Abstract
Biomolecule phosphorylation by protein kinases is a fundamental cell signaling process in all living cells. Following the comprehensive cataloguing of the protein kinase complement of the human genome (Manning, G., Whyte, D. B., Martinez, R., Hunter, T., and Sudarsanam, S. (2002) The protein kinase complement of the human genome. Science 298, 1912–1934), this review will detail the state-of-the-art human and mouse kinase proteomes as provided in the UniProtKB/Swiss-Prot protein knowledgebase. The sequences of the 480 classical and up to 24 atypical protein kinases now believed to exist in the human genome and 484 classical and up to 24 atypical kinases within the mouse genome have been reviewed and, where necessary, revised. Extensive annotation has been added to each entry. In an era when a wealth of new databases is emerging on the Internet, UniProtKB/Swiss-Prot makes available to the scientific community the most up-to-date and in-depth annotation of these proteins with access to additional external resources linked from within each entry. Incorrect sequence annotations resulting from errors and artifacts have been eliminated. Each entry will be constantly reviewed and updated as new information becomes available with the orthologous enzymes in related species being annotated in a parallel effort and complete kinomes being completed as sequences become available. This ensures that the mammalian kinomes available from UniProtKB/Swiss-Prot are of a consistently high standard with each separate entry acting both as a valuable information resource and a central portal to a wealth of further detail via extensive cross-referencing.
In the late 1950s, the role of reversible phosphorylation in enzymatic regulation was recognized by Fischer et al. (1). Phosphorylation events are most commonly mediated by protein kinases, which transfer the γ-phosphate from nucleotides, usually ATP, via a phosphoester bond (O-phosphate) to the hydroxyl side chain of serine, threonine, or tyrosine residues on their protein substrates. Phosphates are bulky, negatively charged groups, and their addition to a protein can result in a profound change in its interactions with other molecules or subcellular location and/or to a conformational change of the protein itself. Kinase-mediated protein phosphorylation can be reversed through dephosphorylation after which the protein switches back to its original charge state and conformation. As protein conformation often determines function, the phosphorylation event may be considered a type of molecular switch, turning the activity of the molecule on or off. These reversible and dynamic phosphorylation events are under tight control, being governed by the opposing activities of protein kinases and protein phosphatases.
Eukaryotic protein kinases (ePKs)1 play a key role in cell communication pathways and in the transmission of information from outside the cell or between subcellular components within the cell. The ePKs constitute one of the largest mammalian gene families comprising ∼1.7–2.5% of genes in eukaryotic genomes. Most protein kinases belong to a single superfamily, containing a conserved ePK catalytic domain that consists of a mainly β-sheet, NH2-terminal subdomain and a larger α-helical COOH-terminal subdomain with the ATP-binding pocket situated between the two subdomains. Serine/threonine protein kinases constitute the majority of kinases (67%) within the human kinome; however, tyrosine protein kinases (17%) also play a key role in signaling mechanisms, particularly in cell-cell communication in multicellular organisms (2). The remainder, the atypical protein kinases (aPKs), lack sequence similarity to the ePK catalytic domain but are known to have catalytic functional activity.
THE CONTENT OF A UniProt KnowledgeBase (UniProtKB)/Swiss-Prot KINASE ENTRY
The UniProtKB consists of two sections (3). UniProtKB/Swiss-Prot contains records combining full manual annotation with computer-assisted, manually verified annotation performed by biologists and biochemists and based on published literature and sequence analysis. UniProtKB/TrEMBL contains records with computationally generated annotation and large scale functional characterization. UniProtKB/Swiss-Prot records provide an integrated presentation of annotations such as protein name and function, taxonomy, enzyme-specific information (catalytic activity, cofactors, metabolic pathway, and regulatory mechanisms), domains and sites, post-translational modifications, subcellular locations, tissue-specific or developmentally specific expression, interactions, and diseases. Literature citations provide evidence from experimental data, which, along with feedback information from contacted authors, are regarded as information of the highest value, and are constantly being added to each record as they become available.
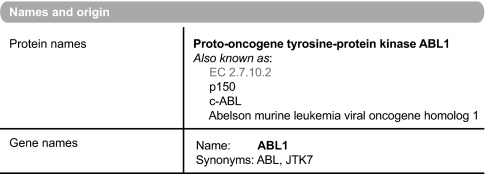
To aid the user and to enable text miners to make maximal use of this wealth of knowledge, this information is added to a UniProtKB/Swiss-Prot record in specific comments fields and, where possible, following a defined syntax and utilizing a controlled vocabulary. It is clearly indicated within the record when experimental evidence has been transferred from an orthologous protein in a closely related species. The references from which the data have been collected are retained within the entry, and the information extracted from that publication is also clearly described (Fig. 1).
Fig. 1.
Extraction from the annotation fields of ABL1 (P00519) in UniProtKB/Swiss-Prot.
As part of the manual curation process to create the UniProt/Swiss-Prot record for each gene product, each related sequence in the database is examined; splice variants and amino acid polymorphisms are identified, and sequencing errors, such as frameshifts or premature stop codons, are corrected. All the information is documented within the Swiss-Prot record such that the user may trace it back to its original source if required. A feature table at the end of each entry maps protein domains, active sites, binding sites, modified residues, and other sequence features onto the given sequence.
THE PROTEIN KINASE CONTENT OF THE HUMAN GENOME
In 2002 Manning et al. (2) predicted the existence of 518 typical protein-coding protein kinase genes within the human kinome based on the then current public and proprietary genomic DNA, complementary DNA, and expressed sequence tag sequences. These 518 kinases were further subdivided into 478 ePKs and 40 aPKs. At the time of the publication of that study, many of the sequences were not available in the public domain, and the sequencing of the human genome was far from complete. We now believe the number of ePKs to be 480 with the divergent isoforms of PRKG1 accessible in UniProtKB release 13.1 (March 18, 2008) from two separate entries (Q13976 and P14619). One sequence described by Manning et al. (2), SgK424, may have been an erroneous prediction in that we can find no firm evidence for its existence, although part of the sequence is identifiable within the current genome build, and a further three proteins were recognized subsequent to the Manning et al. (2) publication. Two gene products have now been identified within a duplicated genomic region that gives rise to two proteins, CDC2L1 (P21127) and CDC2L2 (Q9UQ88), that differ by only 15 amino acids (4). The second of these, CDC2L2, was not on the original list. PAN3 (Q58A45), a subunit of a poly(A)-specific ribonuclease complex (5), also contains a previously unidentified kinase domain, although this appears to be catalytically inactive. Finally PLK5 was originally found only in mouse, but a human homologue (Q496M5) has now been identified.
