Abstract
Glypican-3 (gpc3) is the gene responsible for Simpson-Golabi-Behmel overgrowth syndrome. Previously, we have shown that GPC3 is overexpressed in hepatocellular carcinoma (HCC). In this study, we demonstrated the mechanisms for GPC3-mediated oncogenesis. Firstly, GPC3 overexpression in NIH3T3 cells gave to cancer cell phenotypes including growing in serum-free medium and forming colonies in soft agar, or on the other way, GPC3 knockdown in HuH-7 cells decreased oncogenecity. We further demonstrated that GPC3 bound specifically through its N-terminal proline-rich region to both Insulin-like growth factor (IGF)-II and IGF-1R. GPC3 stimulated the phosphorylation of IGF-1R and the downstream signaling molecule extracellular signal-regulated kinase (ERK) in an IGF-II-dependent way. Also, GPC3 knockdown in HCC cells decreased the phosphorylation of both IGF-1R and ERK. Therefore, GPC3 confers oncogenecity through the interaction between IGF-II and its receptor, and the subsequent activation of the IGF-signaling pathway. This data are novel to the current understanding of the role of GPC3 in HCC and will be important in future developments of cancer therapy.
Introduction
Hepatocellular carcinoma (HCC) is the leading cause of death among cancers in many countries especially in Asia, and the incidence of HCC is rising in many other countries (1,2). The molecular mechanisms for hepatocarcinogenesis are quite complex (3), and despites early detection and aggressive therapies, the outcome of HCC remains grave. We previously discovered that glypican-3 (GPC3, also known as MXR7) is overexpressed in HCC (4). gpc3 mRNA was detectable in 143 of 191 (74.8%) primary and recurrent HCCs, whereas only in 3.2% of the non-tumor part of the livers. GPC3 overexpression correlates to high alpha-fetoprotein, high tumor grade and high tumor aggressiveness in HCC individuals (4), and GPC3 stimulates in vitro and in vivo growth of HCC (5). Overexpression of gpc3 mRNA is also observed in metastatic colorectal carcinomas (6), alpha-fetoprotein-producing gastric carcinoma (7), hepatoblastoma, Wilms’ tumor (8), malignant melanoma (9), yolk sac tumor, choriocarcinoma (10), ovarian carcinoma (11) and in cell lines derived from breast cancer (12) and ovarian cancer (13). These findings suggest that GPC3 plays a role in oncogenesis.
GPC3 is a glycosyl-phosphatidylinositol-anchoring heparan sulfate proteoglycan (14). It functions as a ‘coreceptor’ for heparin-binding proteins like growth factors or adhesion molecules (15,16), and the binding of coreceptors facilitate interactions between the heparin-binding factors and their corresponding receptors (17,18). GPC3 is first processed to the 65 kDa core protein, which can be further cleaved by a furin-like convertase into a 40 kDa protein. GPC3 modulates Wnt signaling and stimulates the growth of HCC cells in a glycosaminoglycan-independent way (5). GPC3 has also been reported to bind Insulin-like growth factor-II (19,20). The IGF-signaling pathway plays a pivotal role in cell proliferation (21), G1 cell cycle progression (22), prevention of apoptosis (23) and the initiation and maintenance of oncogenesis (24–26). Increase in IGF-II expression has been observed in liver cancers including HCC (27–29), and insulin receptor substrate-1, an adapter molecule for IGF-II signaling, is overexpressed in HCC (30,31). GPC3 is also involved in the pathogenesis of Simpson-Golabi-Behmel overgrowth syndrome (19,20).
In order to study the mechanism for GPC3-mediated oncogenesis, we expressed GPC3 in GPC-null NIH3T3 cells and also knocked down GPC3 in GPC3-expressing HCC cells. We demonstrated specific interactions both between GPC3 and IGF-II and between GPC3 and IGF-IR, and we also showed GPC3- and IGF-II-dependent phosphorylation of IGFI-R and extracellular signal-regulated kinase (ERK). Therefore, GPC3 confers oncogenecity through interacting with IGF-II and its receptor and the activation of the IGF-signaling pathway.
Materials and methods
Cell lines
HEK293, HEK293T, HeLa, PLC-PRF-5 (American Tissue Cell Collection), NIH3T3 (Biosources Collection and Research Center, National Health Research Institute, Taiwan), HA22T/VGH (32) and HuH-7 (33) cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. All cell lines were maintained at 37°C in a humidified atmosphere with 5% CO2. Serum starvation was done in 0.5% serum for the HA22T/VGH cells and was in 0% serum for all other cells.
Plasmids
The wild-type GPC3 expression vector pcDNA-gpc3 was constructed by inserting gpc3 complementary DNA into pcDNA3.1 (+) myc-his vector (Invitrogen, San Diego, CA) (4). The C-terminal green fluorescence protein (GFP)-tagged GPC3 (GPC3-GFP) expression vector pgpc3-GFP was constructed by replacing the stop codon with an EcoRI site (using primer T3 and GTGCTTCTTCTTCCTGGTGAAGGCTGGTGAATTCT, with the EcoRI site underlined), and transferred into pcDNA3.1. KpnI/EcoRV fragment of the insert was transferred into KpnI and SmaI site of pEGFP-N1 vector (Clontech, Mountain View, CA). Arginine residues 355 and 358 of the GPC3 protein were mutated to alanine by site-directed mutagenesis to express GPC3 mutant RR → AA (RR → AA) (34); and proline residues 25–29 were mutated to alanine in GPC3 mutant P25-29A (P25-29A). All constructs were confirmed by DNA sequencing.
Transfection and selection for stable lines
Transient transfection was done with Lipofectamine 2000 according to the manufacturer's instruction (Invitrogen). To generate NIH3T3, PLC-PRF-5 and HA22T/VGH stable lines, selection was performed with 800 μg/ml G418 (Invitrogen). Surviving cells were isolated and expanded.
