Abstract
Liquid chromatography combined with electrospray ionization is widely used for direct analysis of polar and labile molecules by LCMS. The on-line coupling in LCMS is a major strength but also causes a principal limitation that each eluting analyte has to be analyzed immediately and is not available for detailed interrogation after the LCMS run. Here we developed a new chromatographic strategy, which removes this limitation. After column separation the flow is split, one portion is analyzed directly, and the other is diverted to a capture capillary. After the direct LCMS run, the flow is switched, and the portion stored in the capillary is analyzed (“replay run”). We describe a setup consisting of an analytical column, a splitting valve, and a focusing column, which performs at full sensitivity and undiminished chromatographic resolution. We demonstrate three principal advantages of this system: nearly continuous MS utilization, duplicate analysis without requirement for additional sample, and targeting of important but undersampled features in the replay run.
The development of electrospray mass spectrometry has allowed analysis directly from the liquid phase (1). This feature of electrospray makes it eminently suitable for the on-line coupling of separation and ionization before MS analysis. In nanoscale LCMS analyte species are concentrated into very small volumes, increasing sensitivity. Furthermore the excellent separation capacity of high performance chromatographic systems is multiplied with the high resolution of modern mass spectrometers, resulting in exceptional combined separation power (2). Small molecules, peptides, and proteins are routinely analyzed by this powerful technology. However, compared with off-line methods such as nanoelectrospray (3) or MALDI (4), on-line coupling also has some inherent drawbacks. The short time during elution of a peak requires a fast and automatic decision on which peak to sequence. In complex mixtures, many peptides co-elute, and some may not be sequenced at all (5). Peptides of special interest, for example, those with regulatory post-translational modifications, should be characterized in depth, but the fact that they are important may only become evident after the analysis.
Some of these drawbacks can in principle be addressed by slowing down the flow (“peak parking” (6)), fraction collection, or repeat injection. However, none of these methods is ideal from an analytical standpoint. Peak parking is of limited utility for complex mixtures because the flow may have to be stopped every few seconds, and the run would be extended to impractical lengths. Fraction collection is useful in many instances, but at very low flow rates it is less practical because of the low volume of fractions. In nanoflow LC peptides typically elute in about 50 nl (based on an elution time of 15 s using a 200 nl/min flow rate). To handle these fractions one would need ∼1 μl of sample, but adding buffer reduces the concentration 20-fold causing a dramatic loss of signal intensity. Repeat injections multiply total required analysis time and may not be optimal if sample is limited because reinjection consumes twice as much sample.
We wished to develop a novel concept in LCMS that would allow targeted measurement of analyte mixtures without compromising sensitivity or chromatographic performance while requiring little or no additional time. Because electrospray is a concentration-dependent process and therefore maintains full signal at decreased flow rates, we and others previously developed splitting systems in which the column effluent was directed to MS analysis as well as to a fraction collector to enable the reanalysis of chromatographic fractions (7, 8). However, at very low flow rates, fraction collection became increasingly difficult, prompting us to explore alternative ways of storing the chromatographically separated sample.
Here we describe a novel setup in which we collect part of a column effluent in a long capture capillary that we reanalyze after the direct run. In this study we describe the new concept and evaluate the system in terms of chromatographic and mass spectrometric performance. Furthermore we show the applicability to a complex proteomics sample and demonstrate a useful application: targeting important peptides that were not characterized in sufficient detail in the direct run. A “head to head” and exhaustive comparison with other possible LCMS setups or MALDI methods is not the subject of this study.