THE PROTEIN KINASE CONTENT OF THE MOUSE GENOME
An initial analysis of the mouse kinome was published by Caenepeel et al. (6) in 2004 in which they identified a complement of 540 gene products, 510 of which are orthologues of the human enzymes. Our analysis suggests a total of 484 ePKs. As with the human kinome, the 97 pseudogenes identified by Caenepeel et al. (6) have not been annotated in the UniProtKB/Swiss-Prot database. Eight kinases were originally identified as being present in human but not mouse; however, one of these, cyclin-dependent kinase 3 (CDK3) (Q80YP0), was identified as a transcribed mouse pseudogene with a single gene within the kinase domain near the T loop that is involved in activation by CDK-activating kinase and deletes motif X known to be required for kinase function (7). Consequently the truncated protein generates a null allele. This mutation is found in laboratory strains but not in wild-mice species such as Mus spretus and Mus mus castaneus. Because of the existence of at least one mRNA (BC116895 + BC119894) and one expressed sequence tag (BY709505) confirming the full-length protein CDK3, an annotated version in the UniProtKB/Swiss-Prot database has been made publicly available.
Finally Caenepeel et al. (6) described a group of microtubule affinity-regulating kinase-related CAMKs that could not be separated because of high sequence similarity and the, then, poor genome assembly. This group of kinases are encoded on the t-complex, a region of 20–30 Mb on the proximal third of mouse chromosome 17. Naturally occurring variant forms of the t-complex, known as complete t-haplotypes, are found in wild mouse populations. The t-haplotypes contain at least four nonoverlapping inversions that suppress recombination with the wild-type chromosome and lock into strong linkage disequilibrium loci affecting normal transmission of the chromosome, male gametogenesis, and embryonic development. To date, 10 protein kinases have now been identified in this region, all appearing to play a role in sperm motility.
ATYPICAL PROTEIN KINASES
Atypical protein kinases lack sequence similarity with the classical ePKs and are often deficient in the usual kinase motifs, although many possess a common kinase-like structural fold described by SUPERFAMILY hidden Markov model (InterPro entry IPR011009) (8), suggesting a shared ancestry for these proteins. A recent study of the structural evolution of the kinase family suggested that these atypical kinases diverged early in evolution to form a distinct phylogenetic group. The study encompassed a broader group of kinases, including enzymes capable of phosphorylating small molecules, such as choline kinase, and lipids. The exception to this appeared to be the α-kinases, such as α-protein kinase 1 (Q96QP1), a small family of enzymes that recognize phosphorylation sites in which the surrounding peptides have an α-helical conformation and that contain a zinc finger motif. This group appears to have arisen fairly recently in eukaryotes, and the authors speculate that this may be due to a single gene event such as a deletion of the COOH-terminal end of the gene or a gene fusion event. Members of this family are identified within a UniProt/Swiss-Prot record by the SIMILARITY comment “Belongs to the protein kinase superfamily.”
The aPKs can only be identified by functional experimentation, and since the publication of the original list by Manning et al. (2), several more proteins that appear to display protein kinase activity have been identified, for example COL4A3BP (Q9Y5P4), CPNE3 (O75131), and GTF2F1 (P35269). For some of these, only a single report exists, and the observation has yet to be confirmed; in these cases, the reported kinase activity may only be added as a CAUTION comment. As a result, our classification of a protein as an aPK is somewhat more conservative than that of Manning et al. (2), including only 24 human proteins and 24 mouse proteins. However, where a related domain has been found within a sequence, this fact is recorded within the entry: many proteins identified by Manning et al. (2) as atypical fall within this category. It is anticipated that more proteins may be found by bench scientists as having an atypical kinase mechanism as time progresses. Manning et al. (2) also identified 106 pseudogenes with similarity to either an ePK or aPK; however, UniProtKB is only concerned with the annotation of proteins, so the existence and the significance of these have not been investigated any further by this particular group.
INACTIVE PROTEIN KINASES
Manning et al. (2) predicted that up to 50 human protein kinases would be catalytically inactive because of the loss of one or more highly conserved amino acids of either the HRD motif (that precedes the catalytic loop at the active site) or DFG motif (in the activation segment) or would contain, in addition to a fully functional protein kinase domain, a second pseudokinase domain that has lost its catalytic function but may have acquired some new but unknown functions (2, 9). Within UniProtKB/Swiss-Prot, those ePK proteins that lack catalytic activity are tagged with the DOMAIN comment “The protein kinase domain is predicted to be catalytically inactive,” and 23 of these proteins are recognized as such within the human kinase set; 22 of these proteins are recognized as such within the mouse. Where the alternative function of these proteins is known, this has been annotated in full. However, it should be noted that there are kinases that would be predicted to be inactive but do in fact retain full catalytic activity, for example members of the WNK (with no lysine) family, so these may yet prove to be overpredictions. Those proteins with an active kinase domain and a second pseudokinase domain, such as JAK1 (P23458), are described as such within the entry.
PROTEIN KINASE NOMENCLATURE
As part of the annotation process, UniProtKB strives to provide a unified name for both gene and protein product that describes the function of the enzyme while retaining a unique identification that is recognizable by the scientific community; it is most commonly based on the gene name (Fig. 2). The primary name follows a series of rules, which have been made public by the UniProtKB consortium, that enable a protein to be assigned a recommended name providing the maximal amount of information about that gene product while still enabling its propagation across orthologues in other organisms. The database works closely with other nomenclature groups, such as the Human Genome Nomenclature Committee (10) and the Mouse Genome Informatics (11), which are cross-referenced from within the appropriate entries. However, many kinases are already known by more than one well accepted name, and where it proves impossible to include these within the primary name, they are retained within the entry as a synonym to allow ease of searching. All entries have a stable accession number and a human readable identification. The latter, however, should not be regarded as stable because these are occasionally updated, for example with change of gene name. The activity of the kinase, as described by the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology, is captured within the protein description as the Enzyme Classification (EC) number (12).
Fig. 2.
Protein nomenclature: ABL1 (P00519).