Reverse transcription-polymerase chain reaction
Total RNA was extracted by the Trizol reagent (Invitrogen) and was reverse transcribed according to the manufacturer's instruction (Invitrogen). Semiquantitative reverse transcription–polymerase chain reaction was used to measure gpc3 mRNA levels using primers GPC3-974-F (TGCTCTTACTGCCAGGGACT) and GPC3-1671-R (TCAAACCCTTCCTCATCCAG). For checking human IGF-1 mRNA, hIGF-1-F (ATGGGAAAAATCAGCAGTCT) and hIGF-1-R (CATCCTGTAGTTCTTGTTTCC) were used. For checking murine IGF-1 mRNA, mIGF-1-F (CATGTCGTCTTCACACCTCTT) and mIGF-1-R (CTTGTGTTCTTCAAATGTA) were used. For checking human IGF-II mRNA, hIGF-II-F (ATGGGAATCCCAATGGGGAA) and hIGF-II-R (CTCGGACTTGGCGGGGGTA) were used. For checking murine IGF-II mRNA, mIGF-II-1255-F (CGCTTAGTTTGTCTGTTCG) and mIGF-II-1597-R (CGTTTGGCCTCTCTGAACTC) were used.
Antibodies and immunoprecipitation assay
Antibodies used included anti-GPC3 (1G12, BioMosaics, Burlington, VT), anti-GFP (Clontech), anti-tubulin, anti-actin (Sigma–Aldrich, St Louis, MO), anti-phosphotyrosine (PY20) (Upstate Cell Signaling Solutions, Lake Placid, NY), anti-IGF-1Rβ, anti-phospho-ERK1/2, anti-phospho-IGF-1Rβ (Tyr1135/1136) (Cell Signaling Technology, Danvers, MA), anti-ERK1/2 and anti-glutathione S-transferase (Santa Cruz Biotechnology, Santa Cruz, CA). The anti-CC3 antibody was raised in rabbit with a peptide corresponding to residues 291–301 of the GPC3 protein. To determine IGF-1R phosphorylation, 600 μg of cell extract was immunoprecipitated with anti-IGF-1Rβ and then blotted with PY20 or blotted with anti-phospho-IGF-1Rβ.
Intact cell IGF-1R phosphorylation
Subconfluent cells in 24-well plates were serum starved in serum-free medium for 24 h and then incubated either with or without IGF-II (20 ng/ml) for 5, 15, 30 or 60 min at 37°C. The cells were washed rapidly with ice-cold phosphate-buffered saline then lysed in the lysis buffer (20 mM HEPES N-(Hydroxyethyl)piperazine-N'-(2-ethanesulfonic acid), pH 7.5, NP40 1%, NaCl 150 mM, NaF 10 mM, glycerol 10%, Na4P2O7 10 mM, sodium vanadate 0.1 mM, β-glycerol phosphate 2 mM and protease inhibitor cocktail). The amount of phospho-IGF-1 receptor was determined by immunoblotting with anti-phospho-IGF-1Rβ and then reprobed by anti-IGF-1Rβ.
Heparitinase digestion
Cell extract was first immunoprecipitated with anti-CC3. The precipitates were washed for three times with digestion buffer [50 mM HEPES, pH 7.0, 100 mM NaCl, 1 mM CaCl2, 50 μg human albumin, 2 mM phenylmethylsulfonyl fluoride, 5 mM ethylenediaminetetraacetic acid and protease inhibitor cocktail (Roche, Basel, Switzerland)] and treated with 2.5 mU of heparitinase (Seikagaku, Tokyo, Japan) for 2.5 h at 37°C. Western blot analysis was then done using 1G12.
RNA interference
For the gpc3 RNA interference, HuH-7 cells were transfected with 1 μg of gpc3 small hairpin RNA (shRNA) (TGCTGTTGACAGTGAGCGACCCAACATGCTGCTCAAGAAATAGTGAAGCCACAGATGTATTTCTTGAGCAGCATGTTGGGCTGCCTACTGCCTCGGA) or an enhanced green fluorescent proteins shRNA (control) in pSM2c vector (purchased from Open Biosystems, Huntsville, AL). For IGF-II RNA interference, chemically synthesized, double-stranded small interfering RNA (siRNA)s, with 19 nt duplex RNA and 2 nt dTdT overhangs were purchased from Ambion. The sequence was sense primer, GGUUCCAUCCCGAAAAUCUtt and antisense primer, AGAUUUUCGGGAUGGAACCtg. The action of IGF-II siRNA was first proved in HEK293 cells. The effect of IGF-II knockdown was done by experiments of HuH-7 cell transfection.
Determination of cell growth and colony formation
NIH3T3 cells (1 × 104) or PLC-PRF-5 stable cells were plated in 35 mm dishes or 12-well plates in regular medium. On the next day, the cultures were changed to serum-free medium. Viable cells were counted at 48 h intervals. shRNA to gpc3 was performed in HuH-7. HuH-7 cells (1 × 105) were transfected and plated in six-well plates and then selected with 1 μg/ml puromycin for 14 days. Colonies were visualized by Giemsa stain in HuH-7. For IGF-II interference, chemically synthesized double-stranded siRNA to IGF-II were transfected into 4 × 104 HuH-7 cells according to the manufacturer's instruction. On the next day, the cultures were changed to serum-free medium. Cells were counted at 48 h intervals for 6 days.
Soft agar assay
Anchorage-independent growth assay was done on three-layer soft agar in six-well flat-bottomed plates for 30 days. Colonies were then stained with 0.05% p-iodonitrotetrazolium violet and counted by inverted microscopy.
Glutathione S-transferase pull-down assay
The IGF-II complementary DNA, cloned by primers IGF-II-F (CGGGATCCATGGGAATCCCAATGGGGAA) and IGF-II-ls-R (GGAATTCCTTCCGATTGCTGGCCATCTCT), was inserted into pGEX4T1 vector for the production of GST-fusion protein. GST-IGF-II was expressed in Escherichia coli and purified. For the pull-down assay, cell extract from pcDNA-gpc3-transfected HEK293T cells was prepared in lysis buffer (10 mM Tris–HCl, pH7.4, 100 mM NaCl and 2.5 mM MgCl2) with 0.1% Triton X-100 and then incubated with GST-IGF-II at 4°C for 1 h. Proteins bound to the GST beads were analyzed by western blot.
Coimmunoprecipitation detection of the GPC3–IGF-1R complex
HEK293T cells were transfected with pcDNA-gpc3. The transfected cells were lysed in HNTG buffer (20mM Hepes, 100mM sodium chloride, 0.05% triton X-100, 10% glycerol) (35) and precleaned. Immunoprecipitation (IP) was done with anti-CC3, and western blot analysis was done with either anti-IGF-1Rβ or 1G12.