MATERIALS AND METHODS
RePlay Setup—
We constructed the “RePlay system” consisting of a six-port splitting valve, a flow sensor, a long capillary serving as a capture capillary, and short capillaries to adjust the split ratio (see Fig. 1). The splitting valve was specially constructed for accurate flow ratios and extremely low dead volumes at nl/min flow rates (Advion BioSystems, Ithaca, NY). LC was performed on a Nano-HPLC 1200 system (Agilent, Waldbronn, Germany) with a 10-cm-long 75-μm-inner diameter IntegraFrit™ ProteoPepII analytical column (5-μm RP-C18 resin, New Objective, Woburn, MA) coupled to the RePlay valve in which the flow was split. The gradients were essentially as described previously (12) with peptides eluting from 13 to 60% solvent B (0.5% acetic acid in 80% acetonitrile). One part of the effluent was directed to a 7-cm in-house pulled 75-μm-inner diameter fused silica emitter packed with ReproSil-Pur C18-AQ 3-μm resin (Dr. Maisch GmbH, Ammerbuch-Entringen, Germany), termed the “focusing column” for direct on-line LCMS analysis on an LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). Meanwhile the second part of the effluent was collected in the fused silica capture capillary of typically 13-m length and 30-μm inner diameter. The capture line (Composite Metal Services Ltd., Shipley, UK) was capable of storing around 10 μl, appropriate for a 100-min gradient at an effective flow rate of 100 nl/min. For the 30-min gradient a 3-m capillary was used. This stored gradient was then directed to the same 7-cm pulled column by valve switching triggered from the MS acquisition software.
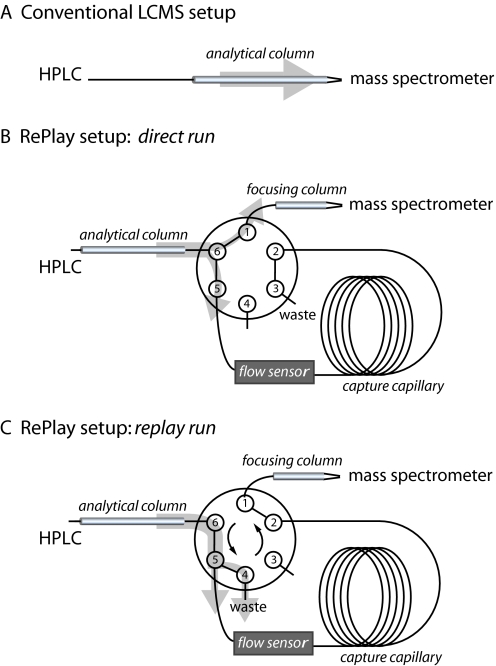
Fig. 1.
Schematics of the conventional LCMS and the RePlay setup. A, conventional LCMS setup using a column with an integrated emitter placed in front of the mass spectrometer. The light gray arrow indicates the direction of the flow. B, in the “direct run” of the RePlay setup the six-port splitting valve is positioned such that part of the effluent of the first nano-LC column (“analytical column”) flows to the mass spectrometer via a short second column (“focusing column”). The other part is stored in a long capture capillary, which has the volume appropriate to hold the complete gradient. C, when the valve is switched the stored gradient is directed to the mass spectrometer (“replay run”). Capture capillaries of different lengths serve as split adjusters, at port 3 in the direct run and port 4 in the RePlay run, and control the split ratio, which is read out by the flow sensor. The split adjuster at port 4 can be replaced by a plug, but a split allows higher flow rates and reduces the time used for washing and loading of the analytical column.
Split adjusters, 10-μm-inner diameter capillaries (Composite Metal Services Ltd.), were cut to the length resulting in the desired split ratio. These split adjusters needed to be fine tuned whenever the focusing column was replaced. We recommend to carefully tighten, not to overtighten, these capillaries because we found that small glass particles easily clog the capillaries with a small inner diameter (used as split adjusters).
For calculation of the theoretical peak broadening we used the “capillary flow calculator” option of the “Molecular Weight Calculator” freely available as open source software (alchemistmatt). This program calculates the width of the peak as a consequence of diffusion when inserting the initial peak width, the length and inner diameter of the capillary, and the flow rate.
