Abstract
Gene therapy is based on the vectorization of genes to target cells and their subsequent expression. Cationic amphiphile-mediated delivery of plasmid DNA is the nonviral gene transfer method most often used. We examined the supramolecular structure of lipopolyamine/plasmid DNA complexes under various condensing conditions. Plasmid DNA complexation with lipopolyamine micelles whose mean diameter was 5 nm revealed three domains, depending on the lipopolyamine/plasmid DNA ratio. These domains respectively corresponded to negatively, neutrally, and positively charged complexes. Transmission electron microscopy and x-ray scattering experiments on complexes originating from these three domains showed that although their morphology depends on the lipopolyamine/plasmid DNA ratio, their particle structure consists of ordered domains characterized by even spacing of 80 Å, irrespective of the lipid/DNA ratio. The most active lipopolyamine/DNA complexes for gene transfer were positively charged. They were characterized by fully condensed DNA inside spherical particles (diameter: 50 nm) sandwiched between lipid bilayers. These results show that supercoiled plasmid DNA is able to transform lipopolyamine micelles into a supramolecular organization characterized by ordered lamellar domains.
In vivo virus-mediated gene transfer is very efficient, but unlike what occurs in nonviral gene transfer, viral-based immunogenicity can significantly limit its efficacy. Nonviral transfer involves procedures in which preformed cationic liposomes or cationic lipids are mixed with plasmid DNA to generate transfecting complexes (1–3).
A variety of these lipids has been synthetized (for review, see ref. 4) with headgroups including a quaternary ammonium such as dioctadecyldiammonium bromide (DODAB), or dioleoyl trimethylammonium propane (DOTAP), or a polycation such as dioctadecylamidoglycylspermine (DOGS). A natural lipid like dioleoyl phosphatidylethanolamine (DOPE) is usually combined with the cationic lipids to form liposomes. The structural characteristics of these DNA/cationic lipid complexes are still poorly understood. Their structure has been studied chiefly by various forms of electron microscopy. Thus, freeze-fracture electron microscopy has shown uncondensed DNA fibers coated with lipid connected to the surface of aggregated fused cationic liposomes of 3-β-[N-(N′,N′-dimethylethane)carbamoyl]cholesterol/DOPE (DC-Chol/DOPE) (5). Electron microscopy performed with negative staining of DNA complexed with the cationic liposome of N-(1-[2,3-bis(oleoyloxy)]propyl)-N,N,N-trimethylammonium chloride/DOPE (DOTMA/DOPE) suggested that cationic liposomes initially form clusters along the DNA molecule. Above a critical liposome density, these clusters coalesce by DNA-induced membrane fusion, leading to DNA encapsulation by the lipids (6). Transmission electron microscopy (TEM) with uranyl acetate staining further indicated that DNA molecules might be condensed and intertwined in the tubules of the lipopolyamine DOGS network, leading to hexagonal packing of DNA (7). Cryo-transmission electron microscopy examination suggested that an excess of cationic lipid led to DNA encapsulation between the lamellae of aggregated multilamellar structures (8). In very recent investigations, a multilamellar organization in which DNA is intercalated between the layers was reported for systems resulting from the interaction of DNA with unilamellar cationic lipid vesicles, i.e., the quaternary ammonium lipids DOTAP (9) and DODAB (10), respectively, combined with DOPE and cholesterol. Both the investigations concerned showed a periodicity of 6.5 nm in the lamellar structure, as determined by x-ray scattering. Because the membrane thickness was 3.9 nm, the thickness of the water layer was 2.6 nm, which is sufficient to include a hydrated DNA double helix with a total diameter of 2.5 nm. A second periodicity was also observed and was attributed to DNA–DNA correlation. It ranged from 2.45 nm to 5.71 nm, as the neutral lipid dioleoyl phosphatidylcholine (DOPC) increased, for a constant DOTAP/DNA ratio (9). Nevertheless, the relevance of this structure to lipopolyamine/DNA complexes remained to be elucidated.
The present experiments were designed to define the conditions under which DNA molecules collapse into oligomolecular complexes rather than forming larger multimolecular aggregates. These types of small complexes should exhibit higher in vivo bioavailability than larger ones.
MATERIALS AND METHODS
Materials.
The plasmid DNA used (6,390 bp) was derived from pBluescript II KS (Stratagene) containing the luciferase gene of Photinus pyralis controlled by the immediate early promoter of the cytomegalovirus. This plasmid was purified from recombinant Escherichia coli DH5α (CLONTECH) by alkaline lysis followed by two cesium chloride ethidium bromide density gradients (11). The purified plasmid DNA (1 mg/ml) was in 10 mM Tris, pH 7.4, 0.1 mM EDTA, and 40 mM NaCl.
