Abstract
Hydrogen peroxide (H2O2) plays a dual role in plants as the toxic by-product of normal cell metabolism and as a regulatory molecule in stress perception and signal transduction. However, a clear inventory as to how this dual function is regulated in plants is far from complete. In particular, how plants maintain survival under oxidative stress via adjustments of the intercellular metabolic network and antioxidative system is largely unknown. To investigate the responses of rice seedlings to H2O2 stress, changes in protein expression were analyzed using a comparative proteomics approach. Treatments with different concentrations of H2O2 for 6 h on 12-day-old rice seedlings resulted in several stressful phenotypes such as rolling leaves, decreased photosynthetic and photorespiratory rates, and elevated H2O2 accumulation. Analysis of ∼2000 protein spots on each two-dimensional electrophoresis gel revealed 144 differentially expressed proteins. Of them, 65 protein spots were up-regulated, and 79 were down-regulated under at least one of the H2O2 treatment concentrations. Furthermore 129 differentially expressed protein spots were identified by mass spectrometry to match 89 diverse protein species. These identified proteins are involved in different cellular responses and metabolic processes with obvious functional tendencies toward cell defense, redox homeostasis, signal transduction, protein synthesis and degradation, photosynthesis and photorespiration, and carbohydrate/energy metabolism, indicating a good correlation between oxidative stress-responsive proteins and leaf physiological changes. The abundance changes of these proteins, together with their putative functions and participation in physiological reactions, produce an oxidative stress-responsive network at the protein level in H2O2-treated rice seedling leaves. Such a protein network allows us to further understand the possible management strategy of cellular activities occurring in the H2O2-treated rice seedling leaves and provides new insights into oxidative stress responses in plants.
Hydrogen peroxide is one of the most abundant reactive oxygen species (ROS)1 in aerobic biological systems, including higher plants, and can be continuously produced through aerobic metabolic processes such as respiration and photosynthesis in mitochondria, chloroplasts, and peroxisomes (1). With both reducing and oxidizing properties, H2O2 has effects on almost all organisms and can influence the life of every single cell (1). On one hand, H2O2 is highly reactive and toxic and can lead to oxidative destruction of cells; on the other hand, it acts as a signaling molecule in regulating cell growth and development, cell proliferation, cell stress response, and signal transduction (2). When accumulated at high enough concentrations, H2O2 can directly or indirectly oxidize enough of the cellular ascorbic acid and glutathione pool to alter the overall redox state of the cells. Such high concentrations of H2O2 can also damage a large variety of biomolecules such as lipids, proteins, and nucleic acids that are essential for the activity and integrity of the cells (3). This situation is in fact oxidative stress or damage, which is of importance for relatively few events such as triggering programmed cell death in living cells (4). The other situation under which H2O2 can function is currently described as H2O2 signaling. Even at low concentrations, H2O2 possibly diffuses into cells and is perceived rapidly and specifically by a limited number of target proteins before it is scavenged by antioxidative defense mechanisms, and then target proteins transmit H2O2 signals to downstream signaling molecules such as Ca2+-binding proteins, G proteins, and phosphatidic acid, which together modulate different developmental, metabolic, and defense pathways in plants (2, 5–8). Because the amount of H2O2 required for this process is low, there would be no significant alteration of the overall redox state in the cells and thus no true oxidative stress. Given the dual function of H2O2 as both toxic by-product and crucial regulator, it is clear that the steady-state level of H2O2 as well as other ROS must be tightly controlled in plant cells.
Under steady-state conditions, the excessive H2O2 is efficiently scavenged by various antioxidative defense mechanisms in plant cells. The major ROS-scavenging enzymes include ascorbate peroxidase (APx), catalase (CAT), superoxide dismutase, glutathione peroxidase, and peroxiredoxin (2). Together with the antioxidants ascorbic acid and glutathione, these enzymes provide plant cells with highly efficient machinery for detoxifying H2O2 and other ROS (2). However, the equilibrium between production and scavenging of H2O2 may be perturbed by various abiotic and biotic stresses, leading to a rapid and transient increase of the intracellular H2O2 levels. Eventually plant tissues under such stresses often suffer from serious oxidative damages when the level of H2O2 reaches the threshold value that triggers the disorder of physiological processes and metabolic pathways in plant cells. In plants, the prolonged period of H2O2 generation that results in oxidative stress has been reported during external adverse stimuli such as chilling (9), drought (10), salinity (11), UV irradiation (12), ozone exposure (13), heavy metal (14), wounding (15), phytohormones including abscisic acid (6) and jasmonic acid (15), and elicitors and pathogen challenges (16). In almost all cases, H2O2 seem to be positively used by plants to activate some stress-responsive genes that help them to cope with environmental changes.
Global gene expression profiling experiments have revealed that a large number of genes in Arabidopsis and tobacco plants are responsive to oxidative stress (17–19). More than 170 non-redundant ESTs are found to be regulated by H2O2. Of these, 113 are induced and 62 are repressed under H2O2 stress. A substantial proportion of these ESTs have predicted functions in cell rescue and defense processes. RNA blot analysis of 14 selected genes demonstrates that other stresses such as wilting, UV irradiation, and elicitor challenge also induce the expression of most of these genes, indicating that H2O2 can mediate cross-tolerance toward other stresses (17). Furthermore another survey at the transcriptome level reveals that 349 transcripts are up-regulated and 88 are down-regulated by high light-induced H2O2 in CAT-deficient Arabidopsis plants (18). In this experiment, H2O2 is inferred to play a key role in regulating the transcriptions of two cluster genes that encode heat shock proteins (HSPs) and that participate in the anthocyanin regulatory and biosynthetic pathway, and several transcription factors and candidate regulatory genes responsive to H2O2 stress are also identified. Likewise 713 ESTs are found to be regulated by high light-induced H2O2 in CAT-deficient tobacco plants, and their transcriptional responses mimic those that have been reported during other abiotic and biotic stresses. Expression profiling corroborated by physiological experiments shows that a short term H2O2 exposure of the CAT-deficient plants can trigger an increased tolerance against a subsequent severely oxidative stress, also indicating that the cross-tolerance is mediated by H2O2 (19). Additionally by comparing the publically available transcriptome data sets of H2O2-stressed Arabidopsis, Mittler et al. (2) established a reactive oxygen gene network in plants. This network is composed of at least 152 genes involving ROS producing and scavenging such as APx, CAT, superoxide dismutase, glutathione peroxidase, peroxiredoxin, thioredoxins, glutaredoxin, monodehydroascorbate reductase, dehydroascorbate reductase, glutathione reductase, alternative oxidase, NADPH oxidase, and ferritin and is involved in managing the level of ROS (2). Although large scale transcriptome analysis has documented the transcriptional dynamics of a large number of antioxidative genes, it should be essential to utilize proteomics and even metabolomics strategies to gain the system-level understanding of plant responses to H2O2 stress.
In the present work, we initiated the first functional proteomics investigation of proteins that are responsive to elevated H2O2 in rice. Using two-dimensional electrophoresis (2-DE) in combination with MS/MS analysis, we characterized proteins whose expression was altered upon exposure to different concentrations of H2O2. Such an oxidative stress challenge resulted in a dramatic proteomic response involving at least 144 proteins. Identification of these proteins combined with their abundance changes as well as the impacts of oxidative stress on leaf photosynthesis reveals a close linkage between the changes in specific protein abundance and the overall defense response to oxidative stress, gives a global view of the ubiquitous cellular changes under oxidative stress, and demonstrates the first possibly intimate protein network elicited by H2O2 in rice seedling leaves. These results presented in this study provide the framework for further functional studies of each member of this network in intracellular redox homeostasis and H2O2 metabolism.
EXPERIMENTAL PROCEDURES
Chemicals—
CHAPS, IPG DryStrip, IPG buffer, and iodoacetamide were purchased from GE Healthcare; thiourea and n-octyl glucopyranoside were from Sigma; and trypsin (MS Gold), urea, and acrylamide were from Promega (Madison, WI). Deionized water (Millipore, Bedford, MA) with resistance greater than 18 megaohms cm was used throughout.
Plant Material and Stress Treatment—
Rice seeds (Oryza sativa L. cv. 93-11, whose genome was sequenced in Beijing Genomics Institute, China) were soaked in distilled water for 24 h and germinated in the dark for 45 h at 37 °C, and then the rice seedlings were grown in a growth chamber at 28/21 °C (16-h day/8-h night), photon flux density of 400 μmol m−2 s−1, and relative humidity of 70%. To provide whole nutrition to rice seedlings, Hogland solution was supplied every 2 days. 12-day-old seedlings were treated with three H2O2 concentrations (0.6, 3.0, and 15.0 mm) for 6 h in three plastic containers, respectively. The middle portions of the second and third leaves were collected and frozen in liquid nitrogen and then stored at −80 °C for protein extraction. Seedlings immersed in double distilled H2O were used as control.
Measurements of Photosynthesis and H2O2 Level—
Net photosynthetic rates, stomatal conductance, intercellular CO2 concentration, and transpiration speed of the second and third rice leaves of each sample were measured using a LI-COR 6400 portable gas analysis system with an light-emitting diode light source (LI-COR Inc., Lincoln, NE). At least nine leaves for each sample were measured.
H2O2 accumulated in treated or control leaves was measured according to Tiwari et al. (20) with modifications. Rice seedling leaves (1.0 g) were ground with a mortar and pestle in liquid nitrogen to a fine powder and added to a 10-ml cuvette containing 8 ml of double distilled H2O and 2 ml of 25 mm titanium sulfate and then incubated for 1 h at room temperature. Oxidation of titanium sulfate was recorded by reading A410. Readings were converted to corresponding concentrations using a standard calibration plot.
Protein Extraction—
Protein extraction was performed according to a method reported recently (21) with modifications. Rice leaves (0.5 g) were ground in liquid nitrogen to a fine powder, and the powder was suspended completely in 10 ml of 10% (w/v) TCA in acetone with 0.5% (w/v) DTT at −20 °C overnight. After centrifugation at 14,000 × g for 30 min at 4 °C, the pellets were washed three times with 10.0 ml of ice-cold acetone. The collected protein pellets were dried with N2 to remove any remaining acetone. The dried powder was resuspended completely in 2 ml of lysis buffer (9 m urea, 4% (w/v) CHAPS, and 2% ampholytes, pH 3–10). After incubation at 25 °C for 1.5 h, the suspension was centrifuged at 16,000 × g for 30 min at 25 °C to remove the insoluble material. The resulting supernatant with predominantly soluble proteins was reduced by adding 5 mm Tris-(β-carboxyethyl)-phosphine hydrochloride. The reduction continued for 1 h at room temperature. Samples were then alkylated by treatment with 16 mm iodoacetamide for 1.5 h at room temperature. This reaction was quenched by the addition of 50 mm DTT. The samples were immediately frozen in liquid nitrogen and then stored at −80 °C in aliquots. Protein contents were quantified according to Yao et al. (22) using BSA as a standard.
2-DE, Gel Staining, and Image Analysis—
First-dimensional electrophoresis was performed on an IPGphor IEF system (GE Healthcare). The protein extract was diluted to a final concentration of 2500 μg/ml with an IEF rehydration solution (2% (w/v) CHAPS, 0.5% IPG buffer, 2 m thiourea, and 6 m urea). After centrifugation for 10 min at 10,000 × g, the 450-μl supernatant was loaded onto a commercially available precast IPG strip with a 24-cm linear pH 4–7 gradient and actively rehydrated at 30 V for 12 h at 20 °C. Then focusing was performed on the IPGphor apparatus under the following conditions: 200 V for 40 min, 500 V for 40 min, 1000 V for 1 h, 4000 V for 2 h, and 8000 V for 8 h achieving ∼73,000 V-h. Before the SDS-PAGE, the strips were equilibrated for 15 min in 10 ml of reducing equilibration buffer (6 m urea, 50 mm Tris-HCl at pH 8.8, 30% (v/v) glycerol, 2% (w/v) SDS, a trace of bromphenol blue, and 1% (w/v) DTT) and for another 15 min in alkylating equilibration buffer that contained 2.5% (w/v) iodoacetamide instead of 1% DTT. The strips were placed on the top of vertical 12.5% SDS-polyacrylamide self-cast gels. The electrophoresis was carried out at 25 °C and 2.5 watts/gel for 30 min and then at 17 watts/gel until the dye front reached about 1 mm from the bottom of the gel using an Ettan™ DALT System (GE Healthcare).
Protein spots in 2-DE gels were detected by a modified colloidal CBB G-250 staining method with blue silver (23). The 2-DE gels were scanned using a UMAX PowerLook 2100XL scanner (UMAX Systems GmbH, Willich, Germany). At least triplicates were applied to each treatment, and a total of 12 CBB-stained 2-DE gels were analyzed using the ImageMaster™ 2-D platinum software version 5.0 (GE Healthcare). Spot detection parameters were set according to the manufacturer's instructions as follows: sensitivity, 8000; operator size, 45; noise factor, 5; and background factor, 8000. The spots were quantified using the percent volume criterion. The match analysis was performed in an automatic mode, and further manual editing was performed to correct the mismatched and unmatched spots. The relative volume of each spot was assumed to represent its expression level. A criterion of p < 0.05 was used to define the significant difference when analyzing the parallel spots between groups with one-way analysis of variance and Student-Newman-Keuls test using the SAS software package version 8.2 (SAS Institute).