Results
Establishment of GPC3-expressing NIH3T3 cell lines
NIH3T3 cells do not have intrinsic GPC3 expression and therefore we expressed GPC3 in these cells to see if GPC3 could confer additional phenotypes (36). For the detection of GPC3 and to trace its posttranslational modification, a GFP tag was added to the C-terminus of GPC3 (GPC3-GFP). GPC3 is a glycosyl-phosphatidylinositol-linked heparan sulfate proteoglycan, and its C-terminal signal sequence will be removed when glycosyl-phosphatidylinositol is added, resulting in a 65 kDa core protein which will be glycanated. The 65 kDa core protein can be further cleaved by a furin-like convertase into a 40 kDa protein.
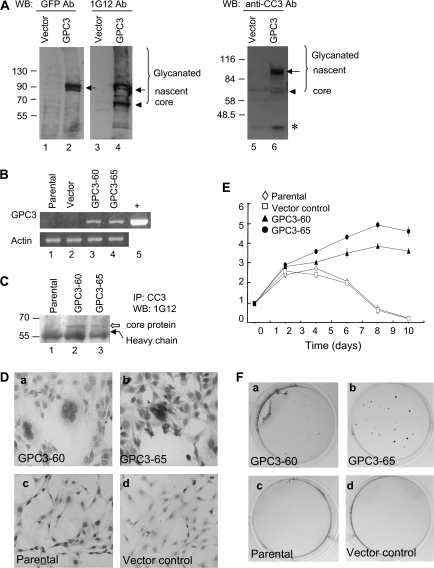
After transient transfection with pgpc3-GFP, nascent GPC3 could be detected by anti-GFP, 1G12, and anti-CC3 (Figure 1A, lanes 2, 4 and 6, arrow), whereas the 65 kDa core protein (arrowhead) and the glycanated forms (bracket) could be detected by 1G12 and anti-CC3 (lanes 4 and 6). The 40 kDa protein (asterisk) could be detected only by anti-CC3 (lane 6) (14). Membrane anchoring of the expressed GPC3 protein was also evident (supplementary Figure, panel B is available at Carcinogenesis Online). Therefore, the GFP tag did not interfere with the C-terminal processing of GPC3. After selection with G418, two stable clones GPC3-60 and GPC3-65 were isolated. gpc3 mRNA was expressed in these positive clones (Figure 1B), and the expression of GPC3 protein could be detected by IP and heparitinase digestion (Figure 1C). Since pEGFP-N1 is toxic to the cells, cells transfected with pcDNA3.1 (+) myc-his were used as vector control (37).
Fig. 1.
GPC3 promotes cell growth in NIH3T3 cells. (A) Transient expression of GPC3 in HEK293T cells. Cells were transfected with pgpc3-GFP, and 48 h later, cytosolic extracts were analyzed by western blot using anti-GFP (lanes 1 and 2), 1G12 (lanes 3 and 4) or anti-CC3 (lanes 5 and 6). Molecular weight markers are indicated on the left. The bracket indicates glycanated GPC3 proteins; the arrow indicates the nascent GPC3 protein; the arrowhead indicates the 65 kDa core protein and the asterisk indicates the 40 kDa convertase-cleaved fragment. (B) Reverse transcription–polymerase chain reaction analysis of gpc3 mRNA in stable lines. Lane 1, the parental NIH3T3 cells; lane 2, vector control cells; lane 3, GPC3-60 cells; lane 4, GPC3-65 cells and lane 5, positive control (+). Mouse actin was served as the RNA loading control. (C) Expression of GPC3 protein in stable lines. Six hundred micrograms of cytosolic extracts were immunoprecipitated with anti-CC3, digested by heparitinase and then blotted with 1G12. Lane 1, the parental cells; lane 2, GPC3-60 cells and lane 3, GPC3-65 cells. The open arrow indicates the GPC3 core protein, and the black arrow indicates the antibody heavy chain. (D) Morphology of the cells. GPC3-60 and GPC3-65 cells revealed increased nucleus-to-cytoplasm (N/C) ratio and multinucleation in Papanicolaou stain (a and b). Parental and vector control cells were shown as a comparison (c and d). (E) Growth rate of the GPC3-expressing cells in serum-free medium. Cells were seeded in 35 mm plates in triplicate and cultured in Dulbecco's modified Eagle's medium without serum. Cells were harvested at 48 h intervals and were counted in triplicates. (F) Colony formation in soft agar was observed for GPC3-expressing line GPC3-60 and GPC3-65 (a and b), but not for the parental or vector control cells (c and d).
Oncogenecity of the GPC3-expressing NIH3T3 cells
Compared with the parental NIH3T3 cells, GPC3-60 and GPC3-65 cells exhibited large nuclei with frequent multinucleation (Figure 1Da and b), but grew at a normal speed in regular medium. However, when cells were grown in serum-free medium, these GPC3-expressing cells kept on proliferating while the control cells died gradually (Figure 1E). Both GPC3-60 and GPC3-65 cells formed colonies on soft agar after 30 days of culture (Figure 1F), but the parental NIH3T3 cells or the vector control cells could not (Table I). Therefore, NIH3T3 cells gain oncogenecity after the expression of GPC3.
Table I.
Anchorage-independent growth of GPC3-expressing NIH3T3 cells
| Lines | Number of colonies per platea | Sizes of colonies (μm) |
| GPC3-60 | 46 ± 9b | ≥40 |
| GPC3-65 | 55 ± 6b | ≥100 |
| NIH3T3 | 0 | — |
| Vector control | 0 | — |
Colonies in soft agar were counted directly using an inverted microscope on day 30.
P < 0.0001 versus the parental and control cells, t-test.
Role of GPC3 in HCC cell oncogenecity
To explore the role of GPC3 in HCC cell oncogenecity, we first overexpressed GPC3 in GPC3-low-expressing PLC-PRF-5 cells (Figure 2A). After gaining the expression of GPC3, PLC-PRF-5 cells grew faster than the parental cells in serum-free medium (Figure 2B). On the other way, we knocked down GPC3 with shRNA in GPC3-high-expressing HuH-7 cells (Figure 2C). The results revealed that HuH-7 cells transfected with gpc3 shRNA showed poorer colony formation than cells transfected with the control shRNA (Figure 2D). These results indicate that the expression of GPC3 increases the oncogenecity of HCC cells.