N-[1-(2,3-Dimyristyloxy)propyl]-N,N-dimethyl-N-(2-hydroxyethyl)ammonium bromide/cholesterol (DMRIE/Cholesterol) was from GIBCO/BRL.
RPR120535 micelles (12) were obtained by sonicating a 10-mM aqueous solution of lipopolyamine for 15 min.
Lipopolyamine/plasmid DNA complexes were prepared by mixing equal volumes of RPR120535 micelles at different concentrations with the plasmid DNA solution at the desired concentration.
Electrophoretic and Dynamic Light Scattering.
Electrophoretic and light scattering measurements were made on a Delsa 440 SX instrument (Coulter) and a Coulter N4Plus instrument, respectively, at 20°C in both cases. Samples of the lipopolyamine/plasmid DNA complex were prepared for zeta potential determination at 0.5 mg DNA/ml in 20 mM NaCl, and diluted 50-fold in 20 mM NaCl. The zeta potential was calculated from the electrophoretic mobility. For dynamic light scattering experiments, samples were prepared at 0.1 mg DNA/ml and four different NaCl concentrations and diluted 10-fold in the same solutions.
In Vitro Transfection.
HeLa cervix carcinoma cells (American Type Culture Collection) were cultured in flasks at 37°C in a 5% CO2 humidified atmosphere, in DMEM (GIBCO/BRL, Life Technologies) supplemented with 20 mM l-glutamine and 10% fetal calf serum (GIBCO/BRL, Life Technologies). Cells were placed in 24-well dishes and transfected at 60–80% confluence with complexes containing 0.5 μg of plasmid DNA. Transfections were performed in 500 μl of DMEM supplemented with 50 μl of lipopolyamine/DNA solution prepared at 150 mM NaCl, per well. Fetal calf serum (10%) was added 2 hr after transfection. Luciferase activity was measured as previously described (12).
Electron Microscopy.
Experiments using cryo-transmission electron microscopy (cryo-TEM) (13) were performed with 7 μl of the different samples, prepared at 0.5 mg DNA/ml in 20 mM NaCl, which were spread over holes in a perforated carbon film supported by 200-mesh copper electron microscope grids. The grids were then quenched in liquid ethane at its freezing point and transferred into a Gatan 626 cryo-holder precooled with liquid nitrogen. For cryo-phosphotungstate fixation and staining, samples were mixed with 1% (wt/vol) phosphotungstate before water vitrification. For uranyl acetate staining, 7 μl samples were added to the grid, blotted, washed, and stained with 1% uranyl acetate. All samples were examined with a transmission electron microscope (Philips CM12). For light microscopy examinations, we used a Zeiss Axiophot microscope equipped with a Differential Interference Contrast function, and a 40× objective.
X-ray Scattering.
Samples were prepared at 0.5 mg DNA/ml in 20 mM NaCl and centrifuged at 109,000 × g (Optima TL, Beckman). Pellets were introduced into a cell comprising two Kapton windows. The x-ray scattering experiments were carried out by using the synchrotron radiation source DCI of LURE (Orsay, France) on beamline D22. A Ge monochromator (111 reflection) allowed the selection of a wavelength of 1.38 Å by using a linear high-resolution Xe-CO2 detector. Results are expressed in arbitrary intensity units as a function of log(q), where q is the momentum transfer q = (4π/λ)sinθ/2, θ, the scattering angle, and λ, the x-ray wavelength. q values ranged between 10−2 Å−1 and 0.4 Å−1.
CD.
All samples were prepared at 50 μg DNA/ml (7.57 × 10−5 M, in base pairs) in 150 mM NaCl. CD spectra were recorded in a 1-cm light-path cell on a Jobin-Yvon CD6 dichrograph (Jobin-Yvon, Longjumeau, France).
RESULTS
Colloidal Stability of Lipopolyamine/Plasmid DNA Complexes.