Liver Proteome Analysis—
Frozen mouse liver tissue was homogenized and in-solution digested by trypsin as described previously (9), desalted and concentrated on in-house prepared StageTips (10), and eluted for LCMS analysis. With a split ratio of 1:1, we ran a 100-min gradient (100 nl/min to MS) and reanalyzed the sample in another 100 min (100 nl/min). We also split 3:1 (direct:replay), used a 100-min direct gradient (200 nl/min to MS), and reanalyzed the sample in 35 min (200 nl/min). During the replay run the LC system loaded the next sample on the analytical column. The run was analyzed by in-house developed MaxQuant software (version 1.07.5) essentially as described previously (11). The data were searched using Mascot (version 2.1.04, Matrix Science Ltd., London, UK) against the mouse International Protein Index database (version 3.37) supplemented with frequently observed contaminants and concatenated with reversed copies of all sequences (2 × 51,467 entries). Enzyme specificity was set to trypsin, allowing for cleavage N-terminal to proline and between aspartic acid and proline (12). Carbamidomethylcysteine was set as a fixed modification, and N-acetylation and methionine oxidation were set as variable modifications. The initial maximum allowed mass deviation (13) was set to 5 ppm for monoisotopic precursor ions and 0.5 Da for MS/MS peaks. Maximally three missed cleavages were allowed. The false positive rate at the peptide level and false discovery rate at the protein level were set to 1%, and the required minimum peptide length was 6 amino acids. If the identified peptide sequence set of one protein was equal to or contained the peptide set of another protein, these two proteins were grouped together by MaxQuant, and they were not counted as independent protein hits. Proteins were considered identified when at least two peptides were identified (of which one was uniquely assignable to the respective sequence).
Determination of Small Ubiquitin-like Modifier-2 Interaction Partners—
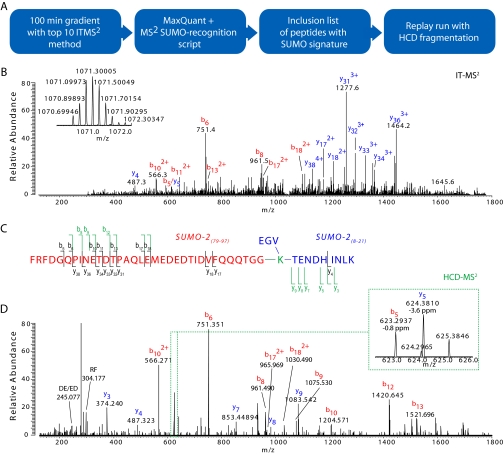
His6-SUMO-21 conjugates were purified as described previously (14). The samples were subsequently digested by trypsin and concentrated on in-house made StageTips (10). After measuring the direct run with a “top 10” method using fragmentation of the 10 most abundant ions in the ion trap, a peak list was created with MaxQuant software (version 1.07.5) using default parameters. This list was screened for precursors that when fragmented showed ions with m/z corresponding to SUMO-2 b-ions in the range of b5–b18. Of the precursors that had more than three fragment matches and a total mass larger than a free SUMO-2 peptide, the highest expected isotope peak was added to an inclusion list. In the replay run only these ions were sequenced by use of higher collision dissociation (HCD) fragmentation (15). We deselected charge state, precursor mass screening, and dynamic exclusion and fragmented all ions matching an inclusion list item with a 30-ppm tolerance and with a minimum intensity of 30,000. These high resolution fragmentation spectra were validated manually.
RESULTS AND DISCUSSION
RePlay Setup—
In a first iteration of our systems we used a storage capillary instead of the fraction collector, and we reversed the flow for reanalysis of the stored gradient. This arrangement inverted the order of the elution and resulted in good resolution of late eluting peaks (those stored in the capillary for a short time) but not of early eluting peaks (supplemental Fig. 1). To enable first in first out analysis, we combined the functions of flow splitting and redirection of the flow into a single device, a low flow, low dead volume splitting valve (Fig. 1). Finally to prevent possible deterioration of chromatographic performance in the second run due to diffusion in the capture capillary, we introduced a second “focusing” column prior to the electrospray emitter. We termed this system “RePlay.” The effluent from the analytical column was split in the appropriate, fixed ratio for direct analysis in the mass spectrometer and for storage in a capture capillary (Fig. 1A). At the end of the gradient, the valve was switched, and the stored gradient separation was eluted through the same focusing column as in the direct mode (Fig. 1B). We tested the performance of the RePlay system (Advion BioSystems, Ithaca, NY) with a BSA standard (Sigma-Aldrich). As shown in Fig. 2, peak intensity and chromatographic resolution were very similar between direct and RePlay analysis with no deterioration over the gradient (Fig. 2C). The fact that the signal in electrospray is concentration-dependent rather than dependent on total analyte amount readily explains this feature of the RePlay setup.