PROTEIN KINASE FUNCTION AND REGULATION
Protein kinases can be said to share a single function: the transfer of a phosphate group from ATP to a protein substrate molecule. However, the range of protein substrates targeted by these several hundred molecules is very broad, and the downstream consequences of each phosphorylation event are correspondingly varied. This level of detail is impossible to predict and must be collected by careful and thorough reading of the literature. In many cases, the exact target of a kinase may not be known, although the process regulated by the enzyme has been identified. Both the target and downstream effect are summarized in the FUNCTION comment in a UniProtKB/Swiss-Prot entry, but this field is currently empty for many kinases, reflecting the huge amount of work that still remains to be undertaken in the laboratory. Additional information such as the tissues in which these proteins are expressed and the subcellular locations at which they are found are also annotated in the appropriate fields.
Additional annotation is included by the incorporation of Gene Ontology cross-references (13), via the Gene Ontology Annotation project (14), which is contributed to by UniProtKB curators as well as by curators from the model organism databases.
However, the generic function of a protein kinase can be described, and indeed this is done within the CATALYTIC ACTIVITY statement added to every enzyme classified by the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology EC system. The regulation of protein kinases tends to be at the level of protein binding to, and/or phosphorylation of, each kinase enzyme in a manner that is often either family- or subfamily-specific rather than via changes in protein expression. The existence and potential significance of these phosphorylation sites are again collected from the experimental literature and are detailed in both the ENZYME REGULATION comment and the feature table. Where a protein is known to be post-translationally modified, for example by amino acid phosphorylation, but either the exact amino acid position and/or the effect of this modification is unknown, this information is stated in a POST-TRANSLATIONAL MODIFICATION comment rather than in the feature table. Further post-translational modifications may be required before a kinase can be activated by phosphorylation; for example both the palmitoylation and myristoylation of some kinases are believed to position the protein at a membrane surface possibly resulting in a required conformation change. In addition, there are secondary mechanisms that can adjust the level of protein kinase activity of a phosphorylated protein and may even target it for destruction in the case of protein ubiquitination.
In any process governed by protein phosphorylation, a fine balance between the activities of protein kinases and protein phosphatases is vital to cellular physiologic function. Dysregulation of this balance may lead to abnormal cell growth giving rise to complex diseases such as cancer. Protein phosphatases are classified into subfamilies such as serine/threonine-specific, tyrosine-specific, and dual specificity phosphatases. Each of these subclasses has a well conserved but distinct catalytic protein domain, all of which are described in the UniProtKB/Swiss-Prot database.
SPLICE VARIANTS
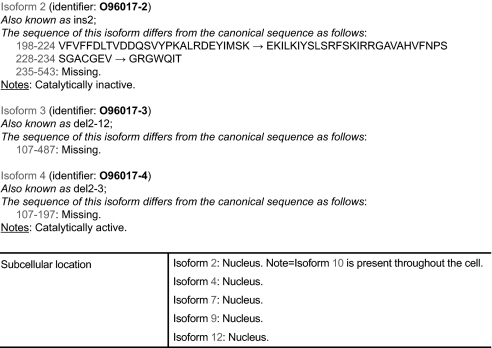
As previously stated, one major task of any UniProtKB curator is the annotation of protein sequences, which includes the identification of splice isoforms. Each of these is given a stable and unique identifier and may be recreated from the feature table of a single UniProtKB entry using the freely available tool VARSPLIC (15) or are directly accessible within a UniProt entry viewed over the Web (Fig. 3). A FASTA-formatted file containing all splice variants annotated in UniProtKB/Swiss-Prot can be downloaded for use with similarity search programs. The differing domain composition of these isoforms can be viewed within InterPro. Protein kinases appear to be highly alternatively spliced with a further 510 isoforms identified, at time of going to press, adding to the 480 human ePKs. This gives an average number of 1.06 additional splice variants per entry for the kinome as opposed to 0.64 for the entire proteome, suggesting that this highly important family of signal transduction proteins requires an increased level of protein variation to maintain a subtle degree of control over cellular processes. It must be noted, however, that this family of proteins has been more intensively studied than the proteome as a whole, and this may account for this higher number of splice isoforms identified. However, as many of the transcripts identified in both sets come from high throughput cDNA sequencing programs, which are generated without bias to a particular protein family, it still seems probable that this family produces an above average number of splice variants. The mouse kinome currently appears to generate a lower level of alternative transcripts, but it is too early to tell whether this represents a true difference between the genomes of the two organisms or merely reflects the lower number of cDNA sequences deposited via the nucleotide databases for the mouse.
Fig. 3.
Representation of splice variants in UniProt/Swiss-Prot: CHEK2 (O96017).
PROTEIN KINASES AND INTERACTIONS WITH OTHER MOLECULES
Eukaryotic protein kinases are sophisticated proteins often with a modular organization containing other domains with additional functionality that may affect the activity of the entire protein. For example, these domains may serve to determine the subcellular location of the protein (PH or FERM (IPR000299) domains) or enable interaction with other molecules (leucine zipper, zinc finger, Cadherin, J, EF hand, WH2, MIRO (IPR013684), CARD (IPR001315), OPR (IPR000270), PAN (IPR003014), NAF (IPR004041), SAM (IPR001660), RWD (IPR006575), FCH (IPR001060), RBD (IPR003116), DH (IPR000219), DEATH (IPR000488), SARAH (IPR011524), CNH (IPR001180), IQ (IPR000048), FHA (IPR000253), CD1, SEMA (IPR001627), SPRY (IPR003877), SH3, CRIB (IPR000095), or UBA (IPR009060) domains) or with cytoskeletal structures (POLO (IPR000959) box, doublecortin, or MIT (IPR007330) domains). Several protein kinases contain domains able to bind double-stranded RNA (DRBM (IPR001159) domain), ribonucleoprotein, or single-stranded DNA (TUDOR (IPR002999) domain). Some protein kinases may encompass domains with GTPase-activating properties, domains that act as a signal sensor (PAS (IPR000014)), or domains that acts as a receptor to secreted wingless glycoproteins (Frizzle or WIF (IPR003306) domains). Others may contain domains involved in cell adhesion (Fibronectin type-III), regulation of vesicular trafficking machinery, and/or growth factor receptor degradation in lysosomes; structural domains such as immunoglobulin-like fold-containing domains (IPT/TIG (IPR002909) or Ig-like C2-type domain); epidermal growth factor-like domains; or simply repeat regions. All the information as to which domain a protein kinase may contain, where it is situated within the amino acid sequence, and exactly which molecules with which each kinase interacts is captured in several ways within a UniProtKB/Swiss-Prot record. The domain architecture is described within the feature table and related to the sequence given at the end of each entry. Further detail about each domain is given by cross-referencing to the appropriate InterPro entry (8) and also to its member databases, such as PROSITE (16), Pfam (17), and (SMART) (18). Additional information may be supplied in a DOMAIN annotation comment if required.