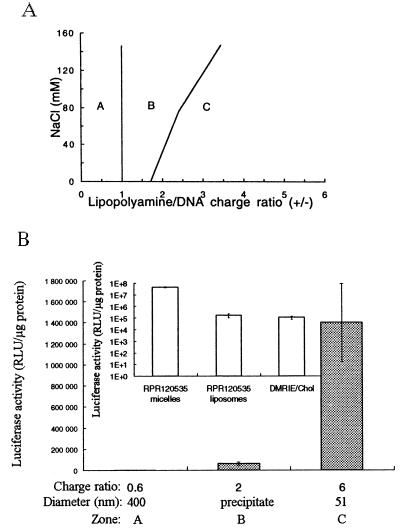
Lipopolyamines are amphiphiles with a self-aggregating hydrocarbon tail linked to a polycation DNA-binding headgroup. The lipopolyamine RPR120535 used (Fig. 1) has a spermine-derived amino acid linked via a glycine to its dioctadecyl hydrophobic entity. Cryo-TEM examination of aqueous solutions of RPR120535 showed the presence of homogeneous densely packed spherical micelles about 5 nm in diameter (Fig. 2A). Addition to plasmid DNA of an ethanolic solution of lipopolyamine, which forms tubular micelles, has been reported to lead to the formation of complexes with a hexagonal structure (7, 14) that promote gene transfer (3, 15). The stability of these complexes is essential for their use as gene delivery systems. Therefore, it is necessary to understand how colloid stability may be affected by important parameters such as the RPR120535/DNA charge ratio (charge ratio) which is expressed as moles of positive charge/moles of negative charge and corresponds to 3/2 of the molar ratio, expressed as moles RPR120535/moles plasmid DNA base pair. This is because at neutral pH, there are three positive charges per cationic lipid molecule, i.e., the weakest secondary amine has a pK of 6 (data not shown) and two negative charges per DNA base pair. Another important parameter is the interaction involving electrolytes, which may be required to adjust tonicity, because electrolytes interact strongly with charge-stabilized colloids. RPR120535/DNA complexes, prepared at different ionic strengths, were found to exhibit a three-zone model of colloidal stability, whatever the initial plasmid DNA concentration (i.e., 0.05, 0.1, or 0.5 mg DNA/ml). For given NaCl and DNA concentrations, the three zones in the complexation, termed A, B, and C (Fig. 3A), were determined by their concentration of RPR120535. In zones A and C (Fig. 3A), RPR120535/DNA complexes were colloidally stable and had a negative theoretical charge ratio in zone A and a positive one in zone C. The mean diameter of the RPR120535/DNA complexes from zone A was around 500 nm, as measured by dynamic light scattering. The complexes in zone C had a mean diameter that decreased from 300 to 50 nm as the RPR120535 concentration was raised. In zone B, the complexes were not colloidally stable, because a visible precipitate was observed within a few minutes of complexation. In this zone, the RPR120535/DNA complexes had a theoretical charge ratio close to neutral.
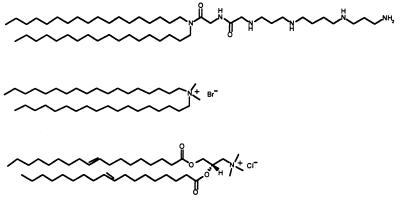
Figure 1.
Structure (from top to bottom) of the lipids lipopolyamine (RPR120535), dioctadecyldiammonium bromide (DODAB), and dioleoyl trimethylammonium propane (DOTAP).
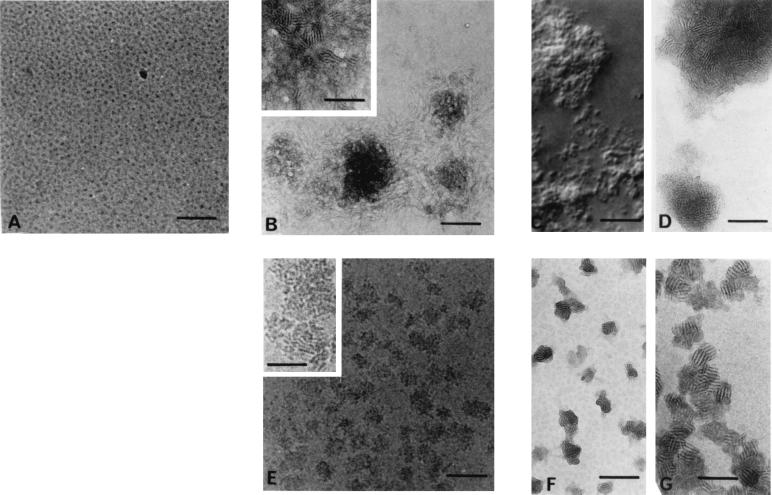
Figure 2.
