0.5 mg/ml. After an additional 40 min incubation, the medium was removed, and cells were dissolved in 50 μl/well 0.5% (wt/vol) SDS, 25 mM HCl in 90% (vol/vol) isopropanol to measure the production of formazan by viable cells. The A570 was read using a microplate reader, and cell viabilities were calculated relative to controls. In similar add-back experiments, cells were treated with two-fold dilutions of LT and fixed concentrations of lactacystin (15 μM), L-PheNH2 (2 mM), or bestatin methyl ester (10 μM) were added to cells at various time points following LT addition. Cell viability by MTT staining was determined 3 h after LT addition. In synergy experiments, cells were treated with lactacystin (950 nM), bestatin methyl ester (75 nM), L-Phe-NH2 (85 μM), and L-Trp-NH2 (225 μM) alone or in all treatment combinations. These concentrations represent approximate EC20 values as determined by protection assays. After a 30-min pretreatment with drugs, LT at a final concentration of 1 μg/ml or DMEM was added to cells. After a further 3-h incubation, viability was determined by MTT as described above.
Cell Lysate Preparation and Western Blot Analyses
RAW264.7 cells and BMDM were plated in 6-well plates and treated with LT at 1 μg/ml for various times in the presence or absence of bestatin methyl ester (10 μM), L-Phe-NH2 (1 mM), lactacystin (20 uM), quinidine (250 μM), TEA (100 mM), or the caspase inhibitors Z-VAD-FMK, Z-WEHD-FMK, Boc-D-CMK, and Ac-YVAD-CMK (all at 100 μM). Cells were pretreated with the above drugs for 25 - 60 min as indicated prior to LT addition. In other experiments, cells were treated with LT in DMEM containing 130 mM KCl or 300 mM sucrose. Cells were lysed with RIPA lysis buffer (1% Nonidet, 0.5% sodium deoxycholate, 0.1% SDS in PBS) containing EDTA-free Complete protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN), and Western blot analyses were performed with anti-c-IAP1 (1:2000), anti-XIAP (1:2000), anti-survivin (1:500), or anti-caspase-1 (1:500) antibodies. Primary antibodies were detected using IR-dye conjugated secondary IgG antibodies (anti-rabbit: 1:5000; anti-goat: 1:5000; anti-mouse: 1:5000) and the Odyssey Infrared Imaging System (LI-COR Biosciences, Lincoln, NE).
Animals
BALB/cJ mice were purchased from Jackson Laboratories (Bar Harbor, ME). 500 μg bestatin methyl ester prepared in 10% DMSO was injected intravenously (i.v.), and LT (100 μg PA plus 100 μg LF/ml in PBS) was injected intraperitoneally (i.p.) 5 min later. Animals were monitored for survival.
Cell fractionation
For cell fractionation, RAW264.7 cells and BALB/cJ BMDM plated in 6-well plates were treated with 1 μg/ml LT for various times. Cells were pelleted (1000 rpm, 4°C) and resuspended in lysis buffer (250 mM sucrose, 70 mM KCl, 137 mM NaCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4,) containing 200 μg/ml digitonin and EDTA-free Complete protease inhibitor cocktail and incubated on ice for 10 min. Complete plasma membrane permeabilization was verified by trypan blue staining. Cells were spun at 10,000 g for 10 min at 4°C, and the resultant supernatant was saved as the cytoplasmic fraction. The mitochondrial-containing pellet was lysed in an equal volume of RIPA lysis buffer. After a 30-min incubation on ice, lysates were spun at 12,000 g for 10 min at 4°C, and supernatants were saved as the mitochondrial fraction. Protein concentrations in the fractions were quantified using a BCA assay. Samples were then subject to SDS-PAGE and probed with anti-cytochrome c (1:1000), MEK1 CT (1:1000), anti-Smac/DIABLO (1:3000), and anti-Omi/HtrA2 (1:3000) antibodies.