In-gel Digestion, MS Analysis, and Database Searching—
Protein spots showing significant changes in abundance during the treatments were selected and excised manually for protein identification. In-gel digestion of protein spots was performed according to Yao et al. (22). All MALDI-TOF/TOF mass spectra were collected with an Ultraflex MALDI-TOF/TOF tandem mass spectrometer and analyzed by the peak list-generating FlexControl™ 2.2 software and the search engine Biotools 2.2 (Bruker Daltonics Inc., Bremen, Germany). The TOF spectra were recorded in the positive ion reflector mode with a mass range from 800 to 4000 Da. Ten subspectra with 30 shots per subspectrum were accumulated to generate one main TOF spectrum. After the search results were assessed automatically by the MASCOT software (Matrix Science, London, UK), the samples not identified by peptide mass fingerprinting (PMF) were automatically submitted to MS/MS analysis based on the potential lift technology. Two of the strongest peaks of the TOF spectra per sample were chosen for MS/MS analysis.
All of the PMFs were searched in the MASCOT version 2.1 (Matrix Science) with the following criteria: National Center for Biotechnology Information non-redundant (NCBInr) (release date September 4, 2006; including 3,658,925 protein entries and 1,257,151,091 residues); species restriction to O. sativa (rice) and Viridiplantae (green plants) only when no proteins matched in rice; p ≤ 0.05; the coverage of protein by matched peptides was >15%; at least five independent peptides matched; at most two missed cleavage sites; MS tolerance was set as ±100 ppm and MS/MS tolerance was set as ±0.7 Da; fixed modification was carbamidomethyl (Cys) and variable modification was oxidation (Met); and cleavage by trypsin was the C-terminal side of Lys and Arg unless the next residue was Pro. If peptides were matched to multiple members of a protein family or a protein appeared under different names and accession numbers, one entry with the highest score was selected. When protein isoforms were observed, these entries were inspected manually, and the presence of each protein isoform was confirmed by the identification of at least two unique peptides. Additionally the theoretical values of molecular weight (Mr) and pI of identified proteins were predicted by using the PeptideMass program (ExPASy).
RESULTS AND DISCUSSION
Morphological Changes and Physiological Responses Induced by H2O2 Stress in Rice—
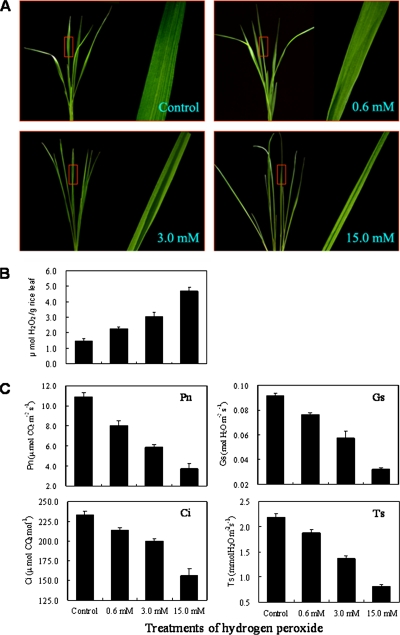
The exposure of 12-day-old rice seedlings to exogenous H2O2 results in dramatic changes in morphological and physiological traits. As presented in Fig. 1A, H2O2 treatment with 0.6 mm concentration for 6 h caused the leaf margins to roll inward, whereas 3.0 and 15.0 mm treatments caused the leaves to become cylindrical as the leaf margins rolled inward over the upper surface to the extent that they became completely curled as the H2O2 concentration was increased. To gain more understanding of the adverse effects of different H2O2 levels, we determined the content of endogenous H2O2 and found increasing H2O2 levels in treated rice leaves (Fig. 1B). The levels of H2O2 in leaves treated with exogenous H2O2 of 0.6, 3.0, and 15.0 mm concentrations were increased by 55.5, 110.7, and 225.5% over control, respectively, thus showing a dose-dependent pattern of accumulation.
Fig. 1.
Effects of H2O2 stress on the Pn, Gs, Ci, and Ts in rice seedlings. 12-day-old seedlings were treated with 0 (control), 0.6, 3.0, and 15.0 mm H2O2 for 6 h. The photographs of the same samples were taken at each treatment concentration point, and the framed regions were enlarged (A). The endogenous accumulation of H2O2 and changes of Pn, Gs, Ci, and Ts are shown in B and C, respectively.
To evaluate the adverse effects of oxidative stress, we measured the photosynthetic responses of rice seedling leaves following the stress treatments. The net photosynthetic rate (Pn) declined significantly from 10.09 to 8.03, 5.89, and 3.75 μmol of CO2 m−2 s−1 upon H2O2 treatments of 0.6, 3.0, and 15.0 mm, respectively (Fig. 1C). Similar patterns of rate decrease were also observed for the stomatal conductance (Gs), the intercellular CO2 concentration (Ci), and the transpiration speed (Ts) (Fig. 1C), suggesting that H2O2 treatment resulted in stomatal closure, further reduced water transpiration, and exoteric CO2 absorption in the treated rice seedling leaves. Similar results were observed in low temperature-treated rice seedlings (24) because adverse stress treatments are often linked to oxidative damage in plants (2, 3).
Rice Leaf Proteome in Response to H2O2 Stress—
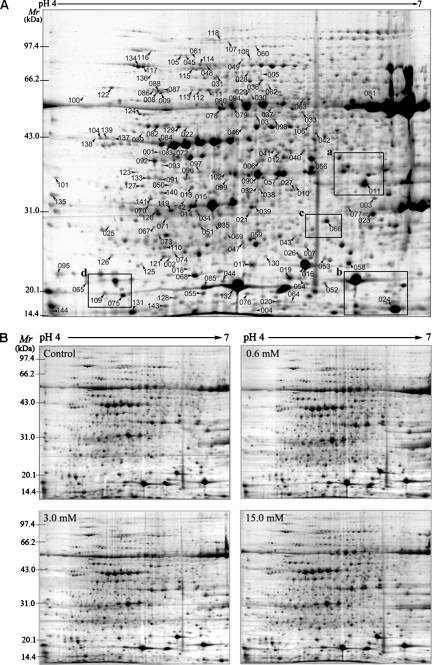
Using the modified TCA/acetone extraction protocol in combination with CBB staining, the average number of detectable spots in this study reached ∼2000 on each 2-DE gel (Fig. 2A), suggesting that we could take full advantage of the proteomics strategy to obtain more abundant information about the effects of oxidative stress on plants. Fig. 2B shows four reproducible gel maps in accordance with control and three different treatments. From a spot-to-spot comparison and statistical analysis, a total of 144 stained spots were found to have significant changes (p ≤ 0.05). Of these, 65 spots (numbered from 001 to 065) were up-regulated, and 79 spots (numbered from 066 to 144) were down-regulated in response to H2O2 stress (Fig. 2A). Most of these spots had a greater than 1.5-fold change in abundance under at least one of the H2O2 treatments, and 30 of these spots exhibited a more than 2-fold change (Table I). Supplemental Fig. S1 shows examples representing the dynamics of different proteins under oxidative stress conditions. It is noteworthy that some spots (spots 061 and 064) were absent from gels for the control sample but were induced by H2O2 treatments on the basis of our comparison of proteome profiles.
Fig. 2.
2-DE image analysis of rice leaf proteome under H2O2 stress. Total leaf proteins were extracted using the method of Parker et al. (21) and separated by IEF/SDS-PAGE. Proteins were stained with CBB G-250 based on the blue silver method (23). An equal amount (1.1 mg) of total proteins was loaded on each gel strip. 2-DE gel profiles of total proteins from control and rice leaves treated with three concentrations (0.6, 3.0 and 15.0 mm) of H2O2 are shown in B. The numbers of 144 differentially expressed protein spots in response to oxidative stress are marked in A. Among them, 129 spots correspond to proteins listed in Tables I–III. The framed regions a, b, c, and d in A are enlarged in supplemental Fig. S1.
Table I.
Differentially expressed proteins identified by PMF
| Spot no. | NCBI accession no. | Protein name | Average -fold changea
|
Score | Mb | Cc | ||
|---|---|---|---|---|---|---|---|---|
| CF-1 | CF-2 | CF-3 | ||||||
| % | ||||||||
| Up-regulated protein spots | ||||||||
| Cell rescue/defense | ||||||||
| 001 | ABA95337 | NB-ARC domain-containing protein | nsd | 1.42 | ns | 84 | 22 | 23 |
| 002 | XP_480115 | Putative MLA1 | 1.62 | ns | 1.51 | 64 | 12 | 19 |
| 003 | AAM91872 | Putative nodulin | 1.40 | ns | ns | 81 | 15 | 35 |
| 004 | XP_470893 | Putative salt-induced protein | 1.90 | ns | ns | 111 | 11 | 79 |
| Redox homeostasis | ||||||||
| 005 | XP_482784 | Putative 12-oxophytodienoate reductase | 1.55 | 1.68 | ns | 70 | 12 | 22 |
| 006 | AAO72663 | Coproporphyrinogen-III oxidase | 2.18 | ns | ns | 72 | 13 | 44 |
| 007 | XP_470193 | Putative glutathione S-transferase | ns | ns | 1.73 | 74 | 10 | 37 |
| 008 | AAX85991 | Protein-disulfide isomerase | ns | 1.45 | ns | 116 | 19 | 37 |
| 009 | AAX85991 | Protein-disulfide isomerase | ns | 1.37 | ns | 74 | 12 | 25 |
| 034 | BAD28547 | Putative glyoxalase I | 2.11 | ns | 1.35 | 98 | 17 | 60 |
| 035 | BAD28547 | Putative glyoxalase I | 1.29 | 1.43 | ns | 103 | 17 | 61 |
| Signal transduction | ||||||||
| 010 | AAA61831 | Small GTP-binding protein | 2.54 | 1.76 | 1.79 | 63 | 9 | 51 |
| 011 | NP_916988 | G protein β subunit-like protein | 1.62 | 1.63 | 1.62 | 72 | 11 | 53 |
| 013 | AAF81798 | Putative protein-tyrosine phosphatase | 1.71 | 1.61 | ns | 90 | 13 | 45 |
| Photosynthesis | ||||||||
| 014 | NP_918587 | Putative 33-kDa oxygen-evolving protein of photosystem II | 1.33 | 1.59 | 1.37 | 110 | 16 | 59 |
| 015 | NP_918587 | Putative 33-kDa oxygen-evolving protein of photosystem II | ns | 1.48 | ns | 96 | 12 | 52 |
| 016 | NP_911136 | Probable photosystem II oxygen-evolving complex protein 2 precursor | 1.51 | ns | ns | 129 | 13 | 65 |
| 017 | NP_911136 | Probable photosystem II oxygen-evolving complex protein 2 precursor | 1.44 | ns | 1.36 | 78 | 9 | 46 |
| 018 | CAG34174 | Rubisco large subunit | 1.22 | 1.82 | 1.82 | 74 | 13 | 31 |
| 019 | CAG34174 | Rubisco large subunit | 1.44 | ns | ns | 75 | 11 | 27 |
| 020 | CAJ42306 | Rubisco large subunit | 2.56 | ns | ns | 82 | 8 | 58 |
| 021 | AAX95285 | Rubisco activase small isoform precursor | 1.43 | ns | 1.24 | 61 | 8 | 35 |
| 022 | AAX95414 | Rubisco activase small isoform precursor | 4.63 | ns | ns | 95 | 15 | 37 |
| 023 | AAS46127 | Rubisco large subunit | 2.08 | 1.54 | 1.64 | 215 | 36 | 47 |
| 024 | YP_052757 | Rubisco large subunit | 2.29 | 2.04 | 2.15 | 83 | 21 | 22 |
| 025 | AAO22559 | Sedoheptulose-1,7-bisphosphatase precursor | 1.84 | ns | ns | 87 | 15 | 43 |
| 026 | BAD68170 | Thylakoid lumenal 20-kDa-like protein | ns | ns | 1.52 | 94 | 11 | 58 |
| Energy pathway | ||||||||
| 027 | NP_914553 | Putative acid phosphatase | ns | 1.59 | 2.05 | 86 | 13 | 33 |
| 028 | ABB47307 | ATP synthase CF1, β subunit | 3.22 | 3.01 | 2.43 | 79 | 20 | 52 |
| 029 | ABB47307 | ATP synthase CF1, β subunit | 2.31 | 1.88 | 1.85 | 159 | 28 | 53 |
| 030 | YP_052756 | ATP synthase β subunit | 1.88 | 1.63 | 2.57 | 260 | 23 | 54 |
| 031 | BAD45853 | Putative vacuolar proton-ATPase | ns | ns | 1.61 | 255 | 41 | 58 |
| Lipid metabolism | ||||||||
| 032 | XP_481639 | Putative enoyl-ACP reductase | ns | ns | 1.86 | 66 | 12 | 30 |
| Carbohydrate metabolism | ||||||||