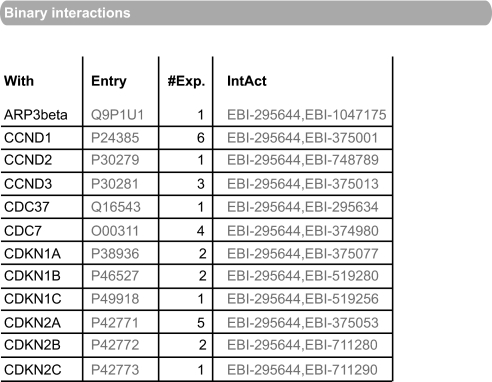
Direct protein interactions made by the protein kinase are stored in the IntAct (19) molecular interaction database and exported to the corresponding UniProtKB records on a monthly basis (Fig. 4). A cross-reference to IntAct allows users to access less direct protein interactions in which the protein may be involved, for example protein complex data, in addition to interactions made with other interactor types such as nucleic acids or small molecules. The SUBUNIT comment allows detail to be given of the functional significance of these interactions and also of stable macromolecular complexes of which the protein may be a member.
Fig. 4.
Binary interactions imported from IntAct: CDK4 (P11802).
PROTEIN KINASES AND HUMAN DISEASE
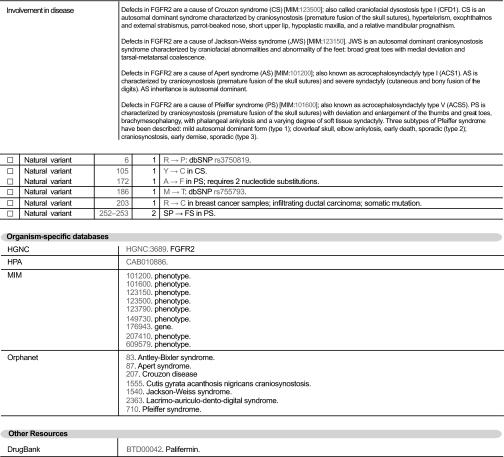
With the advent of the first drugs targeted at specifically inhibiting protein kinase targets and their involvement in many of the signal transduction pathways known to be linked to both physiological and pathological cell growth and development, the pharmaceutical and medical communities have a keen interest in this particular protein family. To aid in the understanding of disease and the development of new pharmaceutical agents, UniProtKB/Swiss-Prot actively strives to supply details about the known role of a protein in the development of illness and disease (Fig. 5). Where a mutation of a protein has been directly linked to a pathological condition, this information is stated within a DISEASE comment. The importance of amino acid polymorphisms to the development of disease conditions is also widely recognized, and these variants are mapped at the amino acid level within the feature table and mapped back to the nucleic acid sequence via the Single Nucleotide Polymorphism database (dbSNP). Finally external resources are referenced, such as the DrugBank database, a bioinformatics and cheminformatics resource that combines detailed drug (i.e. chemical, pharmacological, and pharmaceutical) data with comprehensive drug target information (20), Online Mendelian Inheritance in Man (OMIM) (21), and the Human Protein Atlas (22), which shows the expression and localization of proteins in a large variety of normal human tissues and cancer cells.
Fig. 5.
Disease information in UniProtKB/Swiss-Prot entries: FGFR2 (P21802).
PROTEIN FAMILY MEMBERSHIP
In UniProtKB/Swiss-Prot, protein family membership is indicated by a SIMILARITY comment, and large families, such as the protein kinases, may be further broken down into subfamilies identified by sequence similarity and often sharing common mechanisms of enzymic regulation. Initially a total of seven major classes of eukaryotic protein kinases were originally described by Manning et al. (2) that are represented by multiple subfamilies within UniProtKB/Swiss-Prot. UniProtKB has recognized two additional families not classified previously by Manning et al. (2): the NEK family kinases and the receptor guanylate cyclase kinases.
AGC Group (57 Human and 55 Murine Members)—
The group of AGC protein kinase includes both the subunits of the cAMP-dependent protein kinase, the cGMP-dependent protein kinases, the protein kinase subfamily, the ribosomal protein S6 kinases, G-protein-coupled-receptor kinases, and the myotonic dystrophy protein kinases. Members of this family are identified within a UniProtKB/Swiss-Prot record by the SIMILARITY comment “Belongs to the protein kinase superfamily. AGC Ser/Thr protein kinase family.” Most members of this family are regulated primarily by phosphorylation at two sites: a conserved threonine residue in the activation loop and a serine/threonine residue in a hydrophobic motif near the COOH terminus (23). Additionally some family members also appear to be regulated by phosphorylation of a threonine residue preceding the COOH terminus in the turn motif. At this position, the presence of a phosphate seems to stabilize the kinase core by anchoring the COOH terminus at the top of the upper lobe of the kinase lobe. The turn motif is located just before the hydrophobic region. In protein kinase B, where other mechanisms such as compensating phosphorylation occur, the requirement of this residue is not absolute (23, 24).
It is widely known that the phosphoinositide-dependent protein kinase 1, PDPK1 (O15530), is a sensor of protein conformation because it phosphorylates and contributes to the activation of several protein kinases of the AGC group (including cAMP-dependent protein kinase, cGMP-dependent protein kinase, and protein kinase C) to which PDPK1 also belongs. This protein is able to recognize, interact with, and phosphorylate specific substrate conformations. PDPK1, by itself, does not contain an equivalent hydrophobic motif found at the COOH-terminal extension of the catalytic domain (24).
Tyrosine Kinases (90 Human and 93 Murine Members)—
The largest group of protein kinases, the tyrosine kinases, which are identified within a UniProtKB/Swiss-Prot record by the SIMILARITY comment “Belongs to the protein kinase superfamily. Tyr protein kinase family,” can be further subdivided into two broad classes: the receptor and non-receptor tyrosine kinases, all of which catalyze the transfer of the γ-phosphate of ATP to the hydroxyl of the substrate tyrosine in the presence of a divalent cation. The receptor tyrosine kinases, such as the epidermal growth factor receptor subfamily, comprise an extracellular ligand-binding domain, a transmembrane region, and an intracellular catalytic domain. These molecules frequently form either homo- or heterodimers or oligomers with closely related molecules as part of their regulation process, and transphosphorylation events between adjacent molecules are often an essential part of their activation mechanism. Non-receptor tyrosine kinases, for example the SRC or SYK/ZAP-70 subfamilies, are intracellular molecules capable of binding to transmembrane receptors that lack kinase domains via their molecular interaction regions such as SH2 and SH3 domains and thus participate in signal transduction cascades originating from extracellular signals.
