Abstract
Protein synthesis is one of the most important reactions in the cell. Recent experimental studies indicated that this complex reaction can be achieved with a minimum complement of 36 proteins and ribosomes by reconstituting an Escherichia coli-based in vitro translation system with these protein components highly purified on an individual basis. From the protein-protein interaction (PPI) network of E. coli proteins, these minimal protein components are known to interact physically with large numbers of proteins. However, it is unclear what fraction of E. coli proteins are linked functionally with the minimal protein synthesis system. We investigated the effects of each of the 4194 E. coli ORF products on the minimal protein synthesis system; at least 12% of the entire ORF products, a significant fraction of the gene product of E. coli, affect the activity of this system. Furthermore 34% of these functional modifiers present in the PPI network were shown by mapping to be directly linked (i.e. to interact physically) with the minimal components of the PPI network. Topological analysis of the relationships between modifiers and the minimal components in the PPI network indicated clustering of the minimal components. The modifiers showed no such clustering, indicating that the location of functional modifiers is spread across the PPI network rather than clustering close to the minimal protein components. These observations may reflect the evolutionary process of the protein synthesis system.
The protein translation reaction (1), one of the most important regulators of cell behavior, involves the interactions of a large number of components and can thus be seen as an intermolecular interaction network. It has been demonstrated experimentally that 36 enzymes and the ribosomes are sufficient to carry out protein translation (2). These minimal protein components include the ribosomal proteins; initiation, elongation, and release factors; aminoacyl-tRNA synthetases; and enzymes involved in energy regeneration. This was demonstrated by constructing an Escherichia coli-based reconstituted in vitro translation system with these protein components highly purified on an individual basis.
Although the genome of E. coli contains more than 4000 genes (3), constituting a very large interaction network (4, 5), the number of protein components constituting the minimal protein synthesis system corresponds to only 2.1% of the genes encoded in the genome. Thus, only small subsets of the protein components are required for protein synthesis. On the other hand, a number of previous studies, including protein-protein interaction (PPI)1 network analysis in E. coli (4, 5), indicated that protein components constituting the minimal protein synthesis system interact with a large number of other proteins. To gain a deeper understanding of the protein translation system, it is important to identify not only the proteins that interact physically but also those that interact functionally, i.e. those that affect the activity of the translation machinery. Although there have been many studies to characterize the properties of such individual proteins in detail (e.g. Refs. 6 and 7), there have been no previous attempts to search for such proteins in the entire genome. Hence it is not clear what fraction of gene products of E. coli are linked functionally with the minimal protein synthesis system.
The PPI network shows the physical interactions between the proteins, and such networks from various organisms, including E. coli (4, 5), Saccharomyces cerevisiae (8, 9), Drosophila melanogaster (10), Caenorhabditis elegans (11), and Homo sapiens (12, 13), have been investigated; the results of these studies have indicated that proteins are highly connected with each other. As the protein translation system is embedded in such a large interaction network, we were also interested in the topological relationships between the minimal components and those that are functionally linked with them in the PPI network of E. coli; these may provide insight into the topological structure and evolution of the protein synthesis system.
In the present study, we first performed an experimental search for ORF products of the E. coli genome that affect the activity of the translation system utilizing two resources: ASKA library (a complete set of E. coli K12 ORF archive) and the PURE system (protein synthesis using recombinant elements). The ASKA library is the complete set of cloned E. coli ORF genes (14), and the PURE system is an E. coli-based protein synthesis system composed of the minimal protein components (2). As this is a reconstituted system comprised of defined components, it differs from other conventional cell extract-based in vitro translation systems (15, 16), which contain a number of unidentified components. Therefore, the system described here is highly suited for comprehensive analysis of the effects of each ORF product on the translation system. By measuring the effects of individual ORF products on the green fluorescent protein (GFP) synthesis reaction using the PURE system, we demonstrated that at least 12% of the 4194 ORF products of E. coli can affect the activity of the system. We designated these as functional modifiers of the protein synthesis reaction composed of minimal protein components. We then mapped each of the components involved in the protein synthesis reaction on the PPI network of E. coli (4). Network analyses indicated that functional modifiers seem to be spread across the PPI network rather than clustering close to the minimal protein components. A possible interpretation of this observation in relation to the evolutionary process of the protein synthesis system is discussed.