| 033 | AAT78793 | Putative ADP-glucose pyrophosphorylase | ns | ns | 1.68 | 69 | 15 | 32 |
| 036 | BAD82294 | Putative phosphoglycerate mutase | 1.60 | 1.51 | ns | 192 | 31 | 63 |
| 037 | BAB69069 | UDP-glucose pyrophosphorylase | 1.55 | ns | ns | 70 | 12 | 38 |
| Amino acid metabolism | ||||||||
| 038 | ABA99438 | Cysteine synthase A | 1.55 | 1.78 | 1.67 | 166 | 20 | 81 |
| 039 | ABA99438 | Cysteine synthase A | 1.52 | ns | ns | 72 | 12 | 52 |
| 040 | XP_469854 | Putative dehydrogenase precursor | ns | ns | 1.53 | 63 | 12 | 43 |
| 041 | XP_467663 | Glutamine synthetase shoot isozyme | ns | ns | 1.49 | 103 | 16 | 56 |
| 042 | AAC05590 | S-Adenosyl-l-methionine synthetase | ns | 2.39 | 2.16 | 80 | 14 | 40 |
| Nucleotide metabolism | ||||||||
| 012 | BAD82147 | Phosphoribulokinase/uridine kinase-like | 2.19 | ns | 1.72 | 193 | 27 | 59 |
| 043 | AAU90215 | Putative uracil phosphoribosyltransferase | 1.70 | ns | 1.64 | 68 | 10 | 38 |
| Protein biosynthesis | ||||||||
| 044 | ABA91233 | Putative elongation factor P | 1.37 | ns | ns | 92 | 12 | 38 |
| 045 | S35701 | Translation elongation factor G | ns | ns | 1.45 | 120 | 24 | 38 |
| 046 | XP_466527 | Translation elongation factor Tu | 1.41 | ns | ns | 164 | 29 | 57 |
| % | ||||||||
| Protein folding and assembly | ||||||||
| 048 | XP_463871 | Putative dnaK-type molecular chaperone BiP | ns | 1.79 | ns | 146 | 27 | 37 |
| Protein degradation | ||||||||
| 047 | AAT78811 | 26 S proteasome subunit α-type 2 | 1.33 | 1.38 | 1.74 | 96 | 14 | 54 |
| 049 | XP_468533 | Oligopeptidase A-like | 2.10 | ns | ns | 66 | 16 | 26 |
| Unclassified | ||||||||
| 050 | XP_463882 | Unknown protein | 1.87 | ns | 1.99 | 113 | 16 | 42 |
| Down-regulated protein spots | ||||||||
| Cell rescue/defense | ||||||||
| 066 | AAV43981 | Putative chitinase | −1.28 | ns | −1.52 | 94 | 16 | 37 |
| 067 | AAR26485 | Harpin-binding protein 1 | −1.96 | −1.85 | −1.72 | 81 | 10 | 43 |
| 068 | XP_466697 | Putative temperature stress-induced lipocalin | −1.41 | ns | −1.56 | 61 | 8 | 37 |
| 122 | BAD27963 | Hydroxyproline-rich glycoprotein-like | −1.47 | ns | ns | 69 | 13 | 26 |
| Redox homeostasis | ||||||||
| 069 | XP_479627 | Ascorbate peroxidase | −1.56 | −1.56 | −1.72 | 138 | 19 | 74 |
| Photorespiration | ||||||||
| 070 | BAD29554 | Putative phosphoglycolate phosphatase precursor | −2.86 | −2.22 | −1.64 | 137 | 25 | 50 |
| 071 | BAD29554 | Putative phosphoglycolate phosphatase precursor | −2.22 | −1.39 | −1.64 | 126 | 23 | 50 |
| Photosynthesis | ||||||||
| 072 | XP_462936 | Putative chelatase subunit | −1.79 | −1.43 | −2.63 | 161 | 26 | 55 |
| 073 | AAX95978 | Chlorophyll a/b-binding protein CP26 precursor | −1.64 | −1.72 | −2.27 | 68 | 11 | 43 |
| 074 | XP_464478 | Putative chlorophyll a/b-binding protein type III (PSI) precursor | ns | −2.70 | −2.27 | 62 | 7 | 23 |
| 075 | 2002393A | Oxygen-evolving complex protein 1 | −1.15 | −1.14 | −1.33 | 67 | 9 | 39 |
| 076 | NP_911136 | Probable photosystem II oxygen-evolving complex protein 2 precursor | ns | −1.82 | −1.52 | 77 | 8 | 40 |
| 077 | ABB47308 | Ribulose-1,5-bisphosphate carboxylase | −2.27 | −2.13 | −1.85 | 169 | 27 | 44 |
| 078 | CAG34174 | Rubisco large subunit | ns | −1.59 | −2.04 | 185 | 28 | 53 |
| 079 | CAG34174 | Rubisco large subunit | ns | ns | −1.67 | 186 | 31 | 51 |
| 080 | CAG34174 | Rubisco large subunit | ns | ns | −2.38 | 188 | 30 | 53 |
| 081 | CAG34174 | Rubisco large subunit | ns | −1.59 | −1.47 | 185 | 26 | 51 |
| 082 | AAX95286 | Rubisco activase small isoform precursor | −1.30 | −1.19 | −1.14 | 76 | 16 | 32 |
| 083 | AAX95285 | Rubisco activase small isoform precursor | −1.19 | −1.16 | −1.39 | 82 | 20 | 45 |
| 084 | AAX95285 | Rubisco activase small isoform precursor | −1.20 | −1.16 | −1.35 | 134 | 29 | 56 |
| 085 | AAY64542 | Rubisco large subunit | ns | −1.35 | ns | 76 | 9 | 39 |
| 086 | ABA97087 | Rubisco subunit-binding protein α subunit | −1.43 | −1.21 | −1.19 | 135 | 22 | 46 |
| 087 | ABA97087 | Rubisco subunit-binding protein α subunit | −1.49 | ns | −1.30 | 159 | 25 | 47 |
| 088 | ABA97087 | Rubisco subunit-binding protein α subunit | −1.32 | −1.23 | −1.32 | 172 | 23 | 47 |
| 089 | NP_912361 | Fructose-1,6-bisphosphatase | −3.45 | ns | −1.89 | 111 | 17 | 43 |
| 090 | AAX95073 | Fructose-bisphosphate aldolase class-I | −1.47 | −1.47 | −1.30 | 86 | 14 | 38 |
| 091 | XP_467296 | Ribulose-5-phosphate kinase precursor | −1.27 | −1.64 | −1.59 | 128 | 18 | 53 |
| 092 | AAO22559 | Sedoheptulose-1,7-bisphosphatase precursor | −1.30 | −1.20 | −1.22 | 67 | 13 | 39 |
| 093 | AAO22559 | Sedoheptulose-1,7-bisphosphatase precursor | −1.70 | ns | −1.75 | 102 | 21 | 44 |
| Energy pathway | ||||||||
| 094 | AAA84588 | ATP β gene product | −1.11 | −1.37 | −1.37 | 213 | 33 | 67 |
| 095 | XP_467812 | Putative H+-transporting ATP synthase | ns | −1.52 | −1.85 | 67 | 6 | 23 |
| Carbohydrate metabolism | ||||||||
| 096 | XP_479756 | Putative fructokinase | −1.45 | ns | −1.43 | 167 | 25 | 59 |
| 097 | XP_479756 | Putative fructokinase | −1.22 | −1.54 | −1.45 | 82 | 13 | 46 |
| 098 | XP_466458 | Putative UDP-glucosyltransferase | −1.54 | −1.30 | −1.54 | 73 | 13 | 37 |
| 104 | CAC80698 | N-Acetylglucosaminyltransferase I | −1.61 | ns | ns | 71 | 8 | 75 |
| 119 | XP_467983 | Putative inorganic pyrophosphatase | −1.64 | ns | ns | 155 | 20 | 52 |
| 120 | XP_467983 | Putative inorganic pyrophosphatase | ns | ns | −1.56 | 121 | 16 | 52 |
| Nucleotide metabolism | ||||||||
| 099 | XP_477140 | Putative mRNA-binding protein precursor | ns | −1.56 | ns | 77 | 12 | 32 |
| 100 | CAD27458 | Nucleosome assembly protein 1-like protein 1 | −1.52 | ns | ns | 66 | 8 | 33 |
| 101 | AAL79738 | Putative polyprotein | −2.78 | −2.38 | −1.75 | 62 | 16 | 25 |
| 102 | ABA91072 | Putative retrotransposon protein | −1.39 | −1.85 | −1.47 | 64 | 24 | 17 |
| % | ||||||||
| Protein biosynthesis | ||||||||
| 103 | BAA02152 | Eukaryotic initiation factor 4A | ns | −1.61 | −1.72 | 80 | 15 | 39 |
| 105 | S35701 | Translation elongation factor G | −1.56 | −1.56 | −1.43 | 125 | 24 | 30 |
| 106 | XP_466527 | Translation elongation factor Tu | −1.96 | −1.92 | −1.67 | 165 | 24 | 53 |
| Protein degradation | ||||||||
| 107 | XP_483801 | Putative aminopeptidase N | −1.79 | ns | ns | 139 | 30 | 37 |
| 108 | XP_468533 | Oligopeptidase A-like | ns | −1.41 | ns | 239 | 35 | 53 |
| Protein folding and assembly | ||||||||
| 109 | BAA02151 | 21-kDa polypeptide | −1.49 | −1.61 | −1.39 | 116 | 13 | 78 |
| 110 | BAD36074 | Putative chaperonin 21 precursor | −1.75 | ns | −1.81 | 93 | 12 | 50 |
| 111 | NP_910308 | Putative chaperonin 60 β precursor | −1.22 | −1.32 | −1.39 | 214 | 29 | 53 |
| 112 | NP_910308 | Putative chaperonin 60 β precursor | −1.59 | −1.43 | −1.39 | 151 | 25 | 45 |
| 113 | NP_910308 | Putative chaperonin 60 β precursor | −1.85 | −1.82 | −1.54 | 89 | 16 | 30 |
| 114 | AAB63469 | Endosperm lumenal BiP | −1.52 | ns | ns | 114 | 20 | 28 |
| 115 | XP_470141 | Heat shock protein cognate 70 | ns | ns | −1.70 | 84 | 20 | 27 |
| 116 | CAA82945 | 90-kDa heat shock protein | −2.08 | −1.67 | −2.17 | 98 | 15 | 28 |
| 117 | ABA97211 | Stromal 70-kDa heat shock-related protein | −1.54 | −1.23 | −1.43 | 79 | 23 | 27 |
| 118 | NP_921687 | Putative endoplasmic reticulum membrane fusion protein | ns | −1.52 | −1.54 | 204 | 37 | 42 |
| RNA processing | ||||||||
| 121 | XP_450482 | Putative plastid-specific ribosomal protein 2 precursor | −1.35 | −1.32 | −1.30 | 87 | 12 | 46 |
| Unclassified | ||||||||
| 123 | AAT85778 | Hypothetical protein | −1.37 | ns | ns | 63 | 7 | 45 |
| 124 | NP_921715 | Unknown protein | −1.49 | −1.54 | −1.25 | 109 | 20 | 42 |
| 125 | NP_912350 | Unknown protein | −1.41 | −1.70 | ns | 72 | 10 | 79 |
| 126 | NP_913279 | Unnamed protein product | −2.38 | −1.61 | −2.13 | 67 | 7 | 51 |
Spot abundance is expressed as the ratio of intensities of up-regulated (plus value) or down-regulated (minus value) proteins between stress and control. -Fold changes had p values <0.05. CF-1, CF-2, and CF-3 represent 0.6, 3.0, and 15.0 mm H2O2 treatment concentrations, respectively.
Number of mass values matched.
Sequence coverage.
ns indicates no significant change of spot abundance between stress and control.
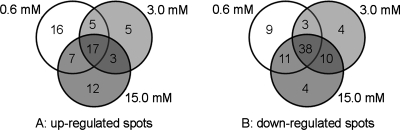
Fig. 3 presents the number of differentially expressed spots under different H2O2 treatments and how these spots overlap using a Venn diagram analysis. Not surprisingly, there were large overlaps between these spots. Of the 65 up-regulated spots, 17 spots were up-regulated under all three different treatments, and 15 spots showed increases in abundance under any two H2O2 treatments (Fig. 3A). Among the 79 down-regulated spots, 38 spots were down-regulated under all three concentration treatments (Fig. 3B). Taken together, spots for more than 65% of differentially expressed proteins exhibited co-regulation under at least two H2O2 treatment concentrations. Additionally 50 spots showed up- or down-regulation under only one H2O2 treatment concentration (Fig. 3). Of these, 26 spots showed significant changes of protein abundance in response to 0.6 mm H2O2 (the lowest concentration), whereas 16 spots were found to be differentially expressed under 15.0 mm H2O2 (the highest concentration) (Table I). The former might represent a specific adaptation of rice seedlings to low H2O2 concentration, but the latter may reflect cellular damage in rice seedlings exposed to high H2O2 concentration. Therefore, these observations document the changes in abundance of these differentially expressed proteins in response to H2O2 stress and further imply that cells in rice seedling leaves are able to monitor the extent of oxidative stress through modulating the expression of the corresponding proteins.
Fig. 3.
Venn diagram analysis of the differentially expressed protein spots in rice seedling leaves treated with 0.3, 0.6, and 1.5 mm H2O2. The numbers of differentially expressed protein spots with up- or down-regulation under a given concentration of H2O2 are shown in the different segments. A, the up-regulated protein spots; B, the down-regulated protein spots.