Residues important for catalytic activity come from the nucleotide binding loop, the catalytic region, and the activation loop of the kinase domain. The nucleotide binding loop contains a conserved GXGXΦG region where X is any amino acid and Φ is Phe or Tyr. The activation region contains a conserved DFG sequence that binds to the divalent cation (25).
Tyrosine-like Kinases (34 Human and 34 Murine Members)—
The tyrosine-like family of protein kinases was first described by Manning et al. (2) as a diverse group of families that resemble both tyrosine and serine/threonine kinases, such as the mixed lineage kinases and the interleukin-1 receptor-associated kinase Pelle subfamily (IRAKs) (2). Members of this family are identified within a UniProtKB/Swiss-Prot record by the SIMILARITY comment “Belongs to the protein kinase superfamily. TKL Ser/Thr protein kinase family.”
Ste-20-related Kinases (55 Human and 54 Murine Members)—
This family includes STE-related protein kinases, namely homologs of yeast Sterile 7, 11, and 20 kinases (STE stands for “sterile,” referring to the fact that enzymes belonging to this group were first identified in genetics analysis of yeast sterile mutants). This group, which may be identified by the SIMILARITY comment “Belongs to the protein kinase superfamily. STE Ser/Thr protein kinase family,” includes many enzymes functioning in MAPK pathways, such as MAP4K1 (Q92918), although the MAPKs themselves belong to the CMGC group.
CMGC Kinases (62 Human and 61 Murine Members)—
This family, identified by the SIMILARITY comment “Belongs to the protein kinase superfamily. CMGC Ser/Thr protein kinase family,” includes the CDKs, MAPKs, glycogen synthase kinases, and CDK-like kinases. Dual specificity tyrosine phosphorylation-regulated kinase, and casein kinase 2 (CK2)-α proteins also belong to this group. All members contain a CMGC-specific region, the COOH-terminal substrate-binding domain (26).
Among members of this group, the regulation process of MAP kinase ERK2 (P28482) and cyclin-dependent kinase CDK2 (P24941) is the best documented. In addition to phosphorylation at the classically conserved activation loop threonine, activation of almost all MAP kinases requires a second phosphorylation at a neighboring tyrosine residue, consensus sequence TXY in MAPK (27), whereas the cyclin-dependent kinases require association with cyclin (28). In dual specificity tyrosine phosphorylation-regulated kinases, the consensus sequence is YXY. Although many kinases undergo phosphorylation by an autocatalytic mechanism, the MAP kinases and CDKs undergo phosphorylation and activation by a heterologous kinase. They display essentially only a core domain whose catalytic properties are controlled by a remodeling of the active site in response to activation loop phosphorylation and/or cyclin binding. Both of these dual modifications are essential for complete activation. In a comparable manner, activation of glycogen synthase kinases occurs via prephosphorylation of a target substrate site that then stabilizes the activation loop for phosphorylation of a second nearby residue (29).
CK1 Kinases (12 Human and 11 Murine Members)—
CK1 was one of the first serine/threonine kinases to be isolated and characterized. Members of this kinase family, identified by the SIMILARITY comment “Belongs to the protein kinase superfamily. CK1 Ser/Thr protein kinase family.” are highly conserved in eukaryotic organisms. The COOH-terminal part of the protein plays a key role in the regulation of these enzyme; for example, CK1 δ (P48730) and ɛ (P49674) are controlled by autophosphorylation, dephosphorylation, and proteolytic cleavage of their COOH termini (30) with the phosphorylated COOH terminus appearing to act as a pseudosubstrate. The autophosphorylation of the COOH-terminal region is associated with loss of enzyme activity and may be reversed by dephosphorylation by protein phosphatases. The catalytic domain has an unusual substrate selectivity, being directed toward phosphate groups rather than unmodified amino acids (31).
CAMK Kinases (74 Human and 80 Murine Members)—
Calcium and calmodulin-dependent serine/threonine protein kinases are enzymes that play a pivotal role in a plethora of calcium signaling pathways. Members of this family are identified within a UniProtKB/Swiss-Prot record by the SIMILARITY comment “Belongs to the protein kinase superfamily. CAMK Ser/Thr protein kinase family.” They may be dedicated CAMKs with strict substrate specificities or multifunctional CAMKs that can phosphorylate multiple protein substrates. In addition to a requirement for calcium and calmodulin, CAMKs autoregulate their own kinase activity. To understand the intricacy of the process, it is interesting to study the complex regulative properties of CAMKII. CAMKII is a multisubunit holoenzyme (32). The structure of the holoenzyme has alternatively been referred to as a hub and spoke pattern (33) or a gear and foot pattern (34) and enables a unique autoregulatory mechanism (33). In resting cells, the average Ca2+ concentration is less than 100 nm, and the calmodulin concentration is ∼10 μm. The autoinhibitory domain (AID) of CAMKII interacts with its own catalytic domain resulting in the autoinhibition of catalytic activity. However, in activated cells, when the intracellular calcium level rises to 10−5 m and four Ca2+ ions bind to calmodulin, the Ca2+-calmodulin complex is able to bind to the NH2-terminal region of the AID (35) and to cause a conformational change of the CAMKII protein. This causes a disruption of the interaction between the AID and the catalytic domain, leading to the activation of the enzyme. Each subunit undergoes autophosphorylation (at Thr-286 for CAMKII α isoform and at Thr-287 for the other subunits) and is then able to phosphorylate the same site on adjacent subunits (36), acting in turn as both a substrate and as a kinase. This Ca2+-dependent, primary autophosphorylation of CAMKII is a key step resulting in an increased affinity of CAMKII for calmodulin. Eventually activation of CAMKII occurs independently of the presence of Ca2+/calmodulin, and CAMKII α undergoes additional autophosphorylation at Thr-305 or Thr-306.