EXPERIMENTAL PROCEDURES
Preparation of DNA Fragments—
The ASKA library was provided by the National BioResource Project (National Institute of Genetics, Shizuoka, Japan). Plasmids of the ASKA library (14) were purified using a MultiScreen Plasmid DNA purification kit (Millipore Corp.) in accordance with the manufacturer's instructions. Individual E. coli ORF DNA fragments were amplified by PCR using each of the 4211 plasmids as a template with the primers pqe2+ (5′-CTCGAGAAATCATAAAAAATTT) and cDNA-lumio-stop2 (5′-TTATTATTAACAACATCCTGGACAACCTTCTCCTTTACTGCGGCCG). Note that only 4194 plasmids gave PCR products. The resulting PCR products encoded E. coli ORF proteins with a tetracysteine tag (17, 18) fused at the carboxyl terminus under the control of the T5 promoter. PCR products were purified using 96-well plates with QIAquick (Qiagen) in accordance with the manufacturer's instructions. Concentrations of the purified PCR products were estimated using PicoGreen double-stranded DNA quantification reagent (Invitrogen) with λ DNA as a standard, and their purity was confirmed by agarose gel electrophoresis.
The GFP DNA fragment was amplified by PCR using pETG5tag (19) as a template with the primers T7F (5′-TAATACGACTCACTATAGGG) and T7R (5′-GCTAGTTATTGCTCAGCGG), and the resulting PCR products were purified and quantified as described for the ASKA library. The GFP used was GFPuv5, which was constructed previously by Ito et al. (20).
Screening of the ASKA Library—
E. coli ORFs were translated using the PURE system (2) (Post Genome Institute). The PURE system reported by Shimizu et al. (2) did not contain release factor 2 and thus consisted of 35 proteins and the ribosomes, whereas the PURE system used here contained release factor 2 and thus consisted of 36 proteins and the ribosomes. Note that with the exception of myokinase and creatine kinase from chicken muscle cDNA and inorganic pyrophosphatase from S. cerevisiae, the protein components included in the present system were from E. coli. For those not derived from E. coli, we used a corresponding protein of E. coli for PPI network analysis. Briefly aliquots of 10 μl of the PURE system containing 0.5 units of E. coli RNA polymerase (EPICENTRE Biotechnologies), 4 units of RNasin (Promega), and 10 nm individual E. coli ORF PCR fragments were incubated at 37 °C for 2 h. Subsequently an additional 10 μl of PURE system containing 4 units of RNasin, 2 units of Tagetin (21) (EPICENTRE Biotechnologies), 100 pmol of oligonucleotide (5′-GTGAGATCCTCTCAT), 100 nm AlexaFluor647 (Invitrogen), and 50 pm GFPuv5 DNA fragment were added to the same tube. Tagetin and oligonucleotide complementary to the initiation codon region of the ORF gene were added to terminate transcription and translation of E. coli ORFs, respectively. Note that both Tagetin and the oligonucleotide used to suppress the reaction from the ORF gene were confirmed to have no influence on the subsequent GFP synthesis reaction (data not shown). AlexaFluor647 was used as an internal dye to normalize the differences in fluorescence intensity among the wells. Real time detection of the GFP synthesis reaction was carried out using a real time PCR system (Mx3005P; Stratagene) at 37 °C for 3 h. Filter sets used for measuring GFP and AlexaFluor647 fluorescence intensities were 492/516 and 635/665 nm (excitation/emission wavelength), respectively.