Identification and Functional Classification of the Differentially Expressed Proteins—
A total of 144 differentially expressed protein spots were analyzed by MALDI-TOF/TOF MS. Of these, 111 spots were successfully identified by PMF as listed in Table I, and 18 spots were identified by MS/MS analysis (Table II). The results for spot 056 are presented in supplemental Fig. S2 as an example. Meanwhile all PMF images and annotated spectra by MS/MS analysis are shown in supplemental Figs. S3 and S4, respectively. Among the 129 identities, 124 (96%) have been deposited in the current database as putative functional proteins, whereas five identities (spots 050 and 123–126) were annotated as either unknown or hypothetical proteins. To annotate these identities, their sequences were used as queries to search for homologues with BLASTP (NCBI). The corresponding homologues with the highest similarity are listed in Table III. All five proteins shared more than 40% positive identity with their homologues at the amino acid level, suggesting that they might have similar functions. Additionally 36 proteins of all the 129 identified proteins showed the characteristic with multiple members of a protein family (supplemental Table S3) in which one entry with the highest score was selected and described in this study. Taken together, the 129 identified proteins represent 89 unique proteins (unipros) (Tables I and II).
Table II.
Differentially expressed proteins identified by MS/MS
| Spot no. | NCBI accession no. | Protein name | Average -fold changea
|
Score | Cb | Sequence | m/z | zc | ||
|---|---|---|---|---|---|---|---|---|---|---|
| CF-1 | CF-2 | CF-3 | ||||||||
| % | ||||||||||
| Up-regulated spots | ||||||||||
| Cell rescue/defense | ||||||||||
| 051 | XP_462957 | Putative hydrolase | nsd | ns | 1.41 | 47 | 3 | AVSAIVSCLLGPDR | 1457.88 | +1 |
| Redox homeostasis | ||||||||||
| 052 | AAG60202 | Putative peptide methionine sulfoxide reductase | 1.71 | ns | ns | 54 | 5 | FYPAEEYHQR | 1339.65 | +1 |
| 053 | NP_916411 | Putative 1,4-benzoquinone reductase | 1.73 | 1.61 | 1.77 | 48 | 6 | VYVVYYSMYGHVAK | 1694.96 | +1 |
| Photosynthesis | ||||||||||
| 054 | CAA44032 | Rubisco large subunit | 1.48 | ns | ns | 93 | 3 | TFQGPPHGIQVER | 1465.88 | +1 |
| 055 | AAR23409 | Rubisco large subunit | 1.34 | 2.17 | 1.60 | 46 | 3 | TFQGPPHGIQVER | 1465.83 | +1 |
| 056 | XP_493811 | Glyceraldehyde-3-phosphate dehydrogenase | ns | ns | 1.44 | 72 | 3 | VVAWYDNEWGYSQR | 1772.63 | +1 |
| Nucleotide metabolism | ||||||||||
| 057 | XP_477140 | Putative mRNA-binding protein precursor | ns | ns | 1.39 | 55 | 2 | DCEEWFFDR | 1303.55 | +1 |
| Protein biosynthesis | ||||||||||
| 058 | XP_478772 | Putative ribosome recycling factor | 1.23 | 1.26 | 1.65 | 78, 107 | 4, 7 | AIETVQNNFNTVR, TIVAANLGVTPSNDGEVIR | 1505.78, 1926.02 | +1, +1 |
| Protein degradation | ||||||||||
| 059 | NP_568427 | Endopeptidase CLPP2 | 1.41 | 1.50 | ns | 40 | 2 | AYDIFSR | 871.51 | +1 |
| Down-regulated spots | ||||||||||
| Signal transduction | ||||||||||
| 127 | XP_483800 | Putative C2 domain-containing protein | −1.37 | −1.27 | −1.35 | 87 | 5 | LPLRDVLDDAGVGAR | 1566.94 | +1 |
| Photosynthesis | ||||||||||
| 128 | AAA62122 | Ribulose-1,5-bisphosphate carboxylase | −1.32 | −1.72 | −1.69 | 92 | 3 | TFQGPPHGIQVER | 1465.83 | +1 |
| 129 | AAC28134 | Rubisco activase | −1.47 | ns | −1.21 | 97, 94 | 3, 4 | IVDSFPGQSIDFFGALR,VPIIVTGNDFSTLYAPLIR | 1868.98, 2089.19 | +1, +1 |
| 130 | AAB82133 | Glyceralehyde-3-phosphate dehydrogenase subunit | ns | −2.13 | ns | 50 | 3 | VIAWYDNEWGYSQR | 1786.88 | +1 |
| Protein biosynthesis | ||||||||||
| 131 | O22386 | 50 S ribosomal protein L12 | −1.25 | −1.37 | −1.43 | 69 | 9 | TEFDVVIEEVPSSAR | 1677.88 | +1 |
| 132 | ABA96140 | Putative ribosomal protein S15 | ns | −1.21 | −1.43 | 90, 51 | 2, 1 | TFQGPPHGIQVER, DTDILAAFR | 1465.75, 1021.50 | +1, +1 |
| 133 | AAO73241 | Putative ribosomal protein S5 | −1.52 | −1.28 | −1.47 | 43 | 5 | VMLRPACPGSGVIAGGAVR | 1868.07 | +1 |
| Protein folding and assembly | ||||||||||
| 134 | ABA97211 | Stromal 70-kDa heat shock-related protein | −1.28 | ns | −1.30 | 60 | 1 | QFAAEEISAQVLR | 1461.91 | +1 |
| RNA processing | ||||||||||
| 135 | BAD46651 | Putative nucleic acid-binding protein | −1.54 | −1.47 | ns | 89 | 4 | IYVGNLPWQVDDSR | 1661.88 | +1 |
Spot abundance is expressed as the ratio of intensities of up-regulated (plus value) or down-regulated (minus value) proteins between stress and control. -Fold changes had p values <0.05. CF-1, CF-2, and CF-3 represent 0.6, 3.0, and 15.0 mm H2O2 treatment concentrations, respectively.
Sequence coverage.
Precursor charge.
ns represents no significant change of spot abundance between stress and control.
Table III.
The corresponding homologues of the five unknown proteins
BLASTP (NCBI) was used to search the homologues of the unknown proteins in Table I. The homologues with the highest homology are shown.
| Spot no. | NCBI accession no.a | Homologue
|
||||
|---|---|---|---|---|---|---|
| NCBI accession no.b | Protein name | Organism | Identitiesc | Positivesd | ||
| % | % | |||||
| 050 | XP_463882 | CAL52837 | AAA+-type ATPase | Ostreococcus tauri | 24 | 43 |
| 123 | AAT85778 | ABA97805 | Retrotransposon protein, putative, Ty3-gypsy subclass | O. sativa | 68 | 72 |
| 124 | NP_921715 | NP_201547 | Hydrolase, hydrolyzing O-glycosyl compounds | Arabidopsis thaliana | 29 | 42 |
| 125 | NP_912350 | ZP_01202146 | 3-Oxoacyl-[acyl-carrier-protein] synthase III | Flavobacteria bacterium | 25 | 41 |
| 126 | NP_913279 | BAA96146 | Putative heme-binding protein 2 | O. sativa | 100 | 100 |
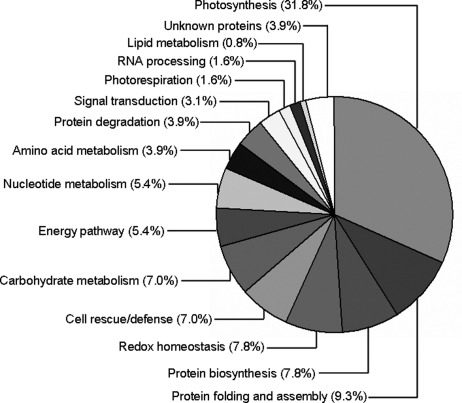
Based on the metabolic and functional features of rice seedling leaves, all of the identities were classified into 14 major categories, including photosynthesis, protein folding and assembly, protein biosynthesis, redox homeostasis, cell rescue/defense, metabolisms of carbohydrate, energy, nucleotide, amino acid and lipid as well as protein degradation, signal transduction, photorespiration, and RNA processing (Fig. 4). An impressive 70.5% of these identified proteins were implicated in the first six functional groups, whereas the largest functional group (31.8%) was proteins involved in photosynthesis that was greatly affected by H2O2 stress (Fig. 1C). Further analysis of the abundance changes in each group revealed that these stress-related proteins in the first six categories were overrepresented, either in number or in expression level, in all the identified proteins, suggesting that these processes were of functional importance for oxidative stress adaptation of rice seedlings.
Fig. 4.
Functional classification and distribution of all 129 identified proteins. Unknown proteins include those whose functions have not been described, but may be deduced based on analysis of sequence homology as listed in Table III.
In general, the apparent Mr value predicted by SDS-PAGE has an error deviation of about ±10% compared with the theoretical value. Among the 129 identified proteins, however, a set of 21 identities were found with observed Mr values much smaller than the theoretical values (Tables I and II), suggesting that these proteins appeared to be the partially degraded products of their intact proteins. Of these, 14 identities were suggested to be related to the photosynthetic process (seven Rubisco large subunits (RLSs), two Rubisco activase small isoform precursors (RASIPs), two glyceralehyde-3-phosphate dehydrogenases (GAPDHs), an oxygen-evolving complex protein 1 (OECP1), a photosystem (PS) II oxygen-evolving complex protein 2 (OECP2), and a sedoheptulose-1,7-bisphosphatase precursor (SBP)), whereas another seven proteins (spots 001–003, 051, 101, 102, and 132) were associated with cell rescue/defense, nucleotide metabolism, and protein biosynthesis (Tables I and II). A particular case was RLS in which the observed molecular mass values of the seven RLSs (spots 018–020, 023, 024, 054, and 055) ranged from 19.14 to 30.86 kDa, much smaller than the theoretical 52.79 kDa. Similar results were found in chilled rice seedling leaves and were further confirmed by Western blot analysis (24). Additionally the enhanced partial degradation of photosynthetic proteins such as RLS, RASIP, GAPDH, and SBP (Table I) might result in impaired photosynthesis in rice seedlings (Fig. 1C). Similar findings were also observed in cold stress rice seedlings (24). Likewise a number of partially degraded products of intact proteins participating in the tricarboxylic acid cycle showed up-regulation in Arabidopsis and pea mitochondria under H2O2 and other abiotic stresses (25, 26), indicating that these adverse stresses impair aerobic respiration in Arabidopsis and pea plants. Thus, a range of adverse stresses impairs physiological processes and metabolic pathways in plants through the accelerative degradation of many key enzymes that may protect plants from further damage triggered by external adverse conditions.
On the other hand, our results also showed that 20 unipros appeared as 47 identities as marked in Fig. 5A. Interestingly 15 unipros representing 35 identities (isoforms) showed that each set of isoforms had the same up- or down-regulated change patterns in abundance in response to H2O2 stress, whereas the other five unipros (ATP synthase CF1, β subunit; translation elongation factors G (EF-G) and Tu (EF-Tu); oligopeptidase A (OPA); and putative mRNA-binding protein precursor) appeared as 12 isoforms that exhibited opposite expression patterns within each set of isoforms (Fig. 5B and supplemental Table S2). Likewise many similar phenomena were also observed in other previously reported proteomics studies (24, 27–30). Based on changes in isoelectric state of protein spots on 2-DE gels and confirmation of phosphatase treatment, Gallie et al. (31) found that the phosphorylation state of several wheat translation initiation factors and the poly(A)-binding protein is subject to alteration during germination and following heat shock of wheat seedlings. This finding allows us to reasonably speculate that the proteomic responses of some of the latter five unipros in this study (e.g. EF-G, EF-Tu, and mRNA-binding protein precursor) are likely involved in posttranslational regulation, such as phosphorylation and dephosphorylation. The posttranslational modification of these unipros in response to H2O2 stress might result in the opposite expression patterns of different isoforms with phosphorylation or dephosphorylation state. These results suggest that isoforms of certain unipros may play the same or different roles in modulating defense responses to H2O2 stress in rice seedlings.
Fig. 5.
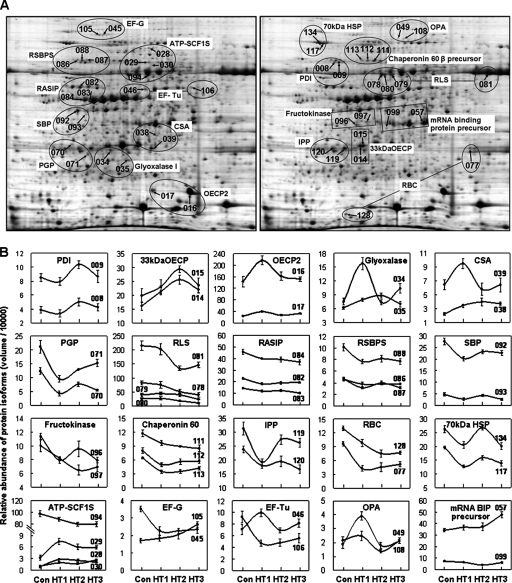
Close-up of possible isoforms detected by 2-DE (A) and their expression profile patterns (B). All 47 differentially expressed protein spots matched to 20 unique proteins are shown. The numbered spots correspond to proteins listed in Tables I and II. 33kDaOECP, 33-kDa oxygen-evolving complex proteins of PS II; ATP-SCF1S, ATP synthase CF1, β subunit; IPP, inorganic pyrophosphatase; RBC, ribulose 1,5-bisphosphate carboxylase; RSBPS, Rubisco subunit binding-protein α subunit; CSA, cysteine synthase A. Con, HT1, HT2, and HT3 represent different H2O2 treatments of 0, 0.6, 3.0, and 15.0 mm, respectively.