In vitro IL-1β cleavage assays
For in vitro assays, RAW264.7 cells were plated in 10-cm plates and grown overnight to confluence. Cells were primed with 1 μg/ml LPS for 2 h to induce the synthesis of pro-IL-1β and then lysed with 300 μl sucrose lysis buffer (250 μM sucrose, 3 mM imidazole, pH 7.4) by passage through a 27 ½ gauge needle. Reactions containing a total volume of 50 μl were prepared, and 1 unit active recombinant caspase-1 was added per 50 μl reaction. Boc-D-CMK, L-Phe-NH2, bestatin methyl ester, and lactacystin were added to some reaction mixtures where indicated. Tubes were incubated at 37°C for 2 h, and reactions were halted by the addition of SDS loading buffer. Caspase-1 activity was monitored by following the cleavage of the caspase-1 substrate pro-IL-1β to its mature 17-kDa form.
ACKNOWLEDGEMENTS
*This research was supported by the Intramural Research Program of the NIH, National Institute of Allergy and Infectious Diseases. We thank Devorah Crown for help with bone marrow isolation.
References
- Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- Baker RT, Varshavsky A. Inhibition of the N-end rule pathway in living cells. Proc Natl Acad Sci U S A. 1991;88:1090–1094. doi: 10.1073/pnas.88.4.1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden ED, Dietrich WF. Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat Genet. 2006;38:240–244. doi: 10.1038/ng1724. [DOI] [PubMed] [Google Scholar]
- Bury M, Mlynarczuk I, Pleban E, Hoser G, Kawiak J, Wojcik C. Effects of an inhibitor of tripeptidyl peptidase II (Ala-Ala-Phe-chloromethylketone) and its combination with an inhibitor of the chymotrypsin-like activity of the proteasome (PSI) on apoptosis, cell cycle and proteasome activity in U937 cells. Folia Histochem Cytobiol. 2001;39:131–132. [PubMed] [Google Scholar]
- Byrd C, Turner GC, Varshavsky A. The N-end rule pathway controls the import of peptides through degradation of a transcriptional repressor. EMBO J. 1998;17:269–277. doi: 10.1093/emboj/17.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clem RJ, Sheu TT, Richter BW, He WW, Thornberry NA, Duckett CS, et al. c-IAP1 is cleaved by caspases to produce a proapoptotic C-terminal fragment. J Biol Chem. 2001;276:7602–7608. doi: 10.1074/jbc.M010259200. [DOI] [PubMed] [Google Scholar]
- Cordoba-Rodriguez R, Fang H, Lankford CS, Frucht DM. Anthrax lethal toxin rapidly activates caspase-1/ICE and induces extracellular release of interleukin (IL)-1beta and IL-18. J Biol Chem. 2004;279:20563–20566. doi: 10.1074/jbc.C300539200. [DOI] [PubMed] [Google Scholar]
- Davydov IV, Varshavsky A. RGS4 is arginylated and degraded by the N-end rule pathway in vitro. J Biol Chem. 2000;275:22931–22941. doi: 10.1074/jbc.M001605200. [DOI] [PubMed] [Google Scholar]
- De Groot RJ, Rumenapf T, Kuhn RJ, Strauss EG, Strauss JH. Sindbis virus RNA polymerase is degraded by the N-end rule pathway. Proc Natl Acad Sci U S A. 1991;88:8967–8971. doi: 10.1073/pnas.88.20.8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveraux QL, Leo E, Stennicke HR, Welsh K, Salvesen GS, Reed JC. Cleavage of human inhibitor of apoptosis protein XIAP results in fragments with distinct specificities for caspases. EMBO J. 1999;18:5242–5251. doi: 10.1093/emboj/18.19.5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveraux QL, Reed JC. IAP family proteins--suppressors of apoptosis. Genes Dev. 1999;13:239–252. doi: 10.1101/gad.13.3.239. [DOI] [PubMed] [Google Scholar]
- Ditzel M, Wilson R, Tenev T, Zachariou A, Paul A, Deas E, et al. Degradation of DIAP1 by the N-end rule pathway is essential for regulating apoptosis. Nat Cell Biol. 2003;5:467–473. doi: 10.1038/ncb984. [DOI] [PubMed] [Google Scholar]