Fig. 2.
Role of GPC3 in HCC cell oncogenecity. (A) GPC3 expression in PLC-PRF-5 cells. Mixed cultures of stable cell line were obtained by transfecting pgpc3-GFP or vector control into PLC-PRF-5 cells. Cell lysates were analyzed by western blot with 1G12. The bracket indicates glycanated GPC3 proteins; the arrow indicates the nascent GPC3 protein and the arrowhead indicates the 65 kDa core protein. (B) Growth rate of the PLC-PRF-5 stable cells. Cells (1 × 104) were seeded in 12-well plates in triplicate and cultured in Dulbecco's modified Eagle's medium without serum. Cells were harvested at 48 h intervals and were counted in triplicates. (C) GPC3 knockdown in HuH-7 cells. Cells were transfected with gpc3 shRNA or control vector (shRNA-EGFP), and 48 h later, cell extracts were analyzed by western blot with 1G12. The bracket indicates glycanated GPC3 proteins, and the arrowhead indicates the 65 kDa core protein. (D) Colony-forming assays for HuH-7 cells. Cells were transfected with either shRNA-EGFP (control) or shRNA-GPC3 cells.
GPC3 is associated with IGF-II and IGF-1R through its proline-rich region
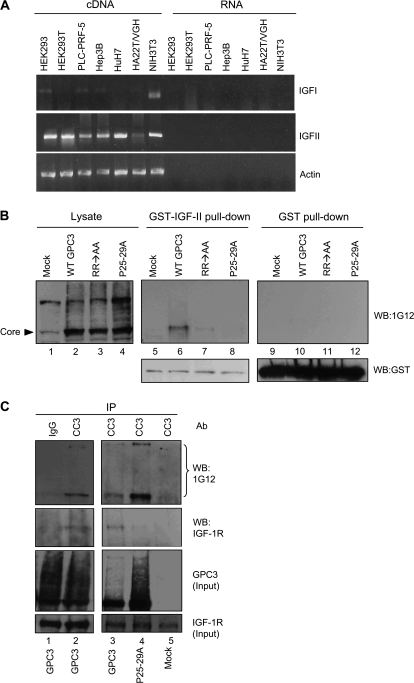
It has been reported that GPC3 binds to IGF-II (19,20). Both NIH3T3 cells and hepatoma cell lines HuH-7 have been shown to produce IGF-II (38,39). Using reverse transcription–polymerase chain reaction method, we found that IGF-II expression was high in cell lines including NIH3T3 and HuH-7 and was low in the HA22T/VGH cells (Figure 3A). Therefore, we tested the interactions between GPC3, IGF-II and IGF-1R (Insulin-like growth factor 1 receptor, one kind of receptor for IGF-II). In GST pull-down assay, GPC3 expressed in HEK293T cells could be pulled down by GST-IGF-II (Figure 3B, lane 6). GPC3 contains a proline-rich region located at residues 25–30, which might be important for protein–protein interaction. GPC3 mutated in these prolines (P25-29A), when expressed in cells, could be localized to the cell surface (supplementary Figure, panel D is available at Carcinogenesis Online) and glycanated (Figure 3B, lane 4). However, P25-29A did not possess the ability to bind IGF-II (Figure 3B, lane 8). We next tested the requirement of convertase digestion for GPC3 interaction with IGF-II. The convertase-resistant mutant RR → AA revealed cell membrane anchorage (supplementary Figure, panel C is available at Carcinogenesis Online) and glycanation (Figure 3B, lane 3) (34), but had very low affinity to GST-IGF-II (Figure 3B, lane 7). Therefore, convertase digestion is required for GPC3 to interact with IGF-II.
Fig. 3.
GPC3 interacts with IGF-II and IGF-1R. (A) Reverse transcription–polymerase chain reaction analysis of IGF-I and IGF-II gene expressions in different cell lines. Total RNA was isolated from HEK293, HEK293T, PLC-PRF-5, HuH-7, Hep3B, HA22T/VGH and NIH3T3 cells and complementary DNA was synthesized using reverse transcriptase. As a negative control, the same RNAs were incubated in the absence of enzyme in the reverse transcription reaction. Polymerase chain reaction was then done with specific primers. Actin was served as the RNA loading control. (B) Interaction of overexpressed GPC3 proteins with IGF-II. Wild-type GPC3 (WT-GPC3), RR → AA or P25-29A expression plasmid was transfected into HEK293T cells. After serum starvation for overnight, cells were harvested and subjected for GST pull-down assay with either GST-IGF-II or GST and then analyzed by western blot with 1G12. Lanes 1–4 represent 5% of cell extract used in the pull-down experiment (lysate). (C) Interaction between GPC3 and IGF-1R. HEK293T cells were transfected with either pcDNA-gpc3 or P25-29A. The cell lysates were then immunoprecipitated with anti-CC3 or IgG and blotted with anti-IGF-1R or 1G12. Five percent of cell extracts were analyzed in parallel (input). The bracket indicates glycanated GPC3.
The interaction between GPC3 and IGF-1R was investigated by co-IP. HEK293T cells were first transfected with pcDNA-gpc3 or P25-29A and serum starved. IP was performed with anti-CC3. The results revealed that IGF-1R could be brought down by anti-CC3 (Figure 3C), whereas P25-29A lost the ability to interact with IGF-1R (Figure 3C, lane 4). Therefore, these results indicate that GPC3 has specific interaction with both IGF-II and IGF-1R.