Differentially Expressed Proteins Are Preferentially Associated with the Photosynthetic Process—
Exposure to exogenous H2O2 stress led to increasing endogenous H2O2 accumulation in rice leaves and impaired the photosynthetic function of rice seedlings (Fig. 1, B and C). Meanwhile 41 differentially expressed identities were found to be associated with the photosynthetic process (Tables I and II), and their dynamics showed the effects of H2O2 stress on photosynthesis at the protein level as shown in Fig. 6. These proteins are implicated in four functional subgroups: 1) chlorophyll-binding proteins, 2) proteins participating in the Calvin cycle, 3) Rubisco activases or -binding proteins, and 4) oxygen-evolving complex proteins of PS II (Fig. 6, A–D).
Fig. 6.
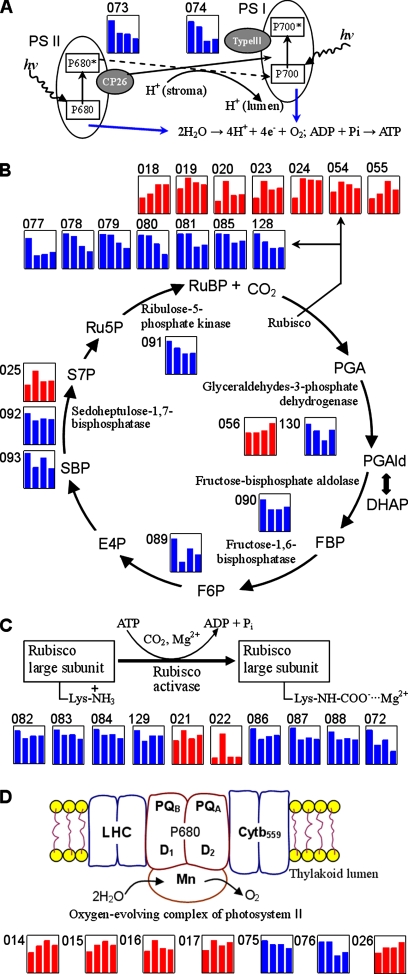
Pathways involved in the light-harvesting reaction (A), the Calvin cycle (B), the activation reaction of Rubisco large subunit (C), and the oxygen-evolving complex of PS II (D) under H2O2 stress in rice seedlings. Only H2O2-responsive proteins are mapped with those up-regulated charted in red and those down-regulated charted in blue. The numbered spots correspond to proteins listed in Tables I and II. RuBP, ribulose 1,5-bisphosphate; PGA, 3-phosphoglyceric acid; PGAld, 3-phosphoglyceraldhyde; DHAP, dihydroxyacetone phosphate; FBP, fructose 1,6-bisphosphate; F6P, fructose 6-phosphate; E4P, erythrose 4-phosphate; S7P, sedoheptulose 7-phosphate; Ru5P, ribulose 5-phosphate; LHC, light-harvesting complex; Cyt, cytochrome; PQ, plastoquinone; hv, light energy harvesting.
In the first subgroup, two precursors of chlorophyll a/b-binding proteins (spot 073, CP26 in PS II; and spot 074, type III in PS I) were down-regulated in response to H2O2 stress (Fig. 6A). CP26 is a component of the light-harvesting complex of PS II in plants and facilitates light absorption and transfer of the excitation energy to reaction centers for charge separation (32). Type III in PS I is a pigment-protein subunit component of the antenna system in plants and combines chlorophyll molecules in sites 1013 and 1023 (33). Therefore, the decreases in abundance of the two proteins might limit the light absorption and energy transfer of PS in the H2O2-treated rice seedling leaves and further affect the photosynthetic process.
In the second subgroup, there were 22 proteins participating in CO2 assimilation, including 12 RLSs, three SBPs, two GAPDHs, two ribulose 1,5-bisphosphate carboxylases, a ribulose-5-phosphate kinase precursor, a fructose-bisphosphate aldolase class-I, and a fructose-1,6-bisphosphatase. Among the 22 identities, 13 proteins showed down-regulation, whereas the other nine proteins increased in abundance and were partially degraded products (Fig. 6B). As mentioned above, seven of the nine degraded fragments are RLSs. Rubisco is a very inefficient enzyme with expression levels of 30–55% in excess under normal conditions (34), and there should be a dynamic balance between intact Rubisco and its degraded fragments in rice leaves (35). Otherwise ROS accumulation in chloroplast induces Rubisco modification and further facilitates subsequent degradation of Rubisco (36). The data presented here show that the down-regulation of the 13 proteins and the up-regulation of the nine degraded fragments together slowed down the Calvin cycle in rice seedling leaves (Fig. 6B) and thus impaired the photosynthetic process (Fig. 1C).
The third subgroup consists of 10 proteins engaged in the activation of Rubisco (five RASIPs, three Rubisco-binding protein α subunits, a Rubisco activase, and a putative chelatase subunit) (Fig. 6C). Besides the two RASIPs, the other eight proteins were down-regulated in response to H2O2 stress. Interestingly the two up-regulated RASIPs were partially degraded products. The fourth subgroup is composed of six oxygen-evolving proteins and a thylakoid lumenal protein. Of them, the five intact proteins, including two 33-kDa oxygen-evolving complex proteins of PS II, two OECP2, and a thylakoid lumenal protein, were up-regulated, whereas the two other degraded products (OECP1 and OECP2) decreased in abundance (Fig. 6D). Remarkably similar results are also observed in proteome analysis of rice seedling leaves in response to salt and low temperature stresses (24, 27, 37), suggesting that abiotic stresses can enhance the expression of oxygen-evolving proteins of PS II. Additionally two isoforms of phosphoglycolate phosphatase precursor (PGP) participating in photorespiration were down-regulated, implying that H2O2 stress might reduce photorespiration in rice seedling leaves (Table I).
Taken together, our results suggest that H2O2 stress seems to impair not only the light absorption and energy transfer of PS, CO2 assimilation, and Rubisco activity but also photorespiration by virtue of the abundance changes and the partial degradation of proteins. These results mirror the conclusion that H2O2 stress ultimately inhibits the photosynthesis and photorespiration in rice seedling leaves and are supported by the photosynthesis measurement (Fig. 1C). These data also provide new insights into the relationship between the impacts of oxidative stress on photosynthetic responses and the regulated expression of proteins (their abundance changes and breakdown) in H2O2-treated rice seedling leaves.
Differentially Expressed Proteins Are Largely Implicated in Inhibition of Biosynthesis and Enhanced Degradation of Proteins—
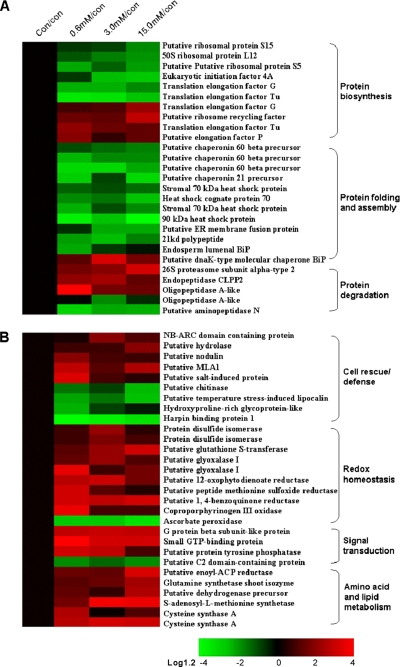
A total of 27 identified proteins (21% in total) in response to H2O2 stress were found to be attributed to protein metabolism (Tables I and II) and could be divided into three functional groups (Fig. 7A). The first group consists of 10 identities functioning in protein biosynthesis with six down-regulated proteins (Fig. 7A), including putative ribosomal protein S15, L12, S5, eukaryotic initiation factor 4A, EF-G, and EF-Tu, which are directly involved in initiation and elongation of the newly growing peptide chains. One of the four up-regulated identities (Fig. 7A) is putative ribosome recycling factor, which has been documented to function in the disassembly of the posttermination complex in protein biosynthesis (38). The three other up-regulated identities include putative elongation factor P and isoforms of EF-G and EF-Tu, which are involved in elongation of the growing peptide chains. Remarkably the opposite change patterns between the two isoforms of both EF-G and EF-Tu may implicate their different roles or compartmentation in the H2O2-treated rice leaves. The second group involves 12 identities related to protein folding and assembly (chaperonin 21, chaperonin 60 with three isoforms, HSP70 with three isoforms, 90-kDa HSP, putative dnaK-type molecular chaperone BiP, endosperm lumenal BiP, 21-kDa polypeptide, and putative endoplasmic reticulum membrane fusion protein). These chaperonins, HSPs, and BiPs have been well studied and are known to be responsible for protein refolding and assembly. Additionally the 21-kDa polypeptide is predicted to function as a molecular chaperone, whereas the endoplasmic reticulum membrane fusion protein is implicated in intercellular protein transport (Gene Ontology). As illustrated in Fig. 7A, our results show an interesting trend in the expression of these proteins (11 in group 2 together with six in group 1) responsible for protein synthesis, folding, and assembly whereby the expression levels are remarkably decreased in response to H2O2 stress, suggesting that protein biosynthesis may be inhibited under oxidative stress conditions.
Fig. 7.
Hierarchical clustering of H2O2-responsive proteins associated with protein metabolism (A) and antioxidative reactions (B). The hierarchical cluster analysis was conducted using the MATLAB image procedure and the log-transformed values of -fold change ratios listed in supplemental Table S1.
Among five degradation-related proteins, three identities (26 S proteasome subunit α, OPA, and endopeptidase CLPP2) were up-regulated in response to H2O2 stress, whereas putative aminopeptidase N and another isoform of OPA were decreased in abundance (Fig. 7A). The 26 S proteasome subunit α is a component of the large proteasome complex that selectively degrades various cellular proteins with specific degradation signals such as a multiubiquitin chain (39). OPA is the major soluble enzyme able to hydrolyze the lipoprotein signal peptide and further degrades the partially degraded portions of the signal peptide (40). CLPP2 is an ATP-dependent enzyme and functions as a master protease (41). Thus, the up-regulation of these three proteins indicates that protein degradation is enhanced in H2O2-treated rice seedling leaves. Additionally aminopeptidase N is known as a proteinase inhibitor and also as an exopeptidase (42). Thus, its down-regulated expression might release the activity of other proteinases and facilitate protein degradation. Taken together, the regulated expression response patterns of all the proteins in the three groups readily mirror the fact that the inhibition of novel protein biosynthesis as well as folding and assembly and the enhancement of protein degradation are required for the survival and growth of rice seedlings under H2O2 stress and that an active quality control system of proteins inside cells plays a crucial role in modulating the accommodation of rice seedlings to oxidative stress.
Differentially Expressed Proteins Were Involved in Leaf Antioxidative Reactions—
A total of 29 identified proteins (22% in total) in response to H2O2 stress were obviously related to leaf antioxidative reactions and could be classified into four functional categories, including cell rescue/defense, redox homeostasis, signal transduction, and amino acid as well as lipid metabolism (Fig. 7B). Of nine cell rescue/defense-related proteins, five (NB-ARC domain-containing protein, putative MLA1, putative nodulin, putative salt-induced protein, and putative hydrolase) were increased, and four (putative chitinase, harpin-binding protein 1, hydroxyproline-rich glycoprotein, and putative temperature stress-induced lipocalin) were deceased in abundance in the H2O2-treated rice leaves (Tables I and II and Fig. 7B). The NB-ARC domain is a component of the disease-resistant nucleotide-binding site leucine-rich repeat (NBS-LRR) proteins and functions as a molecular switch in plant defense systems (43). The MLA1 protein is known to confer race-specific resistance to the powdery mildew fungus in barley (44), and the putative MLA1 identified in this study shares 37% sequence identity with that in barley. Nodulin is also found to be induced in the resistant Medicago truncatula line upon inoculation with Colletotrichum trifolii, indicating a strong correlation between the number of up-regulated genes and the resistance phenotype (45). Otherwise chitinase, harpin-binding protein 1, and hydroxyproline-rich glycoprotein are known to play important roles in plant disease resistance (46), whereas the putative temperature stress-induced lipocalins play a possible biological role in membrane biogenesis and repair as well as in the transport of sterol molecules to the membrane under adverse stress conditions (47). It was not expected that these four proteins were down-regulated in response to H2O2 stress, but a possible explanation may be that there was no ample scope for their abilities especially under chemical oxidative stress. However, the question how these cell rescue/defense-related proteins are implicated in rice antioxidative reactions still remains to be addressed.
Among 10 identities implicated in redox homeostasis, nine were up-regulated in response to H2O2 stress, including GST, protein-disulfide isomerase (PDI) with two isoforms, glyoxalase I with two isoforms, and three reductases (Tables I and II and Fig. 7B). GST is an antioxidative protein, and its expression can be strongly enhanced by abiotic and biotic stresses (48). PDI functions in the thioredoxin-based redox pathway and forms part of the antioxidative defense system (49). Glyoxalase I can convert toxic 2-oxoaldehydes into less reactive 2-hydroxyacids using glutathione as a cofactor (50). The three reductases catalyze the reductions of methoxylated quinine, methionine sulfoxide, and 12-oxophytodienoic acid, respectively (51–53). Thus, the up-regulation of these proteins implies that the antioxidative defense system is provoked in H2O2-treated rice seedling leaves, and such a consistent induction is a likely consequence of antioxidative reactions under oxidative stress in plants. Unexpectedly an APx in this study was found to be down-regulated in response to H2O2 stress (Fig. 7B). Nevertheless similar results are also observed in tobacco and barley plants with different conditions of H2O2 stress (4, 54). Although APx is an important H2O2-scavenging enzyme located in the cytosol, chloroplasts, mitochondria, and peroxisomes of higher plants, the decrease of APx abundance in chemically treated tissues is likely to contribute to an accumulation of H2O2 and the eventual death of plant cells (4, 54).