- Du C, Fang M, Li Y, Li L, Wang X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell. 2000;102:33–42. doi: 10.1016/s0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- Duesbery NS, Webb CP, Leppla SH, Gordon VM, Klimpel KR, Copeland TD, et al. Proteolytic inactivation of MAP-kinase-kinase by anthrax lethal factor. Science. 1998;280:734–737. doi: 10.1126/science.280.5364.734. [DOI] [PubMed] [Google Scholar]
- Erwin JL, DaSilva LM, Bavari S, Little SF, Friedlander AM, Chanh TC. Macrophage-derived cell lines do not express proinflammatory cytokines after exposure to Bacillus anthracis lethal toxin. Infect Immun. 2001;69:1175–1177. doi: 10.1128/IAI.69.2.1175-1177.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedlander AM, Bhatnagar R, Leppla SH, Johnson L, Singh Y. Characterization of macrophage sensitivity and resistance to anthrax lethal toxin. Infect Immun. 1993;61:245–252. doi: 10.1128/iai.61.1.245-252.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geier E, Pfeifer G, Wilm M, Lucchiari-Hartz M, Baumeister W, Eichmann K, et al. A giant protease with potential to substitute for some functions of the proteasome. Science. 1999;283:978–981. doi: 10.1126/science.283.5404.978. [DOI] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- Gonda DK, Bachmair A, Wunning I, Tobias JW, Lane WS, Varshavsky A. Universality and structure of the N-end rule. J Biol Chem. 1989;264:16700–16712. [PubMed] [Google Scholar]
- Gordon JJ, Kelly BK, Miller GA. Actinonin: an antibiotic substance produced by an actinomycete. Nature. 1962;195:701–702. doi: 10.1038/195701b0. [DOI] [PubMed] [Google Scholar]
- Hamilton MH, Cook LA, McRackan TR, Schey KL, Hildebrandt JD. Gamma 2 subunit of G protein heterotrimer is an N-end rule ubiquitylation substrate. Proc Natl Acad Sci U S A. 2003;100:5081–5086. doi: 10.1073/pnas.0831228100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna PC, Kochi S, Collier RJ. Biochemical and physiological changes induced by anthrax lethal toxin in J774 macrophage-like cells. Mol Biol Cell. 1992;3:1269–1277. doi: 10.1091/mbc.3.11.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde R, Srinivasula SM, Zhang Z, Wassell R, Mukattash R, Cilenti L, et al. Identification of Omi/HtrA2 as a mitochondrial apoptotic serine protease that disrupts inhibitor of apoptosis protein-caspase interaction. J Biol Chem. 2002;277:432–438. doi: 10.1074/jbc.M109721200. [DOI] [PubMed] [Google Scholar]
- Herrera B, Fernandez M, Benito M, Fabregat I. cIAP-1, but not XIAP, is cleaved by caspases during the apoptosis induced by TGF-beta in fetal rat hepatocytes. FEBS Lett. 2002;520:93–96. doi: 10.1016/s0014-5793(02)02774-6. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hilbi H, Puro RJ, Zychlinsky A. Tripeptidyl peptidase II promotes maturation of caspase-1 in Shigella flexneri-induced macrophage apoptosis. Infect Immun. 2000;68:5502–5508. doi: 10.1128/iai.68.10.5502-5508.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Yang X. Cellular inhibitor of apoptosis 1 and 2 are ubiquitin ligases for the apoptosis inducer Smac/DIABLO. J Biol Chem. 2003;278:10055–10060. doi: 10.1074/jbc.M207197200. [DOI] [PubMed] [Google Scholar]
- Huang H, Joazeiro CA, Bonfoco E, Kamada S, Leverson JD, Hunter T. The inhibitor of apoptosis, cIAP2, functions as a ubiquitin-protein ligase and promotes in vitro monoubiquitination of caspases 3 and 7. J Biol Chem. 2000;275:26661–26664. doi: 10.1074/jbc.C000199200. [DOI] [PubMed] [Google Scholar]
- Klimpel KR, Arora N, Leppla SH. Anthrax toxin lethal factor contains a zinc metalloprotease consensus sequence which is required for lethal toxin activity. Mol Microbiol. 1994;13:1093–1100. doi: 10.1111/j.1365-2958.1994.tb00500.x. [DOI] [PubMed] [Google Scholar]