This sophisticated regulatory mechanism makes it possible for the enzyme to phosphorylate its substrates even when the intracellular Ca2+ concentration has returned to basal level. By this means, CAMKII is sensitive to the frequency, duration, and amplitude of intracellular Ca2+ variations (37). Positive regulation by phosphorylation is widely called the switch-on mechanism. The switch-off dephosphorylation mechanism is performed by cellular phosphatases. CAMKIs and CAMKIV (Q16566) differ in the way they are regulated; they have an activation loop that requires phosphorylation by an upstream CAMK kinase for maximal activity (38).
NEK Kinases (11 Human and 11 Murine Members)—
The NIMA-related kinases represent a family of serine/threonine kinases implicated in cell cycle control, more specifically in the organization of microtubules (39). A common feature of the COOH-terminal regulatory domains of several NIMA-related kinases is the presence of a coiled-coil motif immediately downstream of the catalytic domain. Recent studies on Nek2 (P51955) have identified key sites of autophosphorylation within the catalytic domain and the requirement for autophosphorylation to prevent the premature activation of Nek2 and thus inappropriate separation of centrosomes in interphase. Members of this family are identified within a UniProtKB/Swiss-Prot record by the SIMILARITY comment “Belongs to the protein kinase superfamily. NEK Ser/Thr protein kinase family.”
Receptor Guanylate Cyclases (Five Human and Six Murine Members)—
The functional transmembrane guanylyl cyclases that synthesize the intracellular second messenger cGMP all contain an intracellular kinase domain (40). In all cases this domain is predicted to be catalytically inactive, and its function in unknown.
PROTEIN KINASES AND UniProtKB/TrEMBL
Although the human and mouse kinomes have been completed within UniProtKB/Swiss-Prot and indeed also that of Saccharomyces cerevisiae, the kinomes of many other organisms are only partially complete. A complete non-redundant set of proteins can be accessed and downloaded via the Integr8 Web portal that provides easy access to integrated information about deciphered genomes and their corresponding proteomes. In many cases, a proportion of the proteins in the set will be computationally maintained within the UniProtKB/TrEMBL database. Protein kinases will be recognized by the protein domain signatures within the InterPro database and will acquire a certain amount of automatic annotation. For example, keywords such as “Kinase,” “Tyrosine protein kinase,” or “ATP binding” will be added as appropriate; nomenclature will be improved including the addition of the EC number; and SIMILARITY statements will be added. This annotation adds value to the sequences as deposited in the nucleotide databases, and the ability to compare sequences across a wide range of species enables an initial analysis of the quality of those yet to be manually curated to be easily made.
DATA DOWNLOADS
The UniProtKB is freely available for both commercial and non-commercial use. The UniProt databases can be accessed on line or downloaded in several formats. New releases are published every 3 weeks. Every human and murine member of each of these kinase subfamilies has been annotated within the UniProtKB/Swiss-Prot database, and the information is available for access via the Web or for download and local installation.
SUMMARY
Protein kinases are a large and evolutionarily conserved family of molecules that play a key role in both inter- and intracellular signaling. Non-physiological levels of protein kinase activation have been associated with a wide range of pathological conditions such as cancer, inflammatory disease, and viral infectivity. Because of this, these enzymes have been widely studied, and a database of reliable sequence data coupled to extensive annotation is a requirement for every laboratory worker. The UniProtKB not only provides this as a public domain service but also allows sequence comparison across a wide range of organisms thus enabling appropriate cellular and disease models to be established.
The annotation in the UniProtKB/Swiss-Prot database is actively maintained and kept up to date by extensive manual sequence curation and literature annotation. As sequences continue to be deposited in the public domain databases, the kinomes of other model organisms such as the rat, cow, and the higher primates will move toward completion allowing a deeper understanding both of the roles these critical enzymes play in the life cycle of a cell and their relationship to each other throughout evolution. Users of the database are strongly encouraged to give feedback from their own specialist knowledge of these molecules, thus contributing to a continual evolution and improvement in this valuable public domain resource.
Acknowledgments
We thank Amos Bairoch and Rolf Apweiler for many helpful discussions. We also thank Elisabeth Gasteiger and Delphine Baratin for implementing the nomenclature index (human and mouse protein kinase classification and index) as well as Claire O'Donovan, Michele Magrane, and Silvia Jimenez for critically reading the manuscript and Louisa Wright for preparing the figures. UniProt is mainly supported by the National Institutes of Health (NIH) Grant 2U01 HG02712-05. Additional support for the European Bioinformatics Institute's involvement in UniProt comes from the European Commission contract FELICS (Grant 021902RII3) and from NIH Grant 2 P41 HG002273. UniProtKB/Swiss-Prot activities at the Swiss Institute of Bioinformatics are supported by the Swiss Federal Government through the Federal Office of Education and Science, by the European Commission contract FELICS (Grant 021902RII3), and by the NIAID, NIH Grant (HHSN 2662040035C ADB Contract N01-AI-40035).