Phosphorylated IGF-1R interacts with GPC3
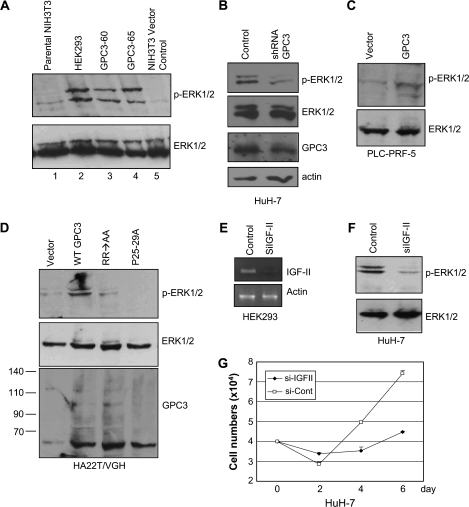
Phosphorylation of IGF-1R occurs upon ligation by growth factors. We then checked if IGF-1R phosphorylation could be activated by GPC3. In GPC3-expressing NIH3T3 cells, IGF-1R immunoprecipitated by anti-IGF-1Rβ could be detected by PY20 (Figure 4A). It was noted that GPC3 overexpression also upregulated the unphosphorylated IGF-1R. Since phosphorylated IGF-1R was present in HuH-7 cells, we checked the status of IGF-1R in these cells after GPC3 knockdown. The results revealed that both total and phosphorylated IGF-1R proteins decreased in amount when cells were treated with gpc3 shRNA (Figure 4B). The mechanism for the downregulation of total IGF-IR by GPC3 knockdown is not known, but this phenomenon suggests both the importance and the complexity of the regulation of the IGF-1R pathway by GPC3.
Fig. 4.
GPC3 enhances the IGF-II-triggered IGF-1R phosphorylation. (A) IGF-1R phosphorylation in GPC3-expressing NIH3T3 stable lines. Cells were serum starved and 600 μg of cell extracts were immunoprecipitated with anti-IGF-1Rβ, followed by western blot with PY20. IGF-1R was phosphorylated. Twenty micrograms of total cell extracts were analyzed with anti-tubulin as an equal loading control. (B) IGF-1R phosphorylation in HuH-7 cells. Cells were transfected with control or gpc3 shRNA and serum starved. Whole-cell extracts were then subjected to immunoblot with anti-phospho-IGF-1R and then reblotted with anti-IGF-1Rβ. Phosphorylated IGF-IR was decreased by gpc3 shRNA in HuH-7 cells. (C) IGF-II-triggered IGF-1R phosphorylation in HEK293 cells. Either wild-type GPC3 (WT-GPC3), RR → AA or P25-29A was expressed in HEK293 cells. Transiently transfected cells were serum starved and then treated with IGF-II (20 ng/ml) for 5, 15, 30 or 60 min at 37°C, followed by western blot probed with anti-phospho-IGF-1Rβ and reprobed with anti-IGF-1R. GPC3 expression was shown in parallel.
We next looked for evidence if GPC3 enhances IGF-II-mediated IGF-1R activation. We transfected GPC3 or its mutants in HEK293 cells and then treated the cells with IGF-II. IGF-II induced IGF-1R phosphorylation, which peaked at the 30th min in control cells, and the phosphorylation declined thereafter (Figure 4C, vector). In pcDNA-gpc3-transfected cells, IGF-II triggered a faster and stronger phosphorylation of IGF-1R and the phosphorylation also sustained longer. RR → AA or P25-29A either demonstrated a later (RR → AA) and/or weaker (RR → AA and P25-29A) responses (Figure 4C). Therefore, GPC3 activates the IGF-signaling pathway in a specific way.
Phosphorylation of ERK by GPC3
Lastly, we checked if GPC3 could activate the downstream mitogenic pathway. In western blot analysis, the levels of total ERK in the parental NIH3T3 cells, 12-O-tetradecanoylphorbol 13-acetate treatment HEK293 cells or GPC3 expression NIH3T3 cells were equal (Figure 5A). 12-O-tetradecanoylphorbol 13-acetate induced prominent ERK phosphorylation in HEK293 cells (Figure 5A, lane 2). Phospho-ERK was barely visible in the parental NIH3T3 cells, but was abundant in GPC3-expressing cell lines GPC3-60 and GPC3-65 (Figure 5A, lanes 3 and 4). In the GPC3-high-expressing HuH-7 cells, ERK phosphorylation was high, but phospho-ERK decreased when GPC3 was knocked down by gpc3 shRNA (Figure 5B). In the GPC3-low-expressing PLC-PRF-5 and HA22T/VGH cells, stable expression of GPC3 caused the elevation of phospho-ERK, but mutants RR → AA or P25-29A had much less effects (Figure 5C and D). Serum starvation for the HA22T/VGH cells was done in 0.5% serum because in 0% serum no ERK phosphorylation could be seen. This could be due to the low IGF-II expression in HA22T/VGH cells (Figure 3A).
Fig. 5.
GPC3 activates ERK phosphorylation. (A) ERK phosphorylation in GPC3-expressing NIH3T3 stable lines. Cells were serum starved for 24 h. Forty micrograms of cell extracts were analyzed by western blot with either anti-phospho-ERK or anti-ERK antibody. Whole-cell extracts from 12-O-tetradecanoylphorbol 13-acetate-treated HEK293 cells (lane 2) were used as a positive control. (B) ERK phosphorylation in GPC3-knocked-down HuH-7 cells. shRNA was transiently transfected into HuH-7 cells. After serum starvation, 35 μg cell extracts were subjected for western blot analysis with anti-phospho-ERK, anti-ERK, 1G12 or anti-actin. ERK phosphorylation decreased after GPC3 knockdown. (C) ERK phosphorylation in PLC-PRF-5 cells. Cells were stably transfected with p gpc3-GFP (GPC3) or control vector (vector) and serum starved for 24 h. Cell extracts were analyzed with either anti-phospho-ERK or anti-ERK. (D) ERK phosphorylation in HA22T/VGH cells. Cells were stably transfected with pcDNA-gpc3 [wild-type GPC3 (WT-GPC3)], RR → AA, P25-29A or control vector (vector) and serum starved at 0.5% fetal calf serum for 24 h. Cell extracts were immunobloted with either anti-phospho-ERK, anti-ERK or 1G12. (E) IGF-II knockdown by siRNA (siIGF-II). Chemically synthesized, double-stranded siRNAs for IGF-II and control were transfected into HEK293 cells. Total RNA was extracted and analyzed by reverse transcription–polymerase chain reaction. Specific primers for IGF-II were used in polymerase chain reaction and actin was served as the RNA loading control. (F) ERK phosphorylation was decreased by IGF-II knockdown. HuH-7 cells were transfected with either control or IGF-II siRNA and serum starved. Cell extracts were analyzed with either anti-phospho-ERK or anti-ERK. (G) Growth rate of HuH-7 cells after IGF-II knockdown. Cells were seeded in 12-well plates in triplicate, transfected with either control or IGF-II siRNA and were serum starved. Cells were harvested at 48 h intervals.