RPR120535 and RPR120535/DNA complexes from zones A, B, and C (see Fig. 3), visualized by light microscopy and transmission electron microscopy (TEM). (A) Cryo-TEM micrographs of an aqueous solution of RPR120535 alone. (B) Electron micrograph of uranyl acetate-stained complexes from zone A (RPR120535/DNA charge ratio: 0.3). Inset shows the same complexes at higher magnification. (C) Complexes from zone B (charge ratio: 1.65) observed by light microscopy. (D) Electron micrograph of the same complexes stained with uranyl acetate. (E) Cryo-TEM micrograph of RPR120535/DNA complexes originating from zone C (charge ratio: 6). Inset shows the visualization by cryo-TEM of the ordered domains in these complexes. (F) Electron micrograph of uranyl acetate-stained complexes from zone C. (G) Cryo-phosphotungstate-TEM micrograph of the same complexes. In this micrograph, the complexes seem to have aggregated, because the thickest part of the vitrified film allows them to be superimposed. The scale bar represents 100 nm in A, B Inset, and D–G; 500 nm in B; and 10 μm in C.
Figure 3.
(A) Colloidal stability of RPR120535/DNA complexes, at 0.1 mg DNA/ml, as a function of the RPR120535/DNA charge ratio (ratios were raised from 0 to 7 by increments of 0.3), in 4, 40, 80, or 150 mM NaCl. RPR120535/DNA complexes were defined as colloidally stable if their mean diameter was below 700 nm for at least 24 hr. Complexes from zones A and C were stable, whereas those from zone B exhibited a visible precipitate. (B) In vitro luciferase activity, measured in transfected HeLa cells, of RPR120535/DNA complexes originating from zones A, B, and C. Inset shows, in transfected NIH 3T3 cells, the in vitro transfection activity of DNA complexes with RPR120535 micelles or liposomes, i.e., RPR120535/DOPE (1/1, M/M), both at a charge ratio of 6, and of DNA complexes with DMRIE/Cholesterol, at a molar ratio of 1.65.
Electrophoretic light scattering measurements showed that the RPR120535/DNA complexes from zones A and C had zeta potentials of −37 and + 29 mV, respectively. These complexes therefore are colloidally stable, because the repulsive electrostatic forces between them are proportional to the square of the zeta potential (16). On the other hand in zone B, the complexes close to neutral aggregated, because Van der Waals forces clumped them together.
In zone B, the complexes were observed to aggregate and became larger as the NaCl concentration was raised (Fig. 3A). This reflected a charge-screening effect, which was probably a result of reduced electrostatic repulsion between complexes that favored aggregation.
Fluorescence experiments were performed by exposing cationic RPR120535/DNA complexes to ethidium bromide, which upon intercalation between the DNA base pairs acts as a fluorescence probe. Plasmid DNA was accessible to the probe in zone A, whereas in zones B and C, the fluorescence level was minimal (data not shown). This indicates that in zones B and C, DNA molecules were condensed by the RPR120535.
Transfection Activities of Lipopolyamine/DNA Complexes.
The luciferase activities measured in transfected HeLa cells (Fig. 3B) showed that the complexes from zone C, with a mean diameter of 50 nm, were by far the most active in promoting transfection. The transfection activity of these complexes was 400 times greater in NIH 3T3 cell lines (Fig. 3B Inset) than the activity of the DNA complexes formed with cationic liposomes of RPR120535/DOPE (1/1; M/M) or of DMRIE/Cholesterol.
Electron Microscopy of Lipopolyamine/DNA Complexes.
An electron micrograph of uranyl acetate-stained complexes from zone A (Fig. 2B) showed either free DNA or loosely bound plasmids near the complexes. However, at higher magnification (Fig. 2B Inset), ordered microdomains with a “fingerprint” shape were observed in these complexes at the center of the particles. Complex size distribution, determined by image observation, was heterogeneous, and the mean diameter was about 500 nm.
Light microscope observation of complexes from zone B (Fig. 2C) showed the aggregation of a large number of particles. One electron micrograph of these complexes stained with uranyl acetate (Fig. 2D) showed that free or loosely bound plasmids had disappeared from the vicinity of the complexes and that the ordered organization seen in the complexes from zone A was also present in zone B complexes.