Footnotes
Published, MCP Papers in Press, April 24, 2008, DOI 10.1074/mcp.R700001-MCP200
The abbreviations used are: ePK, eukaryotic protein kinase; aPK, atypical protein kinase; UniProtKB, UniProt KnowledgeBase; CDK, cyclin-dependent kinase; CAMK, Ca2+/calmodulin-dependent protein kinase; EC, Enzyme Classification; PH, pleckstrin homology; SH, SRC homology; NEK, NIMA-related kinase; MAPK, mitogen-activated protein kinase; MAP, mitogen-activated protein; CK, casein kinase; AID, autoinhibitory domain; FGFR2, fibroblast growth factor receptor 2; DOK1, docking protein 1; SORBS1; sorbin and SH3 domain-containing protein 1; CABLES1, CDK5 and ABL1 enzyme substrate 1; INPPL1, inositol polyphosphate phosphatase-like protein 1; PRKDC, DNA-dependent protein kinase catalytic subunit.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Fischer, E. H., Kent, A. B., and Krebs, E. G. ( 1958) The muscle phosphorylase b kinase reaction. J. Biol. Chem. 231, 73–83 [PubMed] [Google Scholar]
- 2.Manning, G., Whyte, D. B., Martinez, R., Hunter, T., and Sudarsanam, S. ( 2002) The protein kinase complement of the human genome. Science 298, 1912–1934 [DOI] [PubMed] [Google Scholar]
- 3.The UniProt Consortium ( 2007) The universal protein resource (UniProt). Nucleic Acids Res. 35, D193–D197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gururajan, R., Lahti, J. M., Grenet, J. A., Easton, J., Gruber, I., Ambros, P. F., and Kidd, V. J. ( 1998) Duplication of a genomic region containing the Cdc2L1–2 and MMP21–22 genes on human chromosome 1p36.3 and their linkage to D1Z2. Genome Res. 8, 929–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uchida, N., Hoshino, S., and Katada, T. ( 2004) Identification of a human cytoplasmic poly(A) nuclease complex stimulated by poly(A)-binding protein. J. Biol. Chem. 279, 1383–1391 [DOI] [PubMed] [Google Scholar]
- 6.Caenepeel, S., Charydczak, G., Sudarsanam, S., Hunter, T., and Manning, G. ( 2004) The mouse kinome: discovery and comparative genomics of all mouse protein kinases. Proc. Natl. Acad. Sci. U. S. A. 101, 11707–11712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ye, X., Zhu, C., and Harper, J. W. ( 2001) A premature-termination mutation in the Mus musculus cyclin-dependent kinase 3 gene. Proc. Natl. Acad. Sci. U. S. A. 98, 1682–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mulder, N. J., Apweiler, R., Attwood, T. K., Bairoch, A., Bateman, A., Binns, D., Bork, P., Buillard, V., Cerutti, L., Copley, R., Courcelle, E., Das, U., Daugherty, L., Dibley, M., Finn, R., Fleischmann, W., Gough, J., Haft, D., Hulo, N., Hunter, S., Kahn, D., Kanapin, A., Kejariwal, A., Labarga, A., Langendijk-Genevaux, P. S., Lonsdale, D., Lopez, R., Letunic, I., Madera, M., Maslen, J., McAnulla, C., McDowall, J., Mistry, J., Mitchell, A., Nikolskaya, A. N., Orchard, S., Orengo, C., Petryszak, R., Selengut, J. D., Sigrist, C. J. A., Thomas, P. D., Valentin, F., Wilson, D., Wu, C. H., and Yeats, C. ( 2007) New developments in the InterPro database. Nucleic Acids Res. 35, D224–D228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilks, A. F., Harpur, A. G., Kurban, R. R., Ralph, S. J., Zuercher, G., and Ziemiecki, A. ( 1991) Two novel protein-tyrosine kinases, each with a second phosphotransferase-related catalytic domain, define a new class of protein kinase. Mol. Cell. Biol. 11, 2057–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bruford, E. A., Lush, M. J., Wright, M. W., Sneddon, T. P., Povey, S., and Birney, E. ( 2008) The HGNC Database in 2008: a resource for the human genome. Nucleic Acids Res. 36, D445–D448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eppig, J. T., Bult, C. J., Kadin, J. A., Richardson, J. E., Blake, J. A., Anagnostopoulos, A., Baldarelli, R. M., Baya, M., Beal, J. S., Bello, S. M., Boddy, W. J., Bradt, D. W., Burkart, D. L., Butler, N. E., Campbell, J., Cassell, M. A., Corbani, L. E., Cousins, S. L., Dahmen, D. J., Dene, H., Diehl, A. D., Drabkin, H. J., Frazer, K. S., Frost, P., Glass, L. H., Goldsmith, C. W., Grant, P. L., Lennon-Pierce, M., Lewis, J., Lu, I., Maltais, L. J., McAndrews-Hill, M., McClellan, L., Miers, D. B., Miller, L. A., Ni, L., Ormsby, J. E., Qi, D., Reddy, T. B., Reed, D. J., Richards-Smith, B., Shaw, D. R., Sinclair, R., Smith, C. L., Szauter, P., Walker, M. B., Walton, D. O., Washburn, L. L., Witham, I. T., and Zhu, Y. ( 2005) The Mouse Genome Database (MGD): from genes to mice—a community resource for mouse biology. Nucleic Acids Res. 33, D471–D475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fleischmann, A., Darsow, M., Degtyarenko, K., Fleischmann, W., Boyce, S., Axelsen, K. B., Bairoch, A., Schomburg, D., Tipton, K. F., and Apweiler, R. ( 2004) IntEnz, the integrated relational enzyme database. Nucleic Acids Res. 32, D434–D437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris, M. A., Clark, J., Ireland, A., Lomax, J., Ashburner, M., Foulger, R., Eilbeck, K., Lewis, S., Marshall, B., Mungall, C., Richter, J., Rubin, G. M., Blake, J. A., Bult, C., Dolan, M., Drabkin, H., Eppig, J. T., Hill, D. P., Ni, L., Ringwald, M., Balakrishnan, R., Cherry, J. M., Christie, K. R., Costanzo, M. C., Dwight, S. S., Engel, S., Fisk, D. G., Hirschman, J. E., Hong, E. L., Nash, R. S., Sethuraman, A., Theesfeld, C. L., Botstein, D., Dolinski, K., Feierbach, B., Berardini, T., Mundodi, S., Rhee, S. Y., Apweiler, R., Barrell, D., Camon, E., Dimmer, E., Lee, V., Chisholm, R., Gaudet, P., Kibbe, W., Kishore, R., Schwarz, E. M., Sternberg, P., Gwinn, M., Hannick, L., Wortman, J., Berriman, M., Wood, V., de la Cruz, N., Tonellato, P., Jaiswal, P., Seigfried, T., and White, R. Gene Ontology Consortium ( 2004) The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 32, 258–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Camon, E., Magrane, M., Barrell, D., Lee, V., Dimmer, E., Maslen, J., Binns, D., Harte, N., Lopez, R., and Apweiler, R. ( 2004) The Gene Ontology Annotation (GOA) Database: sharing knowledge in UniProt with Gene Ontology. Nucleic Acids Res. 32, D262–D266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kersey, P., Hermjakob, H., and Apweiler, R. ( 2000) VARSPLIC: alternatively-spliced protein sequences derived from Swiss-Prot and TrEMBL. Bioinformatics 11, 1048–1049 [DOI] [PubMed] [Google Scholar]