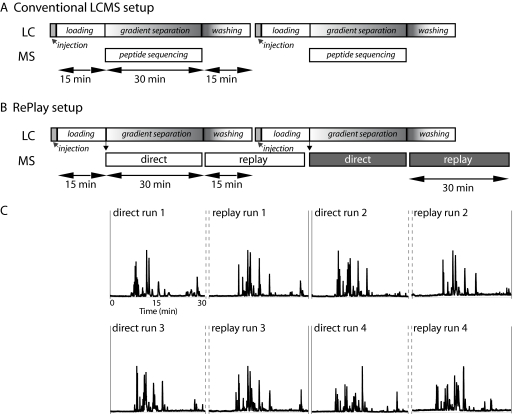
Fig. 2.
Schematics of a conventional LCMS and RePlay setup and base peak chromatograms of four consecutively acquired replay runs with 30-min gradients. A, in the conventional LCMS setup the mass spectrometer sequences effectively only 50% of the time when using short gradients. B, in the RePlay setup the second analysis can be carried out while the LC system is washing and loading the analytical column. In this way no extra time is needed. C, BSA chromatograms of four replay runs measured in a sequence. Chromatographic performance of the direct and RePlay runs was very similar.
Chromatographic Performance—
We tested different split ratios of the total flow (typically 200 nl/min) between the direct and replay runs from 1:4 to 3:1 and found that the chromatographic performance was excellent with splits in this range. For the experiments reported in this study, we chose a split ratio of 1:1 or 3:1 to keep the volume of the capture capillary small and RePlay analysis time short. To test for potential detrimental effects of diffusion, we varied the inner diameter of the capillary between 20 and 40 μm and stored the gradient in the capture capillary for up to 30 min. Theoretically using a flow rate of 100 nl/min, a peak of 20 s measured at the base would diffuse into 33 s in a 20-μm-inner diameter capillary, whereas the same peak would broaden to 45 s in a 30-μm-inner diameter tubing and to 57 s in a capillary of 40-μm inner diameter. All data presented in this study were acquired using a 30-μm inner diameter capture capillary, and for this capillary no significant influence on chromatographic performance was observed. This indicates that the diffusion can still be reversed by the focusing column under these conditions. However, when we did not use the focusing column we indeed observed appreciable peak broadening.
Figs. 2, A and B, and 4A illustrate one of the advantages of the RePlay system compared with the routine work flow in our laboratory. During the replay run, the analytical column is re-equilibrated and loaded with a new sample. Thus, the mass spectrometer is continuously sequencing peptides, and the loading and washing steps do not subtract from the duty cycle of MS utilization. As shown in Fig. 2C, four runs were analyzed in 8 × 30 min with essentially 100% duty cycle, whereas without RePlay, the duty cycle is about 50%. For 100-min gradients we accomplished a duty cycle of 90% by directing one-quarter of the flow to the capture capillary and pushing the gradient out 3 times faster (see Fig. 4). However, to ensure continued high chromatographic resolution we found it advantageous to include a wash and equilibration of the focusing column for a few minutes in all protocols.
Fig. 4.
Timing schedule and base peak chromatogram of a setup using a 3:1 split and a fast replay run demonstrating excellent chromatographic performance in both runs without using additional time. A, timing schedule of two automated replay runs with the nano-LC and MS systems operating asynchronously. After sample loading and before starting the gradient the LC system starts the mass spectrometer. After finishing the gradient, the MS system measures the replay run while the LC system loads the next sample. B, liver digests were run with a direct gradient of effectively 100 min and with a 33-min duration for the replay run. In nearly 90% of the measurement time, the mass spectrometer sequenced peptide ions. C, as shown in the selected ion chromatogram, the elution patterns and intensity of the direct and replay runs are very similar. Every colored symbol indicates a different peptide peak.