- Lee MJ, Tasaki T, Moroi K, An JY, Kimura S, Davydov IV, et al. RGS4 and RGS5 are in vivo substrates of the N-end rule pathway. Proc Natl Acad Sci U S A. 2005;102:15030–15035. doi: 10.1073/pnas.0507533102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppla SH. Bacillus anthracis toxins. In: Alouf JE, Popoff MR, editors. The Comprehensive Sourcebook of Bacterial Protein Toxins. Burlington, MA: Academic Press; 2006. pp. 323–347. [Google Scholar]
- Li X, Yang Y, Ashwell JD. TNF-RII and c-IAP1 mediate ubiquitination and degradation of TRAF2. Nature. 2002;416:345–347. doi: 10.1038/416345a. [DOI] [PubMed] [Google Scholar]
- Lin H, Chen C, Chen BD. Resistance of bone marrow-derived macrophages to apoptosis is associated with the expression of X-linked inhibitor of apoptosis protein in primary cultures of bone marrow cells. Biochem J. 2001;353:299–306. doi: 10.1042/0264-6021:3530299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madura K, Varshavsky A. Degradation of G alpha by the N-end rule pathway. Science. 1994;265:1454–1458. doi: 10.1126/science.8073290. [DOI] [PubMed] [Google Scholar]
- Messmer UK, Pereda-Fernandez C, Manderscheid M, Pfeilschifter J. Dexamethasone inhibits TNF-alpha-induced apoptosis and IAP protein downregulation in MCF- 7 cells. Br J Pharmacol. 2001;133:467–476. doi: 10.1038/sj.bjp.0704093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moayeri M, Haines D, Young HA, Leppla SH. Bacillus anthracis lethal toxin induces TNF-á-independent hypoxia-mediated toxicity in mice. J Clin Invest. 2003;112:670–682. doi: 10.1172/JCI17991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder LC, Muesing MA. Degradation of HIV-1 integrase by the N-end rule pathway. J Biol Chem. 2000;275:29749–29753. doi: 10.1074/jbc.M004670200. [DOI] [PubMed] [Google Scholar]
- Park S, Leppla SH. Optimized production and purification of Bacillus anthracis lethal factor. Protein Expr Purif. 2000;18:293–302. doi: 10.1006/prep.2000.1208. [DOI] [PubMed] [Google Scholar]
- Pellizzari R, Guidi-Rontani C, Vitale G, Mock M, Montecucco C. Anthrax lethal factor cleaves MKK3 in macrophages and inhibits the LPS/IFNgamma-induced release of NO and TNFalpha. FEBS Lett. 1999;462:199–204. doi: 10.1016/s0014-5793(99)01502-1. [DOI] [PubMed] [Google Scholar]
- Ramirez DM, Leppla SH, Schneerson R, Shiloach J. Production, recovery and immunogenicity of the protective antigen from a recombinant strain of Bacillus anthracis. J Ind Microbiol Biotechnol. 2002;28:232–238. doi: 10.1038/sj/jim/7000239. [DOI] [PubMed] [Google Scholar]
- Rao H, Uhlmann F, Nasmyth K, Varshavsky A. Degradation of a cohesin subunit by the N-end rule pathway is essential for chromosome stability. Nature. 2001;410:955–959. doi: 10.1038/35073627. [DOI] [PubMed] [Google Scholar]
- Rose C, Vargas F, Facchinetti P, Bourgeat P, Bambal RB, Bishop PB, et al. Characterization and inhibition of a cholecystokinin-inactivating serine peptidase. Nature. 1996;380:403–409. doi: 10.1038/380403a0. [DOI] [PubMed] [Google Scholar]
- Roy N, Deveraux QL, Takahashi R, Salvesen GS, Reed JC. The c-IAP-1 and c-IAP-2 proteins are direct inhibitors of specific caspases. EMBO J. 1997;16:6914–6925. doi: 10.1093/emboj/16.23.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvesen GS, Duckett CS. IAP proteins: blocking the road to death's door. Nat Rev Mol Cell Biol. 2002;3:401–410. doi: 10.1038/nrm830. [DOI] [PubMed] [Google Scholar]
- Sijts AJ, Pilip I, Pamer EG. The Listeria monocytogenes-secreted p60 protein is an N-end rule substrate in the cytosol of infected cells. Implications for major histocompatibility complex class I antigen processing of bacterial proteins. J Biol Chem. 1997;272:19261–19268. doi: 10.1074/jbc.272.31.19261. [DOI] [PubMed] [Google Scholar]