We further demonstrated that ERK phosphorylation was dependent on the presence of IGF-II. We first selected a proper IGF-II siRNA by transfecting HEK293 cells (Figure 5E). In HuH-7 cells treated with IGF-II siRNA, we could demonstrate that the level of phospho-ERK decreased (Figure 5F) and those treated cells revealed a decreased growth speed in serum-free medium (Figure 5G). Therefore, the activation of intracellular mitogenic pathway requires the presence of both IGF-II and GPC3.
Discussion
In this article, we demonstrate that GPC3 can bind to both IGF-II and IGF-1R through its proline-rich region. GPC3 binding to IGF-II activates IGF-1R and then triggers a phosphorylation cascade including IGF-1R itself and ERK. These processes confer oncogenecity to NIH3T3 and hepatoma cells PLC-PRF-5, whereas the removal of GPC3 decreases the oncogenecity of HuH-7 cell lines. The pathogenesis of HCC has been known to involve p53 (40), β-catenin (41), TGF-β (42) and the retinoblastoma gene (43). p53 mutation occurs in one-third of HCC (40,44); β-catenin mutation is found in 13.1% of HCC (41) and the activation of canonical Wnt-signaling pathway happens in 18% of HCC (45). The involvement of GPC3 in HCC is more recently recognized that overexpression of GPC3 can be found in 70% of HCC (4). And now in this study, we discover the mechanisms that led to GPC3-mediated oncogenesis.
It is very interesting that GPC3 interacts with both IGF-II and its receptor IGF-1R.This may suggests that GPC3 joins a multiprotein complex, which is composed of the ligand, receptor, GPC3, and probably other proteins. The interaction between the proteins in the complex probably involves heparan sulfate. However, here we showed that specific protein–protein interaction through the proline-rich region of the GPC3 protein is also required. Protein–protein interaction through the multiproline residues has been shown for the histidine proline-rich glycoprotein (46). Histidine proline-rich glycoprotein is an abundant plasma protein. It binds with high affinity to FGF-2-stimulated human umbilical vein endothelial cells and immobilizes tropomyosin via the histidine proline-rich domain (46). Moreover, proper processing of GPC3 is required for its function. In the current study, convertase digestion is necessary for GPC3 to activate the IGF-signaling pathway. In a previous study, convertase is required for GPC3-mediated regulation of the Wnt-signaling pathway (34). All these data suggest a specific protein–protein interaction for GPC3 function, besides the non-specific interaction through heparan sulfate.
The activation of IGF-1R and the downstream signaling pathway well explains the role of GPC3 in oncogenesis. The IGF-signaling pathway plays a pivotal role in cell growth. Overexpressions of IGF-II, IGF-1R and insulin receptor substrate-1 have been described in human cancers (30,47–50). ERK is known to mediate IGF-II-induced gene expression, cell invasion and apoptosis protection (51,52). ERK has also been shown to contribute to the multistep hepatocarcinogenesis (53). Although GPC3 knockout mice showed no significant change of IGF-1R or insulin receptor substrate-1 in whole embryo extracts (54), cell growth in GPC3-transgenic mice is promoted in the liver (5). Therefore, GPC3 may have a more prominent role in liver cancers like HCC.
In this study, we not only demonstrate the activation of the IGF-signaling pathway by GPC3 but also more importantly give a direct evidence for GPC3-mediated oncogenesis. GPC3 confers NIH3T3 cells a full cancer cell phenotype including the ability to grow in serum-free medium and to form colonies in soft agar. It is also striking to see the loss of oncogenecity by knocking down GPC3 by shRNA in HCC cells. Therefore, GPC3 could be a new target in cancer therapy in the future.
Supplementary material
Supplementary Figure can be found at http://carcin.oxfordjournals.org/
Funding
Academia Sinica and National Science Council of the Republic of China, Taiwan (NSC-90-2311-B-001-199 to Y.M.L., NSC94-3112-B-002-018 to H.C.H.); National Health Research Institute (NHRI-GT-270 EX89B701L) to H.C.H.; Department of Health, Taiwan (9001, 92101 and 93006) to W.C.