All measurements were carried out in 96-well plates. For each measurement, the tig gene, ybaW gene, and a blank well without any ORF gene were always included as controls. The raw data were then treated as described below. First, the GFP fluorescence intensity value was divided by that of AlexaFluor647 for the same sample (same well) to normalize the differences among wells. Second, the values obtained with the tig gene and without any ORF gene (NoAdd) were used to normalize the differences among different measurements using the equation: FIinormalized − NoAdd0 = (FIi − NoAddi)(tig0 − NoAdd0)/(tigi − NoAddi) where FIinormalized and FIi are the green fluorescence intensity before and after normalization, respectively, at the ith measurement; tig0 and NoAdd0 are the fluorescence intensities obtained with the tig gene and without any ORF gene, respectively, used as standard values; and tigi and NoAddi are the fluorescence intensity obtained at the ith measurement with the tig gene and without any ORF gene, respectively. As a consequence of the second normalization, the values of tigi and NoAddi became constant for all measurements (tig0 and NoAdd0, respectively), and the accuracy of this normalization could be determined from the value of ybaW (see Fig. 1C).
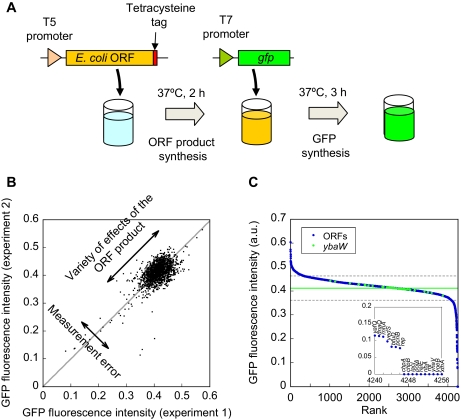
Fig. 1.
Effects of the E. coli ORF products on the protein synthesis reaction consisting of the minimal protein components. A, synopsis of screening strategy. First, ORF products were synthesized in vitro by adding the ORF gene (PCR product) encoded under the control of the T5 promoter. The synthesis of the ORF product can be detected via the tetracysteine tag (17, 18) located at the carboxyl terminus. Subsequently synthesis of the ORF product was specifically suppressed (see “Experimental Procedures” for details), and the gfp gene (PCR product) under the control of the T7 promoter was added. The GFP synthesis reaction was monitored by the increase in green fluorescence intensity, which allowed evaluation of the effect of the presence of the ORF product. B, reproducibility of the measurements. Correlation between the first and second measurements of the effects of 1682 ORFs on the GFP synthesis reaction is shown. Fluorescence intensities after 3 h of GFP synthesis reaction are plotted. The gray line shows the linear regression curve (slope = 0.99). Deviations from the gray line (linear regression curve) represent the errors in the measurements, and the deviation along the gray line represents the variability of the effects of the ORF products. The S.E. (S.D. of the distance of the data from the gray line) was estimated to be 0.016 a.u., whereas the S.D. among different ORF products was 0.042 a.u. C, rank order plot of the fluorescence intensity after 3 h of GFP synthesis reaction in the presence of each of 4194 different ORF products (blue) and in the presence of the ybaW gene measured 62 times independently (green). Numbers on the horizontal axis indicate the ranking of each ORF gene assigned according to its fluorescence intensity among the 4194 ORFs and 62 independent measurements of the ybaW gene. Number 1 is the ORF that gave the highest value. The inset shows the magnification of rank numbers 4237 to 4256, and the gene names are shown above the data. The green line is the average value of ybaW. The gray dashed line is 5 times the S.E. of the results of ybaW.
Synthesis of the ORF product was investigated by adding 5 μm ReAsH (17, 18) (Invitrogen). Details are given in the supplemental notes.
When necessary, the reaction mixtures were subjected to SDS-PAGE followed by Western blotting analysis using anti-GFP monoclonal antibody (Nacalai Tesque) and anti-mouse antibody horseradish peroxidase conjugate (Promega) as the primary and secondary antibodies, respectively. Detection was carried out using an ECL Advance Western Blotting Detection kit (GE Healthcare).