Electron micrographs of complexes from zone C (Figs. 2 E–G) showed that all the DNA was complexed with the RPR120535 to form a homogeneous population of individual particles 50 nm in diameter and fairly round in shape (stereo pairs, not shown). The ordered microdomains covered the entire surface of the particles (Fig. 2E Inset and Fig. 2 F and G). Complexes with the same shape and structure were revealed by cryo-TEM (Fig. 2E and Inset), uranyl acetate TEM (Fig. 2F), and cryo-phosphotungstate TEM (Fig. 2G). Because stained specimens generally display a higher contrast (Fig. 2F) than unstained specimens in vitreous ice (Fig. 2E), we mixed either uranyl acetate or phosphotungstate with a sample from zone C just before water vitrification. Cryo-TEM examination of these samples (Fig. 2G) showed spectacular contrast enhancement. This technique, which is still in its early stages, combines the benefits of vitrification with the increased contrast characterizing negatively stained images (17). However, it was not possible to use either cryo-TEM or cryo-TEM with uranyl acetate or phosphotungstate staining to visualize the RPR120535/DNA complexes from zones A and B, because when the charge ratio was negative, the number of complexes present was limited, and when the charge ratio was neutral, large RPR120535/DNA complexes increased the thickness of the film of the vitreous solution, which made optimal imaging of the structure more difficult.
The morphology of the lipopolyamine/DNA complexes depended on the RPR120535/DNA ratio. However, fingerprint-like ordered domains were present whatever this ratio. They were characterized by even spacing of approximately 80 Å for the microcrystalline domains in zones A, B, and C, as calculated by optical diffraction on micrographs (data not shown).
X-Ray Scattering of Lipopolyamine/DNA Complexes.
X-ray scattering scans of complexes from zones A, B, and C (Fig. 4A) revealed a band with spacings of 80, 83, and 83 Å, respectively, expressed as d = 2Π/q. In clear contrasts, no diffraction maxima were exhibited by either plasmid DNA or RPR120535 alone¶ (Fig. 4B), which indicated that the scattering profile shown in Fig. 4A reflects periodic spacing of DNA and RPR120535, which was observed whatever the RPR120535/DNA ratio. The scattering peak at 80 Å shown in all zones by x-ray scattering was close to the spacing found by micrograph analysis. The half-width of the scattering peak exhibited by the different complexes corresponded to an effective domain size of around 300 Å, which indicated the repetition of approximately four periodicities, in agreement with the structures revealed by TEM.
Figure 4.
X-ray scattering studies. (A) Small-angle x-ray scattering scans of RPR120535/DNA complexes at various cationic lipid-to-DNA ratios. Symbols ○, □, and • correspond to RPR120535/DNA complexes from zones A (RPR120535/DNA charge ratio: 0.33), B (charge ratio: 1.65), and C (charge ratio: 6), respectively. (B) Because complex samples were prepared at 0.5 mg DNA/ml and concentrated 10-fold by ultracentrifugation, small-angle x-ray scattering scans were performed at 5 mg/ml on both DNA alone (○) and RPR120535 aqueous solution (□).
Regarding the low q slopes, which varied significantly between zones A, B, and C, we noted that the low q dependence of the intensity in zone A (Fig. 4A) was the same as in RPR120535-free DNA (Fig. 4B), i.e., its intensity was proportional to q−2.9. This observation suggested that the low q dependence resulted from the presence of excess DNA. In zone B (Fig. 4A), the slope was not as steep as in zone A (q−1.7), but the increase in intensity at low q values still displayed long-range correlations. The latter may have been a result of the aggregation of RPR120535/DNA complexes seen in light microscopy. Last, in zone C, overall intensity was weaker and did not increase at low q values. The curvature in this region was attributed to the shape factor of smaller objects than in zone B. These objects exhibited no significant aggregation. The results of our x-ray scans were consistent with those of the TEM investigations.
Complexed DNA Conformation Studies by CD.
RPR120535/DNA complexes from zone B exhibited a flat CD spectrum. This was because of the sedimentation of the aggregated complexes in the cell. The corresponding spectra of complexes from zones A and C exhibited an attenuated positive lobe at 280 nm, an increased negative lobe at 245 nm, and a red shift of the crossover point (Fig. 5). These spectral changes might indicate a perturbation of the secondary conformation (18), such as less hydration of the DNA molecules. The shape of the CD spectra for all zones revealed that the DNA molecules remained in the conventional double-helix B-type conformation. Therefore, the observation of B-CD signals for high RPR120535/DNA ratios indicated that the DNA was packed between lipid bilayers, so that the DNA molecules were not highly ordered, unlike what we observed with a peptide from the C-terminal domain of histone H1 (19) complexed with plasmid DNA (data not shown). With this peptide, we obtained a nonconservative CD signal called Ψ-DNA, as previously observed under other conditions (20, 21).
Figure 5.
Circular dichroism spectra exhibited by RPR120535/DNA complexes (ɛ, molar ellipticity). Curves 1, 2, and 3 correspond to plasmid DNA alone, and to complexes from zones A (charge ratio: 0.6) and C (charge ratio: 6), respectively. RPR120535/DNA complexes from zone B exhibited a flat CD spectrum, because of the sedimentation of the very large complexes.