Supplementary Material
Acknowledgments
We thank Dr Wuh-Liang Hwu for critical reading of the manuscript and Mr Sui-Tsun Chen, Ken-Fen Lu and Ms Li-Ping Hsiao for valuable technical assistance.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- ERK
extracellular signal-regulated kinase
- GFP
green fluorescence protein
- GPC3
glypican-3
- GST
glutathione S-transferase
- HCC
hepatocellular carcinoma
- IGF
insulin-like growth factor
- shRNA
small hairpin RNA
- siRNA
small interfering RNA
- mRNA
messenger RNA
References
- 1.Befeler AS, et al. Hepatocellular carcinoma: diagnosis and treatment. Gastroenterology. 2002;122:1609–1619. doi: 10.1053/gast.2002.33411. [DOI] [PubMed] [Google Scholar]
- 2.Lodato F, et al. Hepatocellular carcinoma prevention: a worldwide emergence between the opulence of developed countries and the economic constraints of developing nations. World J. Gastroenterol. 2006;12:7239–7249. doi: 10.3748/wjg.v12.i45.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thorgeirsson SS, et al. Molecular pathogenesis of human hepatocellular carcinoma. Nat. Genet. 2002;31:339–346. doi: 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- 4.Hsu HC, et al. Cloning and expression of a developmentally regulated transcript MXR7 in hepatocellular carcinoma: biological significance and temporospatial distribution. Cancer Res. 1997;57:5179–5184. [PubMed] [Google Scholar]
- 5.Capurro MI, et al. Glypican-3 promotes the growth of hepatocellular carcinoma by stimulating canonical Wnt signaling. Cancer Res. 2005;65:6245–6254. doi: 10.1158/0008-5472.CAN-04-4244. [DOI] [PubMed] [Google Scholar]
- 6.Lage H, et al. Expression of the novel mitoxantrone resistance associated gene MXR7 in colorectal malignancies. Int. J. Clin. Pharmacol. Ther. 1998;36:58–60. [PubMed] [Google Scholar]
- 7.Hishinuma M, et al. Hepatocellular oncofetal protein, glypican 3 is a sensitive marker for alpha-fetoprotein-producing gastric carcinoma. Histopathology. 2006;49:479–486. doi: 10.1111/j.1365-2559.2006.02522.x. [DOI] [PubMed] [Google Scholar]
- 8.Toretsky JA, et al. Glypican-3 expression in Wilms tumor and hepatoblastoma. J. Pediatr. Hematol. Oncol. 2001;23:496–499. doi: 10.1097/00043426-200111000-00006. [DOI] [PubMed] [Google Scholar]
- 9.Nakatsura T, et al. Identification of glypican-3 as a novel tumor marker for melanoma. Clin. Cancer Res. 2004;10:6612–6621. doi: 10.1158/1078-0432.CCR-04-0348. [DOI] [PubMed] [Google Scholar]
- 10.Zynger DL, et al. Glypican 3: a novel marker in testicular germ cell tumors. Am. J. Surg. Pathol. 2006;30:1570–1575. doi: 10.1097/01.pas.0000213322.89670.48. [DOI] [PubMed] [Google Scholar]
- 11.Stadlmann S, et al. Glypican-3 expression in primary and recurrent ovarian carcinomas. Int. J. Gynecol. Pathol. 2007;26:341–344. doi: 10.1097/pgp.0b013e31802d692c. [DOI] [PubMed] [Google Scholar]
- 12.Xiang YY, et al. Glypican-3 expression is silenced in human breast cancer. Oncogene. 2001;20:7408–7412. doi: 10.1038/sj.onc.1204925. [DOI] [PubMed] [Google Scholar]
- 13.Lin H, et al. Frequent silencing of the GPC3 gene in ovarian cancer cell lines. Cancer Res. 1999;59:807–810. [PubMed] [Google Scholar]
- 14.Filmus J, et al. Identification of a new membrane-bound heparan sulphate proteoglycan. Biochem. J. 1995;311:561–565. doi: 10.1042/bj3110561. (Pt 2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson RL, et al. Glycosaminoglycans: molecular properties, protein interactions, and role in physiological processes. Physiol. Rev. 1991;71:481–539. doi: 10.1152/physrev.1991.71.2.481. [DOI] [PubMed] [Google Scholar]
- 16.Lander AD. Targeting the glycosaminoglycan-binding sites on proteins. Chem. Biol. 1994;1:73–78. doi: 10.1016/1074-5521(94)90043-4. [DOI] [PubMed] [Google Scholar]
- 17.Lin X, et al. Heparan sulfate proteoglycans are essential for FGF receptor signaling during Drosophila embryonic development. Development. 1999;126:3715–3723. doi: 10.1242/dev.126.17.3715. [DOI] [PubMed] [Google Scholar]
- 18.LaRochelle WJ, et al. Heparan sulfate proteoglycan modulates keratinocyte growth factor signaling through interaction with both ligand and receptor. Biochemistry. 1999;38:1765–1771. doi: 10.1021/bi982092z. [DOI] [PubMed] [Google Scholar]
- 19.Pilia G, et al. Mutations in GPC3, a glypican gene, cause the Simpson-Golabi-Behmel overgrowth syndrome. Nat. Genet. 1996;12:241–247. doi: 10.1038/ng0396-241. [DOI] [PubMed] [Google Scholar]
- 20.Xu Y, et al. Developmental regulation of the soluble form of insulin-like growth factor-II/mannose 6-phosphate receptor in human serum and amniotic fluid. J. Clin. Endocrinol. Metab. 1998;83:437–442. doi: 10.1210/jcem.83.2.4537. [DOI] [PubMed] [Google Scholar]
- 21.Pietrzkowski Z, et al. Inhibition of growth of prostatic cancer cell lines by peptide analogues of insulin-like growth factor 1. Cancer Res. 1993;53:1102–1106. [PubMed] [Google Scholar]
- 22.Baserga R. Oncogenes and the strategy of growth factors. Cell. 1994;79:927–930. doi: 10.1016/0092-8674(94)90023-x. [DOI] [PubMed] [Google Scholar]
- 23.Resnicoff M, et al. The insulin-like growth factor I receptor protects tumor cells from apoptosis in vivo. Cancer Res. 1995;55:2463–2469. [PubMed] [Google Scholar]
- 24.Kaleko M, et al. Overexpression of the human insulinlike growth factor I receptor promotes ligand-dependent neoplastic transformation. Mol. Cell. Biol. 1990;10:464–473. doi: 10.1128/mcb.10.2.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Resnicoff M, et al. Rat glioblastoma cells expressing an antisense RNA to the insulin-like growth factor-1 (IGF-1) receptor are nontumorigenic and induce regression of wild-type tumors. Cancer Res. 1994;54:2218–2222. [PubMed] [Google Scholar]
- 26.Scharf JG, et al. The IGF axis and hepatocarcinogenesis. Mol. Pathol. 2001;54:138–144. doi: 10.1136/mp.54.3.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cariani E, et al. Differential expression of insulin-like growth factor II mRNA in human primary liver cancers, benign liver tumors, and liver cirrhosis. Cancer Res. 1988;48:6844–6849. [PubMed] [Google Scholar]