Cloning, Expression, and Purification of E. coli ORF Proteins—
The ybaW gene was amplified by PCR from the corresponding plasmid of the ASKA library (14) with the primers Eco-cDNA-1 (5′-CCATACGGATCGAATTCTGAGGCCCTGAGG) and cDNA-stop-Hind-1 (5′-AACTCCAGTTAACTTACTTACTTATTAACTGCGGCCGCATAGGCC). PCR was carried out as described for amplification of the GFP DNA fragment. The resultant PCR fragments were digested with EcoRI and HindIII and cloned into the plasmid pASK-IBA35plus (IBA GmbH) digested with EcoRI and HindIII to generate pTet-ybaW. The genes hrpA, orn, phnH, slyD, tig, and trxC were isolated by digesting the corresponding plasmids from the ASKA library (14) with SfiI and cloned into pTet-ybaW digested with SfiI to yield plasmids pTet-hrpA, pTet-orn, pTet-phnH, pTet-slyD, pTet-tig, and pTet-trxC, respectively.
The plasmids thus obtained were transformed into E. coli BL21 (DE3) cells, which were then grown in LB medium at 30 °C. On reaching an A600 of 0.5, anhydrotetracycline (IBA GmbH) was added to a final concentration of 0.2 nm, incubation was continued for a further 4 h, and cells were then harvested. His-tagged ORF proteins were purified using IMAC. With the exception of HrpA, all proteins were purified essentially as described by Shimizu et al. (2) except that the purified proteins were dialyzed against the stock buffer (25 mm HEPES, pH 7.6, 10 mm KCl, 30% glycerol, and 7 mm β-mercaptoethanol) and stored at −80 °C. For purification of HrpA, harvested cells were suspended in 50 ml of buffer (50 mm HEPES-KOH, pH 7.6, 10 mm MgCl2, 0.3 mg/ml lysozyme, 0.1% Triton X-100, 1× protease inhibitor mixture for use in purification of histidine-tagged proteins (Sigma-Aldrich), and 7 mm β-mercaptoethanol) and passed through a French press. The insoluble fraction was collected by centrifugation at 10,000 × g for 30 min at 4 °C. The pellet was resuspended with 50 ml of 50 mm HEPES-KOH, pH 7.6, 1 m NaCl, and 7 mm β-mercaptoethanol. In this way, HrpA protein was solubilized from the pellet, and the soluble fraction was subsequently subjected to IMAC. The supernatant was applied to a Ni2+ precharged 10-ml Hi-Trap chelating column (GE Healthcare) and washed with 100 ml of NaHT buffer (50 mm HEPES-KOH, pH 7.6, 1 m NaCl, and 7 mm β-mercaptoethanol) containing 10 mm imidazole. HrpA protein was eluted with a linear gradient of 10–400 mm imidazole in NaHT buffer. Purified HrpA protein was dialyzed against stock buffer supplemented with 0.5 m NaCl and stored at −80 °C.
To evaluate the effects of these ORF proteins on GFP synthesis, the reaction was carried out with the PURE system in the presence of 50 nm AlexaFluor647 (Invitrogen), 500 pm GFPuv5 DNA, and the purified proteins at concentrations of 10, 2000, 2000, 1000, 750, 1500, and 1000 nm for HrpA, Orn, PhnH, SlyD, Tig, TrxC, and YbaW, respectively (Fig. 2). The reaction mixtures were incubated at 37 °C for 3 h, and the green fluorescence intensity was measured as described above for ASKA library screening.
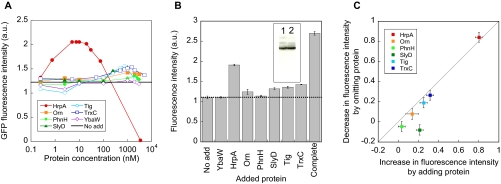
Fig. 2.