DISCUSSION
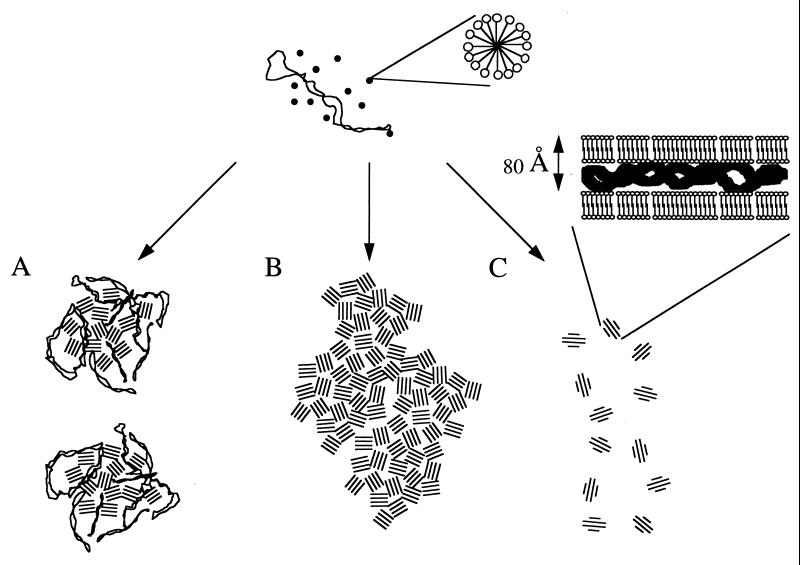
The colloidal stability of the lipopolyamine micelle/DNA complexes, which depended on the charge ratio, determined three main zones—A, B, and C—which corresponded to negatively, neutrally, and positively charged complexes, respectively. The mean diameter of the complexes resulting from the interaction of plasmid DNA with the newly synthetized lipopolyamine RPR120535 was about 500 nm in zone A. In zone C, the mean diameter decreased from 300 to 50 nm as the RPR120535 concentration was raised. In zone B, the complexes were not colloidally stable, because very large particles were formed within a few minutes of complexation. As the NaCl concentration was raised, the boundary between zones B and C shifted toward an increased charge ratio because of charge screening. The morphology of the complexes differed in the three zones, but microdomains shaped like fingerprints, exhibiting regular spacing of 80 Å, were observed in all cases (Fig. 6). There was good agreement between the data from x-ray scattering and transmission electron microscopy experiments, which established that plasmid DNA transforms the initial cationic RPR120535 micelles into a supramolecular organization consisting of ordered microdomains. The spacing and effective domain size were around 80 and 300 Å, respectively, thus corresponding to the repetition of four periodicities. It is not surprising that the second-order reflection of a lamellar symmetry was not observed in the small-angle x-ray scattering experiments, because the first-order reflection peak was fairly broad. However, TEM observations strongly suggested the presence of a lamellar organization consisting of alternating lipid bilayers with plasmid DNA intercalated between them, because angles characteristic of a hexagonal structure (i.e., 120°) were never found among the several hundred complexes examined on many micrographs. In addition, for a not very concentrated RPR120535 solution, i.e., 25% (wt/vol) in 1 M NaCl, we observed, by small-angle x-ray scattering, reflections at positions clearly indicating lamellar symmetry (data not shown). This confirms the very strong ability of the RPR120535 to form a lamellar structure when spermine was charge neutralized. Two series of very recent x-ray scattering experiments (9, 10) also showed that the structure of DNA-cationic liposome complexes consists of a multilamellar membrane in which DNA is intercalated. The cationic lipid used in these experiments bore a single quaternary ammonium, i.e., a single positive charge per molecule, which was DODAB in one series (10) and DOTAP in the other (9), mixed with the colipids DOPE and cholesterol, to form unilamellar vesicles. A spacing of 65 Å was reported in both cases. The difference between this spacing and our spacing of 80 Å may be explained by the intrinsic structure of the cationic lipids, because a cationic lipid bearing a spermine as headgroup should occupy more space than a lipid with a single quaternary ammonium headgroup (Fig. 1).
Figure 6.
Schematic representation of the RPR120535/DNA complexes resulting from the association of cationic micelles and supercoiled DNA, as a function of the RPR120535/DNA ratio. A, B, and C represent negatively, neutrally, and positively charged complexes, respectively. The number of cationic lipid molecules per micelle is arbitrary.