Furthermore, of four signal transduction-related proteins, three identities (G protein β subunit, small GTP-binding protein, and putative protein-tyrosine phosphatase (PTP)) showed an up-regulated expression pattern, whereas the C2 domain-containing protein (C2-P) was decreased in abundance (Tables I and II and Fig. 7B) under H2O2 stress. The G proteins are mediators that transmit the external signals via receptor molecules to effector molecules and participate in multiple signaling pathways such as those involving sphingolipid, jasmonic acid, and pathogen infection in plants (55, 56). PTP is a signaling enzyme that mediates protein phosphorylation events, playing a particularly important role in maintaining ROS balance (57). Remarkably C2-P has multiple roles in negatively regulating defense responses and cell death in Arabidopsis. The loss of the C2-P gene confers an enhanced resistance to pathogens (58). Therefore, the up-regulation of the G proteins and PTP, together with the down-regulation of C2-P, suggests that the stress defense system is up-regulated in H2O2-treated rice seedling leaves. On the other hand, all six identities associated with amino acid and lipid metabolism were increased in abundance (Table I and Fig. 7B), including glutamine synthetase shoot isozyme (GSSI), S-adenosyl-l-methionine synthetase, dehydrogenase, cysteine synthase A with two isoforms, and putative enoyl-ACP reductase. GSSI plays an important role in enhancing rice tolerance to salt and chilling stresses (59). S-Adenosyl-l-methionine synthetase catalyzes the biosynthesis of S-adenosyl-l-methionine and closely correlates with tolerance to cold, salt, and abscisic acid stresses (37, 60). Cysteine synthase A is responsible for the final step in cysteine biosynthesis, the key limiting step in producing glutathione, which is involved in resistance to adverse stresses (61). Enoyl-ACP reductase is a subunit of the fatty-acid synthase complex that catalyzes de novo synthesis of fatty acids, and the lack of this gene in the mol1 mutant of Arabidopsis leads to premature cell death in multiple organs (62). Taken together, the findings reported above largely indicate an intimate balanced correlation between the changes in specific protein abundance and the overall antioxidative defense responses in H2O2-treated rice seedlings.
Preferential Representation of Some H2O2-induced Proteins and Their Cross-talk to Other Abiotic Stresses—
By comparison with previously published rice proteomics data sets, the 12 H2O2-responsive proteins identified in this study (Table IV) were detected in rice seedling leaves under many external stimuli such as cold, O3, SO2, salt, drought, heavy metal, and plant hormones (21, 24, 27–29, 37, 63–69), suggesting their important roles in the defense responses of rice seedlings to various environmental stresses. Some of them show the same expression patterns between H2O2 stress and other adverse stimuli, whereas others do not (Table IV). For example, the expression patterns of eight proteins including GAPDH; GST; EF-Tu; HSP70; PDI; GSSI; ATP synthase CF1, β subunit; and fructose-bisphosphate aldolase (Table IV) in H2O2-treated rice seedling leaves were observed to be similar to those in Arabidopsis mitochondria and yeast subjected to H2O2 stress (25, 30), implying the similar response of different species and/or tissues under H2O2 stress. A particular case was GST: its abundance of protein expression was up-regulated not only by H2O2 stress in this study but also by other abiotic stresses such as cold, drought, jasmonic acid, and brassinolide in rice seedling leaves (29, 37, 64, 65). GST is known to function as a pivotal component in plant defense systems and catalyzes the reduction reaction of toxic organic hydroperoxides by using glutathione as a cosubstrate or coenzyme (48). Thus, these consistent results demonstrate that the expression level of GST is remarkably enhanced under various stress conditions, implying that novel GST synthesis is required for plant tolerance to those stresses and that GST plays a pivotal role in plant stress defense responses.
Table IV.
H2O2-induced proteins and their responses to other abiotic stresses in rice seedling leaves and other species such as Arabidopsis and yeast
ABA, abscisic acid; BL, brassinolide; JA, jasmonic acid; O3, ozone; SO2, sulfur dioxide.
| Protein names | Treatments
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Rice seedling leaves
|
Arabidopsis and yeast: H2O2 (25, 30) | |||||||
| This study H2O2 | Cold (24, 36, 62)a | Hormones (63–65) | Air pollutants (66, 67) | Salt (21, 27) | CuSO4 (28, 68) | Drought (29) | ||
| ATP synthase CF1, β subunit | 3.22 | >1.50 | ↓b (ABA) | ↓ (O3) | 1.90 | ↑ | >3.00 | |
| Fructose-bisphosphate aldolase | −1.47 | >3.00 | ↓ (ABA) | ↓ (O3) | 1.50 | 1.45 | −1.51 | |
| Rubisco activase | −1.47 | <−1.50 | ↓ (ABA) | ↓ (O3) | 1.61 | |||
| Rubisco large subunit | −2.38 | <−1.50 | ↓ (ABA) | ↓ (SO2, O3) | −2.00 | ↓ | ||
| Glyceraldehyde-3-phosphate dehydrogenase | −2.13 | >1.50 | ↑ (BL) | ↑ (O3) | −1.34 | |||
| Ascorbate peroxidase | −1.72 | >1.50 | ↑ (JA) | ↑ (SO2, O3) | ||||
| Glutathione S-transferase | 1.73 | 1.87 | ↑ (JA, BL) | ↓ | 1.67 | >3.00 | ||
| Translation elongation factor Tu | −1.96 | 1.63 | 1.96 | −1.54 | ||||
| Heat shock protein 70 | −1.70 | 2.48 | 1.60 | ↑ | −1.67 | |||
| Protein-disulfide isomerase | 1.45 | <−2.00 | ↓ | >3.00 | ||||
| Glutamine synthetase shoot isozyme | 1.49 | >1.50 | 3.39 | |||||
| PSII oxygen-evolving complex protein | 1.51 | 1.89 | 1.30 | |||||
References are cited in parentheses.
↑ and ↓ represent up-regulated and down-regulated expression of proteins but without the values of -fold change listed.
In contrast, six of the 12 co-detected proteins exhibited different expression patterns between H2O2 and other abiotic stresses (Table IV). APx was down-regulated in response to H2O2 stress but was up-regulated under treatments of cold, SO2, O3, and jasmonic acid (24, 64, 67, 68). Likewise the reverse expression patterns of PDI were also observed at the protein level, showing up-regulation under H2O2 treatment and down-regulation during cold and CuSO4 stresses in rice seedling leaves (28, 63). Nevertheless it is reasonable to believe that the upstream APx-dependent H2O2-scavenging pathway might be disabled, whereas the downstream PDI-based antioxidative defense network is still activated in rice seedling leaves exposed to high doses of H2O2 (4, 70). Relatively small increases of H2O2 during stresses from cold, SO2, O3, CuSO4, and jasmonic acid trigger plant defense responses are in sharp contrast to the large increases of H2O2 resulting from treatment with high doses of H2O2, suggesting that plant cells are able to monitor the levels of H2O2 enhancement induced by different intensities of oxidative stress. This effective strategy allows plants to produce rapid, appropriate, and flexible responses to changing environmental threats (4, 70, 71). In addition, similar patterns in change of abundance were observed for two protein metabolism-related identities (EF-Tu and HSP70) that were down-regulated by H2O2 stress but up-regulated under adverse stresses such as cold, drought, salt, and CuSO4. Similar patterns were also observed for two glycolysis-related proteins (GAPDH and fructose-bisphosphate aldolase) that deceased in abundance during H2O2 stress but were induced by stresses of cold, salt, drought, and brassinolide (Table IV). These results suggest that protein and carbohydrate metabolic reactions may be impaired in rice seedling leaves exposed to high doses of H2O2 but are promoted during abiotic stresses with weakly oxidative intensity. Taken together, the findings presented here suggest that although the cellular responses to abiotic and biotic challenges are rather similar with regard to production of ROS (3, 9–16), the detailed stress defense mechanisms might be different. Therefore, two extremely different types of metabolic changes are involved in the adaptation of plants to different intensities of oxidative stress. Under strongly oxidative situations experienced by plants, the overall reduced substance metabolism may be the result of a series of events in gene expression and oxidative damage that would lead to cell death (70, 71). Nevertheless it may play crucial roles in improving cell fitness and protecting plants from further damage to the continuous ROS production and ultimately maintain plant survival (19). On the other hand, weakly oxidative stresses influence only part of the antioxidative network and modify gene expression in such a way as to strengthen plant defense responses and enhance metabolic systems to maintain homeostasis (70, 71). This finely modulated response strategy in plant cells has evolved over a long period to guarantee plant survival and growth against ever changing environmental threats with a combination of multiple stresses.
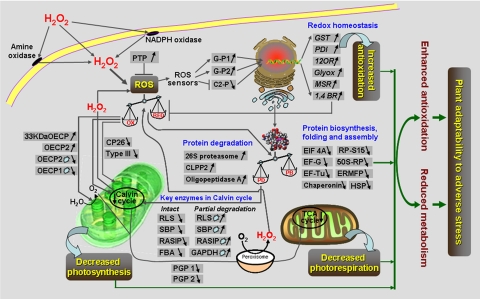
A Possible H2O2 Stress-responsive Protein Network in Rice Seedling Leaves—
A tight regulation of the steady-state levels of ROS (particularly H2O2) in plants is necessary to avoid cellular injury and to maintain a basic level of ROS upon which different developmental and environmental signals can be perceived and transmitted. However, no systemic investigation into how plants maintain survival and growth under oxidative stress through adjustment of their metabolic network and antioxidative system has been conducted to date. In the present study, based on our proteomics and physiological data, an oxidative stress-responsive protein network was proposed with most of the 129 H2O2-responsive proteins identified in rice seedling leaves (Fig. 8 and Tables I and II). This network consists of several functional components, including imbalance between ROS production and scavenging, accelerated degradation and reduced biosynthesis of proteins, impaired photosynthesis and photorespiration, and an enhanced antioxidative defense system (Fig. 8). These changes of metabolic reactions and redox balance eventually lead plants to a new homeostasis that can adapt to and/or resist external adverse stresses. Such a protein network allows us to further understand and describe this possible management strategy of cellular activities occurring in H2O2-treated rice seedling leaves.
Fig. 8.
A putative model of the H2O2 stress responses in rice seedling leaves. Only some of the H2O2-responsive proteins are indicated with those up-regulated marked by ↑ and those down-regulated marked by ↓. The light blue circles outlined in black represent partial degradation of proteins. See the text for details. 1,4 BR, 1,4-benzoquinone reductase; 12OR, putative 12-oxophytodienoate reductase; 33kDaOECP, 33-kDa oxygen-evolving complex proteins of PS II; 50S-RP, 50 S ribosomal protein L12; CP26, chlorophyll a/b-binding protein CP26 precursor; EIF 4A, eukaryotic initiation factor 4A; ERMFP, putative endoplasmic reticulum membrane fusion protein; FBA, fructose-bisphosphate aldolase class-I; Glyox, putative glyoxalase I; G-P1, small GTP-binding protein; G-P2, G protein β subunit; MSR, methionine sulfoxide reductase; RP-S15, putative ribosomal protein S15; Type III, putative chlorophyll a/b-binding protein type III (PS I) precursor; OX, oxidation; RED, reduction; PB, protein biosynthesis; PD, protein degradation.
When rice seedlings are exposed to high doses of H2O2, the exogenous H2O2 as a stable small molecule can diffuse into cells by cross-membrane transport (1) and also activate some enzymes such as membrane-bound NADPH oxidase and apoplast-located amine oxidase (3) to amplify endogenous H2O2, thus resulting in an increase of intracellular H2O2 level in rice leaves (Fig. 1B). The excessive H2O2 levels in rice leaves causes an imbalance of the original redox homeostasis with the characteristics of elevated oxidative intensity, which would lead to changes of biomolecular metabolism.
Synthesis and degradation of biomolecules (particularly proteins) are the most important links in the life processes. Under the above mentioned oxidative stress, the expression of most biosynthesis-, folding- and assembly-related proteins, including eukaryotic initiation factor 4A, EF-G, EF-Tu, putative ribosomal protein S15, 50 S ribosomal protein L12, putative endoplasmic reticulum membrane fusion protein, HSP, and chaperonin, were down-regulated, whereas some degradation-associated proteins such as 26 S proteasome, CLPP2, and OPA increased in abundance in H2O2-treated rice seedling leaves (Figs. 7A and 8). It is not surprising that the elevated oxidative intensity also damaged the equilibrium of protein metabolism between biosynthesis and degradation with a functional characteristic skew into speedup of protein degradation and slowdown of protein biosynthesis (Fig. 8).