Validating the results of comprehensive analysis. A, determination of the optimum concentrations of beneficial components for GFP synthesis reaction. HrpA, Orn, TrxC, Tig, SlyD, PhnH, and YbaW proteins were added at concentrations from 0.25 nm to 5 μm to evaluate their effects on the GFP synthesis reaction. NoAdd (black line) shows the average value of three independent measurements obtained in the absence of the above additional proteins. B, the effects of adding optimum concentrations of purified beneficial proteins to the GFP synthesis. The average and S.E. of three independent measurements are plotted. The values obtained in the absence of additional proteins (No add) and in the presence of the mixture of six proteins (HrpA, Orn, TrxC, Tig, SlyD, and PhnH) added at optimum concentrations (Complete) are also shown. The fluorescence intensities obtained after 3 h of incubation at 37 °C are plotted. Protein concentrations used were 0.01, 2, 2, 1, 0.75, 1.5, and 1 μm for HrpA, Orn, PhnH, SlyD, Tig, TrxC, and YbaW, respectively. The inset shows the results of Western blotting analysis of GFP synthesis reaction. Lane 1, No add; Lane 2, Complete. C, additivity of the effects of the six ORF products on the GFP synthesis reaction. The correlation between the increase in observed fluorescence intensity by adding one of the six proteins to the synthesis reaction (No add in B) and the decrease in observed fluorescence intensity by omitting one of the six proteins from the synthesis reaction containing the other proteins (Complete in B) is shown. The gray line has a slope of 1. The average and S.E. values (shown as error bars) are from three independent measurements.
Protein-Protein Interaction Analysis—
Protein-protein interaction data of E. coli used in this study were obtained previously (4) and consisted of data for 1360 proteins and 6229 interactions. Note that there was no bias in the presence of components involved in the protein synthesis reaction using the largest interconnected network. Calculations were performed using the software R (The R Project for Statistical Computing). Ortholog data were obtained from the Microbial Genome Database for Comparative Analysis (22). All calculated values and data for each ORF are shown in supplemental Table 1.
RESULTS
Strategy for Comprehensive Analysis—
We first established a strategy to investigate the effects of individual ORF products of the E. coli genome on the E. coli-based translation reaction composed of only the minimum number of highly purified protein components, i.e. the PURE system (2) (Fig. 1A). Throughout this study we used the PURE system as an in vitro translation system. Each of 4211 ORF genes was amplified by PCR using the corresponding plasmids (ASKA library) (14) as templates to produce the constructs shown in Fig. 1A. ORF products were synthesized individually in vitro, and the gfp gene (20) was subsequently added to the same test tubes containing synthesized ORF products (Fig. 1A). If the ORF products exhibited beneficial or deleterious effects on the translation and/or transcription machinery (with T7 RNA polymerase), the GFP fluorescence intensity would be expected to increase or decrease, respectively, relative to that without any ORF products. In this way, we screened for the effects of all 4194 ORF products of E. coli on the protein synthesis reaction by the minimal set of protein components.
To evaluate the reliability of our measurements, we examined the effects of 1682 ORFs independently twice. Fig. 1B shows the correlation between the first and second independent measurements (all values are given in supplemental Table 1). The values shown in Fig. 1B are the observed GFP fluorescence intensities after a 3-h reaction corresponding to the stage at which the synthesis reaction was nearly complete. Pearson's correlation coefficient between two independent measurements was found to be r = 0.74. In addition, the deviation from the gray line (linear regression curve) indicating the errors in the measurements was found to be smaller than that along the gray line representing the variety of the effects of the ORF products. We thus concluded that there was a sufficient correlation between the two independent measurements and that the accuracy of our measurements was sufficient to detect the effect of the ORF product on the GFP synthesis reaction.
Comprehensive Analysis—
We then obtained the results with 4194 ORF products using the strategy described above. In Fig. 1C blue circles show a rank ordered plot of the results obtained by analyzing 4194 ORFs (all values are given in supplemental Table 1), and the green circles indicate the effects of YbaW (putative acyl-CoA thioester hydrolase (23)), used as a control, in 62 independent measurements. The high throughput measurements were carried out in 96-well plates, and thus to obtain all 4194 results, the measurements were carried out multiple times. For each measurement, we used the arbitrarily chosen ybaW gene (23) as a control. As shown below (Fig. 2A), the purified product of ybaW (YbaW) had no effect on the GFP synthesis reaction. The value for YbaW was 0.41 ± 0.01 a.u. indicating the accuracy of the measurements. As purified YbaW had no effect on the GFP synthesis reaction (Fig. 2A), we defined the region in the distribution where the ORFs with no effects are located as the region within 5× the S.E. from the average value of YbaW (between gray dashed lines). No ORF will appear outside this region due to measurement error unless 1.7 × 106 or more ORF samples are measured. We defined the ORFs outside this region (above and below the gray line) as modifiers; thus, 8.2 and 3.8% of the entire ORFs were defined as those that increased and decreased the observed GFP fluorescence intensity, respectively. From these results, the fraction of ORFs affecting the GFP synthesis reaction was found to be at least 12%, and very likely more will interact with the minimal system. This is because our assay was performed with one particular protein, GFP, and requires the ORF products to be expressed in their functional form in vitro (see “Discussion” for details). It should also be noted that 12% includes false positives; however, we consider its fraction to be small as described below. Therefore, we concluded that a significant fraction of ORFs encoded in the genome are functionally linked to the protein synthesis reaction carried out by only the minimum number of protein components.