Nonviral gene therapy applications will require rigorous characterization of the complexes formed between plasmid DNA and a gene transfer agent. Our goal here was to control plasmid DNA condensation carefully to obtain virus-sized particles for efficient in vitro and in vivo gene delivery. In particular, the oligomolecular condensation of plasmid DNA with cationic lipids may yield complexes of small size, close to that of viruses. The diameter of the viruses used in most gene transfer protocols ranges from 20 to 24 nm for adeno-associated viruses (22), to 120 nm for adenoviruses (23), and 60 to 90 nm for retroviruses (24). Small transfecting particles 10–12 nm in diameter have been obtained with galactosylated poly(l-lysine) (25). In addition, endocytosis, the recognized route for cell penetration of the complexes (26), can accommodate only particles with a maximal diameter of 150 nm (27). When we used cationic liposomes as starting material, i.e., RPR120535 combined with DOPE at a molar ratio of 1, instead of spherical micelles about 5 nm in diameter, the zone B was larger and the RPR120535/DNA complexes from zones A and C had a bigger mean diameter than that of the complexes obtained with micelles. Our approach implied that the large DNA molecule would nucleate the complex formation rather than the small micelles. These types of conditions probably lead to mono- or oligomolecular complexation of plasmid DNA, as opposed to liposome-mediated complex formation. Cationic lipids currently are mixed with lamellar-forming diacyl lipids such as DOPE, leading to the formation of large complexes when the lipids are mixed with DNA (data not shown; refs. 9 and 28). Lipopolyamine, on the contrary, was found (i) to be able to form micelles in water, (ii) to promote gene transfers without complexation with a co-lipid (3), and (iii) to be as effective for gene transfer as other cationic lipids (15).
Here, highly effective complexes for gene transfer were obtained in zone C. They had 400-fold higher levels of gene expression in NIH 3T3 cells than the DNA complexes formed with cationic liposomes of DMRIE/Cholesterol. They were characterized by their small diameter (50 nm) and high colloidal stability. The number of plasmid molecules per complex from zone C was estimated by assuming that the lipid bilayer thickness was 40 Å (9) and that, because the periodicity was 80 Å, the DNA was contained in half of the volume of a 50-nm spherical particle (3.27 × 10−17 cm3). The volume occupied by one DNA molecule was 4.2 × 10−18 cm3, according to the formula (Mr/N)/dDNA, where dDNA is DNA density (1.66 g/cm3), N is Avogadro’s number, and Mr is 4.21 × 106 g/mol. Therefore, at best, these complexes contained seven plasmids, thus confirming that few plasmids were condensed. The plasmid DNA sandwiched between lipopolyamine bilayers exhibited the same CD spectra as the conventional double-helix B-type conformation, unlike the long-range helical organization, i.e., a Ψ-type CD spectrum, observed when the plasmid DNA was complexed with a peptide from the C-terminal domain of histone H1 (19).
Better understanding of complex formation might make it possible to obtain monomolecular plasmid DNA complexes of a smaller size. Further studies are required to obtain the colloidal stabilization of the complexes from zone B, which, because their charge ratio was close to neutral, might exhibit different transfecting properties from those of the zone C complexes.
Acknowledgments
We thank J.-C. Daniel, J.-B. LePecq, and C. Hélène for their constant support and interest. We also thank B. Cabane and D. Roux for helpful discussions, A. Sanson for giving us access to the dichrograph, F. Blanche for critical reading of the manuscript, M. Ollivier for the plasmid preparation, and V. Thuillier, M.-A. Ivanov, and B. Schwartz for stimulating discussions. This work was supported by the Bioavenir Program financed by Rhône-Poulenc, the French Ministry of Research, and the Ministry of Industry.
ABBREVIATIONS
- DOPE
dioleoyl phosphatidylethanolamine
- DOTAP
dioleoyl trimethylammonium propane
- DODAB
dioctadecyldiammonium bromide
- TEM
transmission electron microscopy
Footnotes
No scattering peak from cationic lipopolyamine micelles was observed, possibly because of the low contrast of the lipid molecule.