The excessive H2O2 in rice leaves not only strongly affects the protein metabolism-related proteins but also acts intensively on carbohydrate metabolism-associated proteins. On the one hand, it is clear from the 2-DE gel images that the abundances of many intact key enzymes involved in the Calvin cycle declined under oxidative stress (Fig. 6, B and C). On the other hand, the protein degradation system provoked by oxidative stress might accelerate the partial degradation of these key enzymes participating in the Calvin cycle in rice leaves, thus increasing protein abundances of the partially degraded products (Fig. 6, B and C). These key enzymes include RLS, SBP, RASIP, fructose-bisphosphate aldolase class-I, and GAPDH (Fig. 8). As a result, the photosynthetic rate occurring in rice seedlings is impaired by H2O2 stress (Fig. 1C). Moreover the elevated H2O2 levels still inhibit the expression of light-harvesting antenna proteins such as CP26 and Type III but increase the abundance of PS II oxygen-evolving proteins like 33-kDa OECP and OECP2 (Figs. 6, A and D, and 8). The down-regulation of CP26 and Type III, together with the up-regulation of 33-kDa OECP and OECP2, leads to the reduced light harvesting capability of rice leaves and eventually impaired photosynthesis (Fig. 1C).
Physiological analysis showed the attenuation of photorespiration in H2O2-stressed rice leaves (Fig. 1C). The photorespiratory reactions are involved in a cooperative interaction between three subcellular organelles: chloroplasts, peroxisomes, and mitochondria. Besides the enhanced partial degradation of the photosynthetic components and the relative attenuation of the light harvesting capability, two key enzymes, PGP1 and PGP2, were also found to be down-regulated under oxidative stress (Fig. 8 and Table I). The changes of these critical proteins eventually result in a reduction of the photorespiration rate in H2O2-treated rice seedling leaves (Fig. 1C). In addition, some proteins related to glycolysis (putative fructokinase with two isoforms) and aerobic respiration (inorganic pyrophosphatase with two isoforms) (Table I) were down-regulated under H2O2 stress. Taken together, these basic metabolisms such as photosynthesis, photorespiration, aerobic respiration, glycolysis, and protein biosynthesis in rice seedling leaves were without exception inhibited by H2O2 stress. It is thus believed that plants must be required to make economical use of their substance and energy to adequately deal with severely adverse environments.
On the other hand, the gradually elevated H2O2 is sensitively perceived by cells of rice seedlings via different proteins, enzymes, or receptors and modulates different stress defense pathways. The up-regulated G proteins (G-P1 and G-P2), together with the down-regulated negative regulatory factor (C2-P protein), may cooperatively induce a number of genes with antioxidative functions such as GST, PDI, putative 12-oxophytodienoate reductase, putative glyoxalase I, methionine sulfoxide reductase, and 1,4-benzoquinone reductase in this study (Fig. 8). The abundances of these gene products were significantly increased on 2-DE gel images (Fig. 2 and supplemental Table S1) and led to the enhancement of the cellular antioxidant levels. The activated antioxidative systems in cells of rice seedlings possess a stronger capability for removing H2O2, which can reduce the intracellular H2O2 levels and attenuate the oxidative damage, thus ultimately establishing a new redox homeostasis. Such antioxidative systems play an important role in maintaining the survival and growth of rice seedlings under strong and sustained oxidative stress (Fig. 8).
Conclusions—
To investigate changes of global proteins under oxidative stress, we performed a comparative proteome analysis of rice seedling leaves using H2O2 as a model for oxidative stress. The present proteomics study revealed 144 H2O2-responsive spots on the 2-DE gel image containing about 2000 reproducible spots from which 129 differentially expressed proteins were successfully identified in H2O2-treated rice seedling leaves. These proteins not only include many well known H2O2-induced proteins such as HSP70, GST, PDI, APx, and ATP synthase but also 42 novel H2O2-responsive proteins, especially some proteins related to redox homeostasis and signal transduction (supplemental Table S3). It should be noted that these novel proteins did not appear in previous proteomics data sets obtained from H2O2-stressed Arabidopsis mitochondria (25) and yeast (30) as well as other abiotic-stressed rice seedling leaves (21, 24, 27–29, 37, 63–69). The discovery of such new proteins from 2-DE gels provides new insights into the oxidative stress defense mechanism in plants.
Based on the putative functions and abundance changes of the 129 identified proteins, together with previously published results, we proposed an H2O2 stress-responsive protein network including a series of events occurring in rice seedling leaves under higher doses of H2O2 (Fig. 8). These results depict a panoramic view of the adaptation strategies of rice seedlings to oxidative challenge and deepen our understanding of the sophisticated functional network for adaptation to oxidative stress in plants. It is also expected that this network can mirror the management of cellular activities in plants under oxidative stress and provide a basis for further functional research of each member of this network in intracellular redox homeostasis and H2O2 metabolism.
Supplementary Material
Acknowledgments
We are grateful to Prof. Jian-guo Ji, College of Life Sciences, Peking University, Beijing, China, for kind assistance in MS/MS analysis. We also thank Saleem A. Bokhari, Yi-Wei Yang, and Shao-Jun Du in our laboratory for excellent technical help.
Footnotes
Published, MCP Papers in Press, April 11, 2008, DOI 10.1074/mcp.M700488-MCP200
The abbreviations used are: ROS, reactive oxygen species; 2-DE, two-dimensional electrophoresis; APx, ascorbate peroxidase; C2-P, C2 domain-containing protein; CAT, catalase; EF-G, translation elongation factor G; EF-Tu, translation elongation factor Tu; GAPDH, glyceralehyde-3-phosphate dehydrogenase; GSSI, glutamine synthetase shoot isozyme; HSP, heat shock protein; OECP1, oxygen-evolving complex protein 1; OECP2, oxygen-evolving complex protein 2; OPA, oligopeptidase A; PDI, protein-disulfide isomerase; PGP, phosphoglycolate phosphatase precursor; PMF, peptide mass fingerprinting; PS, photosystem; PTP, protein-tyrosine phosphatase; RASIP, Rubisco activase small isoform precursor; RLS, Rubisco large subunit; SBP, sedoheptulose-1,7-bisphosphatase precursor; unipro, unique protein; Rubisco, ribulose-bisphosphate carboxylase/oxygenase; EST, expressed sequence tag; CBB, Coomassie Brilliant Blue; Pn, net photosynthetic rate; Gs, stomatal conductance; Ci, intercellular CO2 concentration; Ts, transpiration speed; ACP, acyl carrier protein; BiP, binding protein.
This work was supported by the State Key Basic Research and Development Plan of China (Grants 2006CB101706 and 2004CB117303), the Specialized Research Fund for the Doctoral Program of Higher Education (Grant 20050003066) (to J.-Y. L.), and the China Postdoctoral Science Foundation (Grant 20060400464) (to X.-Y. W.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Bienert, G. P., Schjoerring, J. K., and Jahn, T. P. ( 2006) Membrane transport of hydrogen peroxide. Biochim. Biophys. Acta 1758, 994–1003 [DOI] [PubMed] [Google Scholar]
- 2.Mittler, R., Vanderauwera, S., Gollery, M., and Breusegem, F. V. ( 2004) Reactive oxygen gene network of plants. Trends Plant Sci. 9, 490–498 [DOI] [PubMed] [Google Scholar]
- 3.Mittler, R. ( 2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 7, 405–410 [DOI] [PubMed] [Google Scholar]
- 4.de Pinto, M. C., Paradiso, A., Leonetti, P., and Gara, L. D. ( 2006) Hydrogen peroxide, nitric oxide and cytosolic ascorbate peroxidase at the crossroad between defence and cell death. Plant J. 48, 784–795 [DOI] [PubMed] [Google Scholar]
- 5.Stone, J. R. ( 2004) An assessment of proposed mechanisms for sensing hydrogen peroxide in mammalian systems. Arch. Biochem. Biophys. 422, 119–124 [DOI] [PubMed] [Google Scholar]
- 6.Pei, Z. M., Murata, Y., Benning, G., Thomine, S., Klusener, B., Allen, G. J., Grill, E., and Schroeder J. I. ( 2000) Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406, 731–734 [DOI] [PubMed] [Google Scholar]
- 7.Baxter-Burrell, A., Yang, Z. B., Springer, P. S., and Bailey-Serres, J. ( 2002) RopGAP4-dependent Rop GTPase rheostat control of Arabidopsis oxygen deprivation tolerance. Science 296, 2026–2028 [DOI] [PubMed] [Google Scholar]
- 8.Anthony, R. G., Henriques, R., Helfer, A., Meszaros, T., Rios, G., Testerink, C., Munnik, T., Deak, M., Koncz, C., and Bogre, L. ( 2004) A protein kinase target of a PDK1 signalling pathway is involved in root hair growth in Arabidopsis. EMBO J. 23, 572–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prasad, T. K., Anderson, M. D., Martin, B. A., and Stewart, C. R. ( 1994) Evidence for chilling-induced oxidative stress in maize seedlings and a regulatory role for hydrogen peroxide. Plant Cell 6, 65–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loggini, B., Scartazza, A., Brugnoli, E., and Navari-Izzo, F. ( 1999) Antioxidative defense system, pigment composition, and photosynthetic efficiency in two wheat cultivars subjected to drought. Plant Physiol. 119, 1091–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valderrama, R., Corpas, F. J., Carreras, A., Gomez-Rodriguez, M. V., Chaki, M., Pedrajas, J. R., Fernandez-Ocana, A., Del Rio, L. A., and Barroso, J. B. ( 2006) The dehydrogenase-mediated recycling of NADPH is a key antioxidant system against salt-induced oxidative stress in olive plants. Plant Cell Environ. 29, 1449–1459 [DOI] [PubMed] [Google Scholar]
- 12.A.-H.-Mackerness, S., Surplus, S. L., Blake, P., John, C. F., Buchanan-Wollaston, V., Jordan, B. R., and Thomas, B. ( 1999) Ultraviolet-B induced stress and changes in gene expression in Arabidopsis thaliana: role of signaling pathways controlled by jasmonic acid, ethylene and reactive oxygen species. Plant Cell Environ. 22, 1413–1423 [Google Scholar]
- 13.Pellinen, R., Palva, T., and Kangasjärvi, J. ( 1999) Subcellular localization of ozone-induced hydrogen peroxide production in birch (Betula pendula) leaf cells. Plant J. 20, 349–356 [DOI] [PubMed] [Google Scholar]
- 14.Schutzendubel, A., and Polle, A. ( 2002) Plant responses to abiotic stresses: heavy metal-induced oxidative stress and protection by mycorrhization. J. Exp. Bot. 53, 1351–1365 [PubMed] [Google Scholar]
- 15.Liu, Y., Huang, W., Zhan, J., and Pan, Q. ( 2005) Systemic induction of H2O2 in pea seedlings pretreated by wounding and exogenous jasmonic acid. Sci. China Ser. C Life Sci. 48, 202–212 [DOI] [PubMed] [Google Scholar]
- 16.Levine, A., Tenhaken, R., Dixon, R., and Lamb, C. ( 1994) H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 79, 583–593 [DOI] [PubMed] [Google Scholar]
- 17.Desikan, R., A.-H.-Mackerness, S., Hancock, J. T., and Neill, S. J. ( 2001) Regulation of the Arabidopsis transcriptome by oxidative stress. Plant Physiol. 127, 159–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vanderauwera, S., Zimmermann, P., Rombauts, S., Vandenabeele, S., Langebartels, C., Gruissem, W., Inze, D., and Van Breusegem, F. ( 2005) Genome-wide analysis of hydrogen peroxide-regulated gene expression in Arabidopsis reveals a high light-induced transcriptional cluster involved in anthocyanin biosynthesis. Plant Physiol. 139, 806–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vandenabeele, S., Van Der Kelen, K., Dat, J., Gadjev, I., Boonefaes, T., Morsa, S., Rottiers, P., Slooten, L., Van Montagu, M., Zabeau, M., Inzé, D., and Van Breusegem, F. ( 2003) A comprehensive analysis of hydrogen peroxide-induced gene expression in tobacco. Proc. Natl. Acad. Sci. U. S. A. 100, 16113–16118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tiwari, B. S., Belenghi, B., and Levine, A. ( 2002) Oxidative stress increased respiration and generation of reactive oxygen species, resulting in ATP depletion, opening of mitochondrial permeability transition, and programmed cell death. Plant Physiol. 128, 1271–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parker, R., Flowers, T. J., Moore, A. L., and Harpham, N. V. J. ( 2006) An accurate and reproducible method for proteome profiling of the effects of salt stress in the rice leaf lamina. J. Exp. Bot. 57, 1109–1118 [DOI] [PubMed] [Google Scholar]