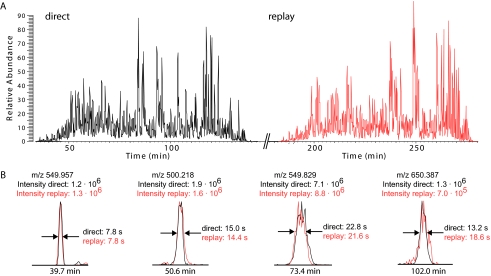
RePlay Analysis of Complex Proteomics Samples—
Next we characterized the performance of the RePlay system for complex peptide mixtures typical of proteomics experiments. We loaded slightly less than 1 μg of mouse liver tryptic digest, corresponding to less than 20,000 hepatocyte cells, onto the RePlay system. Analysis was performed with a 100-min gradient by a standard “top 5” method on the LTQ-Orbitrap as routinely used in our laboratory (16). The base peak chromatogram in Fig. 3 shows that the peptide pattern is largely indistinguishable between the direct and replay runs. Peaks eluted on average within 15 s in the direct run and 16 s in the replay run (with standard deviations of 5 and 7 s, respectively; see Fig. 3B). This is as good or better a performance than our standard one-column setup for proteomics experiments. At a false discovery rate of 1%, 6,535 fully tryptic peptides were identified in the direct run, and 5,936 were identified in the replay run, documenting the high reproducibility between the runs. A total of 8,383 unique peptides were identified in the combined analysis. Although this experiment was meant to investigate reproducibility, the replay run nevertheless added 1,848 peptides (28%) not identified in the direct run. On the protein level, the RePlay system identified 1,093 proteins with two unique peptides and at a false discovery rate of 1%, demonstrating that it is well suited to complex proteomics samples. This performance will likely further improve if the mass spectrometer is programmed to sequence only peptides in the replay run that have not been sequenced in the direct run.
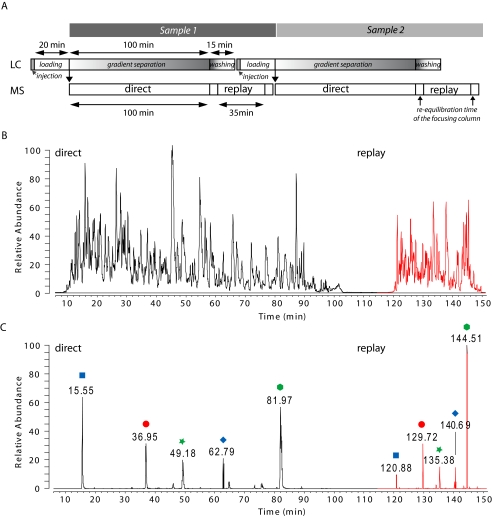
Fig. 3.
Chromatographic performance in complex proteome analysis with the RePlay system. A, typical base peak chromatogram of a RePlay analysis consisting of 100-min direct (black) and 100-min replay runs (red) indicating the similarity of the chromatography performance. One microgram of tryptic digest of liver homogenate was measured with the RePlay system using a 1:1 split (100 nl/min direct:100 nl/min RePlay). B, selected ion chromatograms (0.01 m/z units wide) of four peptide peaks selected over the length of the gradient showing essentially identical signal intensities and peak widths at half-height in direct and replay runs.
To demonstrate the ability of reanalyzing a complex mixture without adding to total analysis time, we split one-quarter of the flow into the capture capillary (Fig. 4). The total flow was 270 nl/min during direct analysis (200 nl/min to the focusing column) that lasted for 100 min. During the replay run the flow to the focusing column was also 200 nl/min, “squeezing” the reanalysis into 35 min. Fig. 4, B and C, show that this regime preserved peak intensities and slightly sharpened chromatographic peaks. Note that total analysis time was 150 min, very close to the 140-min standard cycle used in our laboratory in which peptides elute for about 100 min.