Validating the Results of Comprehensive Analysis—
To verify that the beneficial components defined in Fig. 1C are effective when added to the reaction as purified proteins, HrpA (24) (ATP-dependent RNA helicase), Orn (25) (oligoribonuclease), TrxC (26) (thioredoxin 2), Tig (6, 27) (trigger factor), SlyD (28) (FK506-binding protein-type peptidyl-prolyl cis-trans isomerase), and PhnH (29) (carbon-phosphorus lyase complex subunit) proteins (ranked 1, 7, 3, 2, 9, and 4, respectively) were overexpressed and purified from E. coli. In addition to the six purified proteins, YbaW was purified as a control. The effects of these proteins on the GFP synthesis reaction were investigated by adding different concentrations of purified proteins to the reaction at concentrations ranging between 0.25 nm and 5 μm (Fig. 2A). Although the optimum concentration for each protein differed (Fig. 2A), addition of six purified proteins resulted in an increase in the fluorescence intensity, but there was no increase with the control protein YbaW (Fig. 2B). Furthermore when all six proteins (HrpA, Orn, PhnH, SlyD, Tig, and TrxC) were added at the optimum concentrations, the fluorescence intensity increased by 2.44-fold (Fig. 2B, Complete). We also investigated whether the mixture of these six proteins was effective in increasing the yields of the synthesized proteins and found no detectable differences in the yield (inset). SlyD (28) and Tig (6, 27) are known as chaperones and indeed were effective in increasing the yield of functional GFP (Fig. 2B). HrpA (ATP-dependent RNA helicase), which showed the maximum effect among the six, has been reported to be involved in mRNA processing (30) but has not been shown to increase the fraction of functional proteins during the translation reaction. The roles of Orn, TrxC, and PhnH remain to be determined. Nevertheless it is important to note that we were able to verify the results obtained through comprehensive analysis using purified proteins.
We then evaluated whether the effects of these proteins were additive (Fig. 2C). That is, we examined whether the mechanisms responsible for the increase in GFP fluorescence by each protein were independent. Each of the six proteins was omitted from the mixture of the six (Fig. 2B, Complete), and the decrease in fluorescence intensity relative to that with the mixture of all six proteins was measured. Fig. 2C shows the correlation between the increase in observed fluorescence intensity with addition of one of the six proteins and the decrease in the observed fluorescence intensity with the omission of one of the six proteins during GFP synthesis reaction. If the effects were additive, the plots would be expected to align on the line with a slope of 1 (Fig. 2C, gray line). This was almost the case with five proteins identified (HrpA, Orn, Tig, PhnH, and TrxC) indicating that their mechanisms of action on the GFP synthesis reaction are independent. The additivity observed here and its relationship with the protein-protein interaction network are discussed below.