- 22.Yao, Y., Yang, Y. W., and Liu, J. Y. ( 2006) An efficient protein preparation for proteomic analysis of developing cotton fibers by two-dimensional gel electrophoresis. Electrophoresis 27, 4559–4569 [DOI] [PubMed] [Google Scholar]
- 23.Candiano, G., Bruschi, M., Musante, L., Santucci, L., Ghiggeri, G. M., Carnemolla, B., Orecchia, P., Zardi, L., and Righetti, P. G. ( 2004) Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis 25, 1327–1333 [DOI] [PubMed] [Google Scholar]
- 24.Yan, S. P., Zhang, Q. Y., Tang, Z. C., Su, W. A., and Sun, W. N. ( 2006) Comparative proteomic analysis provides new insights into chilling stress responses in rice. Mol. Cell. Proteomics 5, 484–496 [DOI] [PubMed] [Google Scholar]
- 25.Sweetlove, L. J., Heazlewood, J. L., Herald, V., Holtzapffel, R., Day, D. A., Leaver, C. J., and Millar, A. H. ( 2002) The impact of oxidative stress on Arabidopsis mitochondria. Plant J. 32, 891–904 [DOI] [PubMed] [Google Scholar]
- 26.Taylor, N. L., Heazlewood, J. L., Day, D. A., and Millar, A. H. ( 2005) Differential impact of environmental stresses on the pea mitochondrial proteome. Mol. Cell. Proteomics 4, 1122–1133 [DOI] [PubMed] [Google Scholar]
- 27.Kim, D. W., Rakwal, R., Agrawal, G. K., Jung, Y. H., Shibato, J., Jwa, N. S., Iwahashi, Y., Iwahashi, H., Kim, D. H., Shim, I. S., and Usui, K. ( 2005) A hydroponic rice seedling culture model system for investigating proteome of salt stress in rice leaf. Electrophoresis 26, 4521–4539 [DOI] [PubMed] [Google Scholar]
- 28.Ahsan, N., Lee, D. G., Lee, S. H., Kang, K. Y., Lee, J. J., Kim, P. J., Yoon, H. S., Kim, J. S., and Lee, B. H. ( 2007) Excess copper induced physiological and proteomic changes in germinating rice seeds. Chemosphere 67, 1182–1193 [DOI] [PubMed] [Google Scholar]
- 29.Salekdeh, G. H., Siopongco, J., Wade, L. J., Ghareyazie, B., and Bennett, J. ( 2002) Proteomic analysis of rice leaves during drought stress and recovery. Proteomics 2, 1131–1145 [DOI] [PubMed] [Google Scholar]
- 30.Weeks, M. E., Sinclair, J., Butt, A., Chung, Y. L., Worthington, J. L., Wilkinson, C. R. M., Griffiths, J., Jones, N., Waterfield, M. D., and Timms, J. F. ( 2006) A parallel proteomic and metabolomic analysis of the hydrogen peroxide- and Sty1p-dependent stress response in Schizosaccharomyces pombe. Proteomics 6, 2772–2796 [DOI] [PubMed] [Google Scholar]
- 31.Gallie, D. R., Le, H., Caldwell, C., Tanguay, R. L., Hoang, N. X., and Browning, K. S. ( 1997) The phosphorylation state of translation initiation factors is regulated developmentally and following heat shock in wheat. J. Biol. Chem. 272, 1046–1053 [DOI] [PubMed] [Google Scholar]
- 32.Takahashi, H., Iwai, M., Takahashi, Y., and Minagawa, J. ( 2006) Identification of the mobile light-harvesting complex II polypeptides for state transitions in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U. S. A. 103, 477–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mozzo, M., Morosinotto, T., Bassi, R., and Croce, R. ( 2006) Probing the structure of Lhca3 by mutation analysis. Biochim. Biophys. Acta 1757, 1607–1613 [DOI] [PubMed] [Google Scholar]
- 34.Makino, A., Nakano, H., Mae, T., and Shimada, T., and Yamamoto, N. ( 2000) Photosynthesis, plant growth and N allocation in transgenic rice plants with decreased Rubisco under CO2 enrichment. J. Exp. Bot. 51, 383–389 [DOI] [PubMed] [Google Scholar]
- 35.Zhao, C. F., Wang, J. Q., Cao, M. L., Zhao, K., Shao, J. M., Lei, T. T., Yin, J. N., Hill, G. G., Xu, N. Z., and Liu, S. Q. ( 2005) Proteomic changes in rice leaves during development of field-grown rice plants. Proteomics 5, 961–972 [DOI] [PubMed] [Google Scholar]
- 36.Marin-Navarro, J., and Moreno, J. ( 2003) Modification of the proteolytic fragmentation pattern upon oxidation of cysteines from ribulose 1,5-bisphosphate carboxylase/oxygenase. Biochemistry 42, 14930–14938 [DOI] [PubMed] [Google Scholar]
- 37.Cui, S. X., Huang, F., Wang, J., Ma, X., Cheng, Y. S., and Liu, J. Y. ( 2005) A proteomic analysis of cold stress responses in rice seedlings. Proteomics 5, 3162–3172 [DOI] [PubMed] [Google Scholar]
- 38.Janosi, L., Mottagui-Tabar, S., Isaksson, L. A., Sekine, Y., Ohtsubo, E., Zhang, S. J., Goon, S., Nelken, S., Shuda, M., and Kaji, A. ( 1998) Evidence for in vivo ribosome recycling, the fourth step in protein biosynthesis. EMBO J. 17, 1141–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tanahashi, N., Tsurumi, C., Tamura, T., and Tanaka, K. ( 1993) Molecular structures of 20S and 26S proteasomes. Enzyme Protein 47, 241–251 [DOI] [PubMed] [Google Scholar]
- 40.Novak, P., and Dev, I. K. ( 1988) Degradation of a signal peptide by protease IV and oligopeptidase A. J. Bacteriol. 170, 5067–5075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maurizi, M. R., Clark, W. P., Katayama, Y., Rudikoff, S., Pumphrey, J., Bowers, B., and Gottesman, S. ( 1990) Sequence and structure of ClpP, the proteolytic component of the ATP-dependent Clp protease of Escherichia coli. J. Biol. Chem. 265, 12536–12545 [PubMed] [Google Scholar]
- 42.Tu, C. J., Park, S. Y., and Walling, L. L. ( 2003) Isolation and characterization of the neutral leucine aminopeptidase (LapN) of tomato. Plant Physiol. 132, 243–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takken, F. L. W., Albrecht, M., and Tameling, W. I. L. ( 2006) Resistance proteins: molecular switches of plant defence. Curr. Opin. Plant Biol. 9, 383–390 [DOI] [PubMed] [Google Scholar]
- 44.Zhou, F. S., Kurth, J., Wei, F. S., Elliott, C., Valè, G., Yahiaoui, N., Keller, B., Somerville, S., Wise, R., and Schulze-Lefert, P. ( 2001) Cell-autonomous expression of barley Mla1 confers race-specific resistance to the powdery mildew fungus via a Rar1-independent signaling pathway. Plant Cell 13, 337–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Torregrosa, C., Cluzet, S., Fournier, J., Huguet, T., Gamas, P., Prospéri, J. M., Esquerré-Tugayé, M. T., Dumas, B., and Jacquet, C. ( 2004) Cytological, genetic, and molecular analysis to characterize compatible and incompatible interactions between Medicago truncatula and Colletotrichum trifolii. Mol. Plant-Microbe Interact. 17, 909–920 [DOI] [PubMed] [Google Scholar]
- 46.Deepak, S., Shailasree, S., Kini, R. K., Hause, B., Shetty, S. H., and Mithofer, A. ( 2007) Role of hydroxyproline-rich glycoproteins in resistance of pearl millet against downy mildew pathogen Sclerospora graminicola. Planta 226, 323–333 [DOI] [PubMed] [Google Scholar]
- 47.Charron, J. B. F., Breton, G., Badawi, M., and Sarhan, F. ( 2002) Molecular and structural analysis of a novel temperature stress-induced lipocalin from wheat and Arabidopsis. FEBS Lett. 517, 129–132 [DOI] [PubMed] [Google Scholar]
- 48.Dixon, D. P., Lapthorn, A., and Edwards, R. ( 2002) Plant glutathione transferases. Genome Biol. 3, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gilbert, H. F. ( 1997) Protein disulfide isomerase and assisted protein folding. J. Biol. Chem. 272, 29399–29402 [DOI] [PubMed] [Google Scholar]
- 50.Espartero, J., Sanchez-Aguayo, I., and Pardo, J. M. ( 1995) Molecular characterization of glyoxalase-I from a higher plant; upregulation by stress. Plant Mol. Biol. 29, 1223–1233 [DOI] [PubMed] [Google Scholar]
- 51.Akileswaran, L., Brock, B. J., Cereghino, J. L., and Gold, M. H. ( 1999) 1,4-Benzoquinone reductase from Phanerochaete chrysosporium: cDNA cloning and regulation of expression. Appl. Environ. Microbiol. 65, 415–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rouhier, N., Vieira Dos Santos, C., Tarrago, L., and Rey, P. ( 2006) Plant methionine sulfoxide reductase A and B multigenic families. Photosynth. Res. 89, 247–262 [DOI] [PubMed] [Google Scholar]
- 53.Schaller, F., Biesgen, C., Müssig, C., Altmann, T., and Weiler, E. W. ( 2000) 12-Oxophytodienoate reductase 3 (OPR3) is the isoenzyme involved in jasmonate biosynthesis. Planta 210, 979–984 [DOI] [PubMed] [Google Scholar]
- 54.Fath, A., Bethke, P. C., and Jones, R. L. ( 2001) Enzymes that scavenge reactive oxygen species are down-regulated prior to gibberellic acid-induced programmed cell death in barley aleurone. Plant Physiol. 126, 156–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Coursol, S., Fan, L. M., Stunff, H. L., Spiegel, S., Gilroy, S., and Assmann, S. M. ( 2003) Sphingolipid signalling in Arabidopsis guard cells involves heterotrimeric G proteins. Nature 423, 651–654 [DOI] [PubMed] [Google Scholar]
- 56.Trusov, Y., Rookes, J. E., Chakravorty, D., Armour, D., Schenk, P. M., and Botella, J. R. ( 2006) Heterotrimeric G proteins facilitate Arabidopsis resistance to Necrotrophic pathogens and are involved in Jasmonate signaling. Plant Physiol. 140, 210–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Luan, S. ( 1998) Protein phosphatases and signaling cascades in higher plants. Trends Plant Sci. 3, 271–275 [Google Scholar]
- 58.Yang, H. J., Li, Y. Q., and Hua, J. ( 2006) The C2 domain protein BAP1 negatively regulates defense responses in Arabidopsis. Plant J. 48, 238–248 [DOI] [PubMed] [Google Scholar]
- 59.Hoshida, H., Tanaka, Y., Hibino, T., Hayashi, Y., Tanaka, A., Takabe, T., and Takabe, T. ( 2000) Enhanced tolerance to salt stress in transgenic rice that overexpresses chloroplast glutamine synthetase. Plant Mol. Biol. 43, 103–111 [DOI] [PubMed] [Google Scholar]
- 60.Espartero, J., Pintor-Toro, J. A., and Pardo, J. M. ( 1994) Differential accumulation of S-adenosyl-methionine synthetase transcripts in response to salt stress. Plant Mol. Biol. 25, 217–227 [DOI] [PubMed] [Google Scholar]
- 61.May, M. J., Vernoux, T., Leaver, C., Van Montagu, M., and Inze, D. ( 1998) Glutathione homeostasis in plants: implications for environmental sensing and plant development. J. Exp. Bot. 49, 649–667 [Google Scholar]
- 62.Mou, Z., He, Y., Dai, Y., Liu, X., and Li, J. Y. ( 2000) Deficiency in fatty acid synthase leads to premature cell death and dramatic alterations in plant morphology. Plant Cell 12, 405–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hashimoto, M., and Komatsu, S. ( 2007) Proteomic analysis of rice seedlings during cold stress. Proteomics 7, 1293–1302 [DOI] [PubMed] [Google Scholar]
- 64.Rakwal, R., Agrawal, G. K., and Yonekura, M. ( 1999) Separation of proteins from stressed rice (Oryza sativa L.) leaf tissues by two-dimensional polyacrylamide gel electrophoresis: induction of pathogenesis-related and cellular protectant proteins by jasmonic acid, UV irradiation and copper chloride. Electrophoresis 20, 3472–3478 [DOI] [PubMed] [Google Scholar]
- 65.Konishi, H., and Komatsu, S. ( 2003) A proteomic approach to investing promotive effects of brassinolide on lamina inclination and root growth in rice seedling. Biol. Pharm. Bull. 26, 401–408 [DOI] [PubMed] [Google Scholar]
- 66.Rakwal, R., and Komatsu, S. ( 2004) Abscisic acid promoted changes in the protein profiles of rice seedling by proteome analysis. Mol. Biol. Rep. 31, 217–230 [DOI] [PubMed] [Google Scholar]
- 67.Agrawal, G. K., Rakwal, R., Yonekura, M., Kubo, A., and Saji, H. ( 2002) Proteome analysis of differentially displayed proteins as a tool for investigating ozone stress in rice (Oryza sativa L.) seedlings. Proteomics 2, 947–959 [DOI] [PubMed] [Google Scholar]
- 68.Rakwal, R., Agrawal, G. K., Kubo, A., Yonekura, M., Tamogami, S., Saji, H., and Iwahashi, H. ( 2003) Defense/stress responses elicited in rice seedlings exposed to the gaseous air pollutant sulfur dioxide. Environ. Exp. Bot. 49, 223–235 [Google Scholar]
- 69.Hajduch, M., Rakwal, R., Agrawal, G. K., Yonekura, M., and Pretova, A. ( 2001) High-resolution two-dimensional electrophoresis separation of proteins from metal-stressed rice (Oryza sativa L.) leaves: drastic reductions/fragmentation of ribulose-1,5-bisphosphate carboxylase/oxygenase and induction of stress-related proteins. Electrophoresis 22, 2824–2831 [DOI] [PubMed] [Google Scholar]
- 70.Pastori, G. M., and Foyer, C. H. ( 2002) Common components, networks, and pathways of cross-tolerance to stress. The central role of “redox” and abscisic acid-mediated controls. Plant Physiol. 129, 460–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Trewavas, A. ( 2005) Green plants as intelligent organisms. Trends Plant Sci. 10, 413–419 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.