Targeting of SUMO-2 Substrates in the Replay Run—
On-line reanalysis also allows targeting important peptides that were missed or unidentified in the direct analysis. We demonstrate this principle by targeted reanalysis of peptides that are extremely difficult to identify by tandem mass spectrometry. In our work investigating the conjugation of substrates with ubiquitin family proteins, we looked for specific proteins that are in vivo sumoylated by SUMO-2 (14). Unlike ubiquitin, SUMO-2 leaves a large 32- or 34-amino acid peptide on the substrate peptide after tryptic digest, making it highly charged. The complex fragmentation spectra of the cross-linked peptides are notoriously difficult to identify with low resolution data (17). For confident identification of the sumoylated peptide sequence, confirmation of high resolution and high accuracy data is required. To identify these conjugated SUMO-2 peptides in a streamlined fashion we purified His6-SUMO-2 conjugates from HeLa lysate and measured them in the standard way in the direct run. Low resolution fragmentation spectra from the linear ion trap that indicated possible SUMO-2-conjugated peptides were then targeted in the replay run. In this run we only targeted these potential substrate peptides, fragmented them by HCD (15), and analyzed them with high resolution and mass accuracy in the orbitrap instead of in the ion trap. Note that this analysis is slower and less sensitive and therefore could not have been performed efficiently in the direct run. Fig. 5 shows the setup of the experiment as well as results for a SUMO-2-conjugated peptide. In the replay run, but not in the direct run, charge states and fragment masses are determined with very high accuracy (Fig. 5D, inset). Interestingly the ion trap CID did not fragment the crucial substrate part of the cross-linked peptide (Fig. 5B); therefore reanalysis with a different fragmentation method (HCD) was especially beneficial in this case (Fig. 5D). The RePlay analysis produced an essentially complete run of y-ions of the substrate peptide.
Fig. 5.
Targeted experiment to identify SUMO-2 substrates with high confidence. A, after a 100-min gradient with an MS method sequencing the 10 most abundant peaks in the ion trap (IT) by CID, a peak list of MS2 fragments was generated. This peak list was checked for peptides that contained multiple b-ions of the SUMO-2 C-terminal peptide, and the corresponding peptide precursor masses were inserted in an inclusion list. These peptides were then specifically targeted by HCD and detected in the orbitrap to obtain high resolution and high mass accuracy data. B, annotated ion trap fragmentation spectrum of a SUMO-2-SUMO-2 conjugate, the orbitrap precursor of which is shown as an inset. Because of low resolution, charge states cannot be assigned, and masses are inaccurate. C, sequence of SUMO-2-SUMO-2 peptide, indicating fragments observed in the ion trap (black and at half-height) and in the orbitrap (green and at full height). D, HCD fragmentation spectrum of the same ion with orbitrap resolution set to 7,500. Charge state can easily be assigned, and the average absolute mass accuracy was in the low ppm range. The inset shows excellent quality of the fragmentation spectrum. Given the identification from the spectrum (D), the quintuply charged precursor ion in B matches within 1.35 ppm.
This experiment was performed with a 3:1 split ratio where the RePlay analysis was kept entirely within the time usually needed for washing and loading but that would suffice for characterizing a handful of interesting peptides. In this way, the mass spectrometer could, instead of standing idle, acquire targeted and very high quality data “for free.”
Conclusion—
In summary, we have demonstrated that the RePlay system enables very efficient use of the mass spectrometer, which is an important advantage given the high demand on sequencing time in large scale proteomics laboratories and the costs of high resolution mass spectrometers. We also demonstrated a “targeted analysis” in which the replay run enabled the collection of crucial, complementary data, thereby dramatically enhancing the information obtained in the direct run.