- 16.Hulo, N., Bairoch, A., Bulliard, V., Cerutti, L., Cuche, B. A., Castro, E. D., Lachaize, C., Langendijk-Genevaux, P. S., and Sigrist, C. J. ( 2008) The 20 years of PROSITE. Nucleic Acids Res. 36, D245–D249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finn, R. D., Mistry, J., Schuster-Bockler, B., Griffiths-Jones, S., Hollich, V., Lassmann, T., Moxon, S., Marshall, M., Khanna, A., Durbin, R., Eddy, S. R., Sonnhammer, E. L., and Bateman, A. ( 2006) Pfam: clans, web tools and services. Nucleic Acids Res. 34, D247–D251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Letunic, I., Copley, R. R., Pils, B., Pinkert, S., Schultz, J., and Bork, P. ( 2006) SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 34, D257–D260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kerrien, S., Alam-Faruque, Y., Aranda, B., Bancarz, I., Bridge, A., Derow, C., Dimmer, E., Feuermann, M., Friedrichsen, A., Huntley, R., Kohler, C., Khadake, J., Leroy, C., Liban, A., Lieftink, C., Montecchi-Palazzi, L., Orchard, S., Risse, J., Robbe, K., Roechert, B., Thorneycroft, D., Zhang, Y., Apweiler, R., and Hermjakob, H. ( 2007) IntAct—open source resource for molecular interaction data. Nucleic Acids Res. 35, D561–D565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wishart, D. S., Knox, C., Guo, A. C., Shrivastava, S., Hassanali, M., Stothard, P., Chang, Z., and Woolsey, J. ( 2006) DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 34, D668–D672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamosh, A., Scott, A. F., Amberger, J. S., Bocchini, C. A., and McKusick, V. A. ( 2005) Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 33, D514–D517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Persson, A., Hober, S., and Uhlen, M. ( 2006) A human protein atlas based on antibody proteomics. Curr. Opin. Mol. Ther. 8, 185–190 [PubMed] [Google Scholar]
- 23.Frödin, M., Antal, T. L., Dümmler, B. A., Jensen, C. J., Deak, M., Gammeltoft, S., and Biondi, R. M. ( 2002) A phosphoserine/threonine-binding pocket in AGC kinases and PDK1 mediates activation by hydrophobic motif phosphorylation. EMBO J. 21, 5396–5407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biondi, R. M., Kieloch, A., Currie, R. A., Deak, M., and Alessi, D. R. ( 2001) The PIF-binding pocket in PDK1 is essential for activation of S6K and SGK, but not PKB. EMBO J. 20, 4380–4390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Till, J. H., Ablooglu, A. J., Frankel, M., Bishop, S. M., Kohanski, R. A., and Hubbard, S. R. ( 2001) Crystallographic and solution studies of an activation loop mutant of the insulin receptor tyrosine kinase: insights into kinase mechanism. J. Biol. Chem. 276, 10049–10055 [DOI] [PubMed] [Google Scholar]
- 26.Kannan, N., and Neuwald, A. F. ( 2004) Evolutionary constraints associated with functional specificity of the CMGC protein kinases MAPK, CDK, GSK, SRPK, DYRK, and CK2α. Protein Sci. 13, 2059–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burack, W. R., and Sturgill, T. W. ( 1997) The activating dual phosphorylation of MAPK by MEK is nonprocessive. Biochemistry 36, 5929–5933 [DOI] [PubMed] [Google Scholar]
- 28.Harper, J. W., and Adams, P. D. ( 2001) Cyclin-dependent kinases. Chem. Rev. 101, 2511–2526 [DOI] [PubMed] [Google Scholar]
- 29.ter Haar, E., Coll, J. T., Austen, D. A., Hsiao, H. M., Swenson, L., and Jain, J. ( 2001) Structure of GSK3β reveals a primed phosphorylation mechanism. Nat. Struct. Biol. 8, 593–596 [DOI] [PubMed] [Google Scholar]
- 30.Gietzen, K. F., and Virshup, D. M. ( 1999) Identification of inhibitory autophosphorylation sites in casein kinase Iɛ. J. Biol. Chem. 274, 32063–32070 [DOI] [PubMed] [Google Scholar]
- 31.Meggio, F., Perich, J. W., Marin, O., and Pinna, L. A. ( 1992) The comparative efficiencies of the Ser(P)-, Thr(P)- and Tyr(P)-residues as specificity determinants for casein kinase-1. Biochem. Biophys. Res. Commun. 182, 1460–1465 [DOI] [PubMed] [Google Scholar]
- 32.Sacchetto, R., Bovo, E., Donella-Deana, A., and Damiani, E. ( 2005) Glycogen- and PP1c-targeting subunit GM is phosphorylated at Ser48 by sarcoplasmic reticulum-bound Ca2+-calmodulin protein kinase in rabbit fast twitch skeletal muscle. J. Biol. Chem. 280, 7147–7155 [DOI] [PubMed] [Google Scholar]
- 33.Kanaseki, T., Ikeuchi, Y., Sugiura, H., and Yamauchi, T. ( 1991) Structural features of Ca2+/calmodulin-dependent protein kinase II revealed by electron microscopy. J. Cell Biol. 115, 1049–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kolodziej, S. J., Hudmon, A., Waxham, M. N., and Stoops, J. K. ( 2000) Three-dimensional reconstructions of calcium/calmodulin-dependent (CaM) kinase IIα and truncated CaM kinase IIα reveal a unique organization for its structural core and functional domains. J. Biol. Chem. 275, 14354–14359 [DOI] [PubMed] [Google Scholar]
- 35.Ishida, A., and Fujisawa, H. ( 1995) Stabilization of calmodulin-dependent protein kinase II through the autoinhibitory domain. J. Biol. Chem. 270, 2163–2170 [DOI] [PubMed] [Google Scholar]
- 36.Soderling, T. R., Chang, B., and Brickey, D. ( 2001) Cellular signaling through multifunctional Ca2+/calmodulin-dependent protein kinase II. J. Biol. Chem. 276, 3719–3722 [DOI] [PubMed] [Google Scholar]
- 37.De Koninck, P., and Schulman, H. ( 1998) Sensitivity of CaM kinase II to the frequency of Ca2+ oscillations. Science 279, 227–230 [DOI] [PubMed] [Google Scholar]
- 38.Chatila, T., Anderson, K. A., Ho, N., and Means, A. R. ( 1996) A unique phosphorylation-dependent mechanism for the activation of Ca2+/ calmodulin-dependent protein kinase type IV/GR. J. Biol. Chem. 271, 21542–21548 [DOI] [PubMed] [Google Scholar]
- 39.O'Regan, L., Blot, J., and Fry, A. M. ( 2007) Mitotic regulation by NIMA-related kinases. Cell Div. 2, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Potter, L. R. ( 2005) Domain analysis of human transmembrane guanylyl cyclase receptors: implications for regulation. Front. Biosci. 10, 1205–1220 [DOI] [PubMed] [Google Scholar]