References
- 1.Felgner P L, Gadek T R, Holm M, Roman R, Chan H W, Wenz M, Northrop J P, Ringold G M, Danielsen M. Proc Natl Acad Sci USA. 1987;84:7413–7417. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao X A, Huang L. Biochem Biophys Res Commun. 1991;179:280–285. doi: 10.1016/0006-291x(91)91366-k. [DOI] [PubMed] [Google Scholar]
- 3.Behr J P, Demeneix B, Loeffler J P, Perez-Mutul J. Proc Natl Acad Sci USA. 1989;86:6982–6986. doi: 10.1073/pnas.86.18.6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lasic D D, Templeton N S. Adv Drug Delivery Rev. 1996;20:221–266. [Google Scholar]
- 5.Sternberg B, Sorgi F, Huang L. FEBS Lett. 1994;356:361–366. doi: 10.1016/0014-5793(94)01315-2. [DOI] [PubMed] [Google Scholar]
- 6.Gershon H, Ghirlando R, Guttman S B, Minsky A. Biochemistry. 1993;32:7143–7151. doi: 10.1021/bi00079a011. [DOI] [PubMed] [Google Scholar]
- 7.Labat-Moleur F, Steffan A-M, Brisson C, Perron H, Feugas O, Furstenberger P, Oberling F, Brambilla E, Behr J-P. Gene Ther. 1996;3:1010–1017. [PubMed] [Google Scholar]
- 8.Gustafsson J, Arvidson G, Karlsson G, Almgren M. Biochim Biophys Acta. 1995;1235:305–312. doi: 10.1016/0005-2736(95)80018-b. [DOI] [PubMed] [Google Scholar]
- 9.Radler J O, Koltover I, Salditt T, Safinya C R. Science. 1997;275:810–814. doi: 10.1126/science.275.5301.810. [DOI] [PubMed] [Google Scholar]
- 10.Lasic D, Strey H, Stuart M, Podgornik R, Frederik P M. J Am Chem Soc. 1997;119:833. [Google Scholar]
- 11.Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. pp. 1.21–1.52. [Google Scholar]
- 12.Byk, G., Dubertret, C., Schwartz, B. & Scherman, D. (1995) Eur. Patent Appl. WO 97,18185.
- 13.Dubochet J, Adrian J-J, Homo J-C, Lepault J, McDowwal A W, Schultz P. Q Rev Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- 14.Behr J P. Tetrahedron Lett. 1986;27:5861–5864. [Google Scholar]
- 15.Remy J-S, Sirlin C, Behr J-P. In: Liposomes as Tools in Basic Research and Industry. Philippot J-R, Schuber F, editors. Boca Raton, FL: CRC; 1995. pp. 159–170. [Google Scholar]
- 16.Washigton C. Adv Drug Delivery Rev. 1996;20:131–145. [Google Scholar]
- 17.Harris J R. Negative Staining and Cryoelectron Microscopy: The Thin Film Techniques. Oxford: BIOS Scientific; 1997. pp. 171–183. [Google Scholar]
- 18.Saenger W. In: Principles of Nucleic Acid Structure. Cantor C R, editor. New York: Springer; 1984. pp. 368–384. [Google Scholar]
- 19.Erard M, Lakhdar-Ghazal F, Amalric F. Eur J Biochem. 1990;191:19–26. doi: 10.1111/j.1432-1033.1990.tb19088.x. [DOI] [PubMed] [Google Scholar]
- 20.Jordan C F, Lerman L S, Venable J H. Nature (London) 1972;236:67. doi: 10.1038/newbio236067a0. [DOI] [PubMed] [Google Scholar]
- 21.Reich Z, Levin-Zaidman S, Gutman S B, Rard T, Minsky A. Biochemistry. 1994;33:14177–14184. doi: 10.1021/bi00251a029. [DOI] [PubMed] [Google Scholar]
- 22.Muzyka N. Curr Top Microbiol Immunol. 1992;158:97–129. doi: 10.1007/978-3-642-75608-5_5. [DOI] [PubMed] [Google Scholar]
- 23.Shenk T. In: Virology. Fields B, editor. New York: Raven; 1996. pp. 2111–2148. [Google Scholar]
- 24.Coffin J. In: Virology. Fields B, editor. New York: Raven; 1996. pp. 1767–1847. [Google Scholar]
- 25.Perales J C, Ferkol T, Beegen H, Ratnoff O, Hanson R W. Proc Natl Acad Sci USA. 1994;91:4086–4090. doi: 10.1073/pnas.91.9.4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zabner J, Fasbender A A, Moninger T, Poellinger K A, Welsh M J. J Biol Chem. 1995;270:18997–19007. doi: 10.1074/jbc.270.32.18997. [DOI] [PubMed] [Google Scholar]
- 27.Watts C, Marsh M. J Cell Sci. 1992;103:1–8. doi: 10.1242/jcs.103.1.1a. [DOI] [PubMed] [Google Scholar]
- 28.Gao X, Huang L. Biochemistry. 1996;35:1027–1036. doi: 10.1021/bi952436a. [DOI] [PubMed] [Google Scholar]