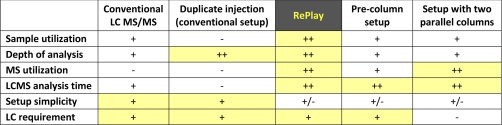
The great value of the RePlay system lies within the combination of several conceptual features that are otherwise only available individually in separate setups. Table I illustrates the features of RePlay compared with several other possible formats for LCMS/MS experiments. Compared with a standard LCMS setup, RePlay allows a second analysis using no or little extra analysis time. A duplicate injection in a standard setup would also allow investigating a sample to greater depth, but in that case it comes at the expense of more sample usage and doubled analysis time. Compared with a setup with two parallel columns and LC systems, RePlay achieves nearly the same time utilization of the mass spectrometer without requiring an additional pump or LC system. Similarly in the case of performing dual injections with dual precolumn setup one would require twice the sample amount and an extra (or more complex) LC system. Not included in the table are fraction collection and peak parking, which are impractical for high throughput proteomics for reasons described in the Introduction.
Table I.
Comparison of conceptual parameters for different LCMS setups
The normal nanoflow LCMS/MS setup is taken as a reference. The most favorable settings are highlighted in yellow.
The only analytical cost of the RePlay system is a higher complexity of the setup. The split needs to be fine tuned whenever pressure changes after the split occur. However, we found the system to be robust, it did not lead to any loss of signal, and the chromatographic performance was better than or equivalent to our standard setup.
Most of the measurement time in standard LCMS/MS is currently spent on obtaining information that later on turns out to be uninteresting. For example, one may be interested in only the peak pairs with unequal ratios in isotope-based quantitative proteomics, or one may only be interested in modified peptides. We envision that the RePlay system will be mainly used to analyze such interesting features in a focused way. This was demonstrated here with the example of the SUMO-cross-linked peptides. However, dedicated acquisition software should make a multitude of interesting features accessible to detailed proteomics analysis. For this purpose, the system will probably be configured to perform the replay analysis in a short time during washing and loading of the analytical column.
Beyond these applications, the system opens up a number of attractive possibilities, including ultrasensitive nanoscale proteomics. In this application, the flow would be split asymmetrically, allowing a very long analysis time in the replay run. Importantly the LC system would still work at normal nanoflow rates (∼200–500 nl/min), but the effective flow to the MS can be reduced by a factor of 5–10 without wasting any sample. This enables two analyses with ultrahigh sensitivity of a single injection and may be very beneficial for samples where the amount is limiting, for example in studies where cells or tissue samples are obtained by laser capture microdissection. In conclusion, we predict that the RePlay system will become a powerful and universal addition to the LCMS tool chest.
Supplementary Material
Acknowledgments
Ivan Matic and Alfred C. O. Vertegaal kindly provided the SUMO-2 conjugates and help with SUMO-2 analysis, and Yong Zhang provided software assistance for targeting SUMO-2 substrates. We thank Peter Bandilla for technical assistance and constructive comments as well as other members of the department for Proteomics and Signal Transduction and of Advion Biosciences.
Footnotes
Published, MCP Papers in Press, April 29, 2008, DOI 10.1074/mcp.M800141-MCP200
The abbreviations used are: SUMO, small ubiquitin-like modifier; HCD, higher energy collision dissociation.