- 28.Nardone G, et al. Activation of fetal promoters of insulinlike growth factors II gene in hepatitis C virus-related chronic hepatitis, cirrhosis, and hepatocellular carcinoma. Hepatology. 1996;23:1304–1312. doi: 10.1002/hep.510230602. [DOI] [PubMed] [Google Scholar]
- 29.Scharf JG, et al. Analysis of the IGF axis in preneoplastic hepatic foci and hepatocellular neoplasms developing after low-number pancreatic islet transplantation into the livers of streptozotocin diabetic rats. Lab. Invest. 2000;80:1399–1411. doi: 10.1038/labinvest.3780147. [DOI] [PubMed] [Google Scholar]
- 30.Tanaka S, et al. Biological effects of human insulin receptor substrate-1 overexpression in hepatocytes. Hepatology. 1997;26:598–604. doi: 10.1002/hep.510260310. [DOI] [PubMed] [Google Scholar]
- 31.Nishiyama M, et al. Cloning and increased expression of an insulin receptor substrate-1-like gene in human hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 1992;183:280–285. doi: 10.1016/0006-291x(92)91640-c. [DOI] [PubMed] [Google Scholar]
- 32.Lin YM, et al. [A new human hepatoma cell line: establishment and characterization] Zhonghua Min Guo Wei Sheng Wu Ji Mian Yi Xue Za Zhi. 1982;15:193–201. [PubMed] [Google Scholar]
- 33.Nakabayashi H, et al. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 1982;42:3858–3863. [PubMed] [Google Scholar]
- 34.De Cat B, et al. Processing by proprotein convertases is required for glypican-3 modulation of cell survival, Wnt signaling, and gastrulation movements. J. Cell Biol. 2003;163:625–635. doi: 10.1083/jcb.200302152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vecchione A, et al. The Grb10/Nedd4 complex regulates ligand-induced ubiquitination and stability of the insulin-like growth factor I receptor. Mol. Cell. Biol. 2003;23:3363–3372. doi: 10.1128/MCB.23.9.3363-3372.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruddon RW. Cancer Biology. Oxford University Press, Inc; 1995. [Google Scholar]
- 37.Liu HS, et al. Is green fluorescent protein toxic to the living cells? Biochem. Biophys. Res. Commun. 1999;260:712–717. doi: 10.1006/bbrc.1999.0954. [DOI] [PubMed] [Google Scholar]
- 38.Arbuthnot PB, et al. In vitro and in vivo hepatoma cell-specific expression of a gene transferred with an adenoviral vector. Hum. Gene Ther. 1996;7:1503–1514. doi: 10.1089/hum.1996.7.13-1503. [DOI] [PubMed] [Google Scholar]
- 39.Abdallah BM. Osteoblast differentiation of NIH3T3 fibroblasts is associated with changes in the IGF-I/IGFBP expression pattern. Cell. Mol. Biol. Lett. 2006;11:461–474. doi: 10.2478/s11658-006-0038-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsu HC, et al. Expression of p53 gene in 184 unifocal hepatocellular carcinomas: association with tumor growth and invasiveness. Cancer Res. 1993;53:4691–4694. [PubMed] [Google Scholar]
- 41.Hsu HC, et al. Beta-catenin mutations are associated with a subset of low-stage hepatocellular carcinoma negative for hepatitis B virus and with favorable prognosis. Am. J. Pathol. 2000;157:763–770. doi: 10.1016/s0002-9440(10)64590-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kitisin K, et al. Disruption of transforming growth factor-beta signaling through beta-spectrin ELF leads to hepatocellular cancer through cyclin D1 activation. Oncogene. 2007;26:7103–7110. doi: 10.1038/sj.onc.1210513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang X, et al. Deletions of chromosome 13q, mutations in Retinoblastoma 1, and retinoblastoma protein state in human hepatocellular carcinoma. Cancer Res. 1994;54:4177–4182. [PubMed] [Google Scholar]
- 44.Hsu HC, et al. Mutations of p53 gene in hepatocellular carcinoma (HCC) correlate with tumor progression and patient prognosis: a study of 138 patients with unifocal HCC. Int. J. Oncol. 1994;4:1341–1347. doi: 10.3892/ijo.4.6.1341. [DOI] [PubMed] [Google Scholar]
- 45.Giles RH, et al. Caught up in a Wnt storm: Wnt signaling in cancer. Biochim. Biophys. Acta. 2003;1653:1–24. doi: 10.1016/s0304-419x(03)00005-2. [DOI] [PubMed] [Google Scholar]
- 46.Guan X, et al. Histidine-proline rich glycoprotein (HPRG) binds and transduces anti-angiogenic signals through cell surface tropomyosin on endothelial cells. Thromb. Haemost. 2004;92:403–412. doi: 10.1160/TH04-02-0073. [DOI] [PubMed] [Google Scholar]
- 47.Caro JF, et al. Insulin-like growth factor I binding in hepatocytes from human liver, human hepatoma, and normal, regenerating, and fetal rat liver. J. Clin. Invest. 1988;81:976–981. doi: 10.1172/JCI113451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scharf JG, et al. Characterization of the insulin-like growth factor axis in a human hepatoma cell line (PLC) Carcinogenesis. 1998;19:2121–2128. doi: 10.1093/carcin/19.12.2121. [DOI] [PubMed] [Google Scholar]
- 49.Su Q, et al. Expression of insulin-like growth factor II in hepatitis B, cirrhosis and hepatocellular carcinoma: its relationship with hepatitis B virus antigen expression. Hepatology. 1994;20:788–799. doi: 10.1002/hep.1840200404. [DOI] [PubMed] [Google Scholar]
- 50.Werner H, et al. The role of the insulin-like growth factor system in human cancer. Adv. Cancer Res. 1996;68:183–223. doi: 10.1016/s0065-230x(08)60354-1. [DOI] [PubMed] [Google Scholar]
- 51.Kim HJ, et al. IGF-II-mediated COX-2 gene expression in human keratinocytes through extracellular signal-regulated kinase pathway. J. Invest. Dermatol. 2004;123:547–555. doi: 10.1111/j.0022-202X.2004.23317.x. [DOI] [PubMed] [Google Scholar]
- 52.Sciacca L, et al. In IGF-I receptor-deficient leiomyosarcoma cells autocrine IGF-II induces cell invasion and protection from apoptosis via the insulin receptor isoform A. Oncogene. 2002;21:8240–8250. doi: 10.1038/sj.onc.1206058. [DOI] [PubMed] [Google Scholar]
- 53.Ito Y, et al. Activation of mitogen-activated protein kinases/extracellular signal-regulated kinases in human hepatocellular carcinoma. Hepatology. 1998;27:951–958. doi: 10.1002/hep.510270409. [DOI] [PubMed] [Google Scholar]
- 54.Song HH, et al. The loss of glypican-3 induces alterations in Wnt signaling. J. Biol. Chem. 2005;280:2116–2125. doi: 10.1074/jbc.M410090200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.