- Singh Y, Leppla SH, Bhatnagar R, Friedlander AM. Internalization and processing of Bacillus anthracis lethal toxin by toxin-sensitive and -resistant cells. J Biol Chem. 1989;264:11099–11102. [PubMed] [Google Scholar]
- Sohn D, Totzke G, Essmann F, Schulze-Osthoff K, Levkau B, Janicke RU. The proteasome is required for rapid initiation of death receptor-induced apoptosis. Mol Cell Biol. 2006;26:1967–1978. doi: 10.1128/MCB.26.5.1967-1978.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Leppla SH. Proteasome activity is required for anthrax lethal toxin to kill macrophages. Infect Immun. 1999;67:3055–3060. doi: 10.1128/iai.67.6.3055-3060.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieku S, Hooper NM. Inhibition of aminopeptidases N, A and W. A re-evaluation of the actions of bestatin and inhibitors of angiotensin converting enzyme. Biochem Pharmacol. 1992;44:1725–1730. doi: 10.1016/0006-2952(92)90065-Q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner GC, Du F, Varshavsky A. Peptides accelerate their uptake by activating a ubiquitin-dependent proteolytic pathway. Nature. 2000;405:579–583. doi: 10.1038/35014629. [DOI] [PubMed] [Google Scholar]
- Umezawa H, Aoyagi T, Suda H, Hamada M, Takeuchi T. Bestatin, an inhibitor of aminopeptidase B, produced by actinomycetes. J Antibiot. 1976;29:97–99. doi: 10.7164/antibiotics.29.97. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. The N-end rule: functions, mysteries, uses. Proc Natl Acad Sci U S A. 1996;93:12142–12149. doi: 10.1073/pnas.93.22.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. The N-end rule pathway of protein degradation. Genes Cells. 1997;2:13–28. doi: 10.1046/j.1365-2443.1997.1020301.x. [DOI] [PubMed] [Google Scholar]
- Varughese M, Chi A, Teixeira AV, Nicholls PJ, Keith JM, Leppla SH. Internalization of a Bacillus anthracis protective antigen-c-Myc fusion protein mediated by cell surface anti-c-Myc antibodies. Mol Med. 1998;4:87–95. [PMC free article] [PubMed] [Google Scholar]
- Verhagen AM, Ekert PG, Pakusch M, Silke J, Connolly LM, Reid GE, et al. Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell. 2000;102:43–53. doi: 10.1016/s0092-8674(00)00009-x. [DOI] [PubMed] [Google Scholar]
- Vitale G, Bernardi L, Napolitani G, Mock M, Montecucco C. Susceptibility of mitogen-activated protein kinase kinase family members to proteolysis by anthrax lethal factor. Biochem J. 2000;352:739–745. [PMC free article] [PubMed] [Google Scholar]
- Wickliffe KE, Leppla SH, Moayeri M. Anthrax lethal toxin-induced inflammasome formation and caspase-1 activation are late events dependent on ion fluxes and the proteasome. Cell Microb. 2007 doi: 10.1111/j.1462-5822.2007.01044.x. doi:10.1111/j.1462-5822.2007.01044.x. (Epub ahead of print Sep 2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang QH, Church-Hajduk R, Ren J, Newton ML, Du C. Omi/HtrA2 catalytic cleavage of inhibitor of apoptosis (IAP) irreversibly inactivates IAPs and facilitates caspase activity in apoptosis. Genes Dev. 2003;17:1487–1496. doi: 10.1101/gad.1097903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang QH, Du C. Smac/DIABLO selectively reduces the levels of c-IAP1 and c- IAP2 but not that of XIAP and livin in HeLa cells. J Biol Chem. 2004;279:16963–16970. doi: 10.1074/jbc.M401253200. [DOI] [PubMed] [Google Scholar]
- Yang Y, Fang S, Jensen JP, Weissman AM, Ashwell JD. Ubiquitin protein ligase activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli. Science. 2000;288:874–877. doi: 10.1126/science.288.5467.874. [DOI] [PubMed] [Google Scholar]