This work was supported by the Max Planck Society. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Fenn, J. B., Mann, M., Meng, C. K., Wong, S. F., and Whitehouse, C. M. ( 1989) Electrospray ionization for mass spectrometry of large biomolecules. Science 246, 64–71 [DOI] [PubMed] [Google Scholar]
- 2.Hernandez-Borges, J., Aturki, Z., Rocco, A., and Fanali, S. ( 2007) Recent applications in nanoliquid chromatography. J. Sep. Sci. 30, 1589–1610 [DOI] [PubMed] [Google Scholar]
- 3.Wilm, M., and Mann, M. ( 1996) Analytical properties of the nanoelectrospray ion source. Anal. Chem. 68, 1–8 [DOI] [PubMed] [Google Scholar]
- 4.Karas, M., and Hillenkamp, F. ( 1988) Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal. Chem. 60, 2299–2301 [DOI] [PubMed] [Google Scholar]
- 5.Kuster, B., Schirle, M., Mallick, P., and Aebersold, R. ( 2005) Scoring proteomes with proteotypic peptide probes. Nat. Rev. Mol. Cell Biol. 6, 577–583 [DOI] [PubMed] [Google Scholar]
- 6.Davis, M. T., Stahl, D. C., Hefta, S. A., and Lee, T. D. ( 1995) A microscale electrospray interface for on-line, capillary liquid chromatography/tandem mass spectrometry of complex peptide mixtures. Anal. Chem. 67, 4549–4556 [DOI] [PubMed] [Google Scholar]
- 7.Staack, R. F., Varesio, E., and Hopfgartner, G. ( 2005) The combination of liquid chromatography/tandem mass spectrometry and chip-based infusion for improved screening and characterization of drug metabolites. Rapid Commun. Mass Spectrom. 19, 618–626 [DOI] [PubMed] [Google Scholar]
- 8.Corso, T. N., Van Pelt, C. K., Li, J., Ptak, C., and Huang, X. ( 2006) Ultralow-volume fraction collection from NanoLC columns for mass spectrometric analysis of protein phosphorylation and glycosylation. Anal. Chem. 78, 2209–2219 [DOI] [PubMed] [Google Scholar]
- 9.Shi, R., Kumar, C., Zougman, A., Zhang, Y., Podtelejnikov, A., Cox, J., Wisniewski, J. R., and Mann, M. ( 2007) Analysis of the mouse liver proteome using advanced mass spectrometry. J. Proteome Res. 6, 2963–2972 [DOI] [PubMed] [Google Scholar]
- 10.Rappsilber, J., Ishihama, Y., and Mann, M. ( 2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 [DOI] [PubMed] [Google Scholar]
- 11.Graumann, J., Hubner, N. C., Kim, J. B., Ko, K., Moser, M., Kumar, C., Cox, J., Schoeler, H., and Mann, M. ( 2007) Stable isotope labeling by amino acids in cell culture (SILAC) and proteome quantitation of mouse embryonic stem cells to a depth of 5,111 proteins. Mol. Cell. Proteomics 7, 672–683 [DOI] [PubMed] [Google Scholar]
- 12.Olsen, J. V., Ong, S. E., and Mann, M. ( 2004) Trypsin cleaves exclusively C-terminal to arginine and lysine residues. Mol. Cell. Proteomics 3, 608–614 [DOI] [PubMed] [Google Scholar]
- 13.Zubarev, R., and Mann, M. ( 2007) On the proper use of mass accuracy in proteomics. Mol. Cell. Proteomics 6, 377–381 [DOI] [PubMed] [Google Scholar]
- 14.Matic, I., van Hagen, M., Schimmel, J., Macek, B., Ogg, S. C., Tatham, M. H., Hay, R. T., Lamond, A. I., Mann, M., and Vertegaal, A. C. ( 2008) In vivo identification of human small ubiquitin-like modifier polymerization sites by high accuracy mass spectrometry and an in vitro to in vivo strategy. Mol. Cell. Proteomics 7, 132–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olsen, J. V., Macek, B., Lange, O., Makarov, A., Horning, S., and Mann, M. ( 2007) Higher-energy C-trap dissociation for peptide modification analysis. Nat. Methods 4, 709–712 [DOI] [PubMed] [Google Scholar]
- 16.Olsen, J. V., de Godoy, L. M., Li, G., Macek, B., Mortensen, P., Pesch, R., Makarov, A., Lange, O., Horning, S., and Mann, M. ( 2005) Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics 4, 2010–2021 [DOI] [PubMed] [Google Scholar]
- 17.Pedrioli, P. G., Raught, B., Zhang, X. D., Rogers, R., Aitchison, J., Matunis, M., and Aebersold, R. ( 2006) Automated identification of SUMOylation sites using mass spectrometry and SUMmOn pattern recognition software. Nat. Methods 3, 533–539 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.