As shown in Fig. 1C, inset, we found 10 ORF products that completely suppressed the GFP synthesis reaction in addition to many that decreased the efficiency to a lesser extent. These genes included those encoding the transcriptional repressor LacI (31) (GFP DNA construct used contained the lacO region), toxins (32–34) (ChpA, ChpB, RelE, and YoeB) known to exhibit ribosome-dependent nuclease activity, and the nuclease Rnt (35) (supplemental Fig. 1A). Thus, our data were consistent with previous observations. Note that we confirmed suppression of the protein synthesis reaction of several proteins by SDS-PAGE (supplemental Fig. 1B). On the other hand, a functionally uncharacterized protein (YhaV), DNA polymerase I (PolA (36)), and GTP cyclohydrolase II (RibA (37)) were also found to be lethal for the reaction. YhaV protein has been reported to exhibit distant but significant similarity to RelE toxin protein (38), and based on this observation, YhaV protein was proposed to be a toxin member of the toxin-antitoxin system. We found that YhaV protein completely inhibited the translational machinery, similar to other toxin proteins. Our results together with those reported recently by Schmidt et al. (39) represent experimental evidence that YhaV protein may be a new toxin member of the toxin-antitoxin system. However, the bases of the inhibitory effects of PolA, RibA, and many other proteins have yet to be determined.
Functional Classification of the Modifiers—
Approximately 60% of E. coli ORF products have experimentally assigned functions (3). Therefore, we investigated the functional classification of the modifiers (Fig. 3). We found that ∼40% of the modifiers do not have experimentally assigned functions (Fig. 3A). Note that we included those assigned as “predicted functions” by Riley et al. (3) as “unknown.” Nevertheless ∼15% remained unknown even after classifying the predicted products based on their predicted functions. These results indicate that our data will be useful in future studies on individual proteins.
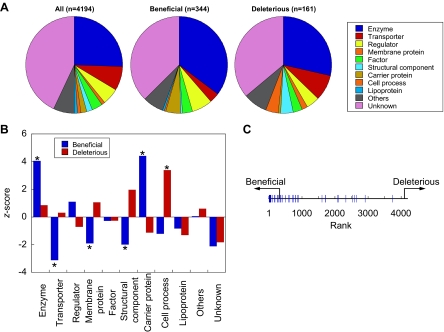
Fig. 3.
Functional classification of the modifiers. A, functional classification of the entire ORF (left), beneficial (middle), and deleterious (right) components. The fractions were calculated from the data shown in supplemental Table 1. The group “Others” consisted of “leader peptide,” “pseudogenes in common between strains,” “phage in common between strains,” and “predicted phage in common between strains” depicted in supplemental Table 1. Unknown consisted of “partial information,” “unidentified protein,” “W3110-specific protein”, and those with predicted functions (3) as depicted in supplemental Table 1. B, differences in the fraction of each functional class between the entire ORF products and beneficial (blue) or deleterious (red) components. The horizontal axis shows the z-score = (Ni − npi)/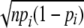 , where Ni, n, and pi are the number of ORFs classified as functional class i, total number of beneficial or deleterious components, and fraction of functional class i of the entire ORF products, respectively. Those with significant differences (p < 0.05) verified by the binomial test are marked with a star. The p values were (from the left): 9.4 × 10−7, 8.4 × 10−8, 4.3 × 10−6, 0.015, 2.8 × 10−7, and 0.006, respectively. C, rank order plot of the entire ORF (horizontal black line) and the locations of 40 Fe-S cluster-containing proteins (blue line). ORFs were aligned according to their rank given in Fig. 1C. The arrows indicate the regions where the beneficial and deleterious genes are located.
, where Ni, n, and pi are the number of ORFs classified as functional class i, total number of beneficial or deleterious components, and fraction of functional class i of the entire ORF products, respectively. Those with significant differences (p < 0.05) verified by the binomial test are marked with a star. The p values were (from the left): 9.4 × 10−7, 8.4 × 10−8, 4.3 × 10−6, 0.015, 2.8 × 10−7, and 0.006, respectively. C, rank order plot of the entire ORF (horizontal black line) and the locations of 40 Fe-S cluster-containing proteins (blue line). ORFs were aligned according to their rank given in Fig. 1C. The arrows indicate the regions where the beneficial and deleterious genes are located.
We also investigated the ratio of proteins belonging to each functional classification and compared the differences between the entire data set and the modifiers (Fig. 3B). We found that with the beneficial component fraction those classified as enzyme and carrier proteins were significantly increased, whereas those classified as transporters, membrane proteins, and structural components were significantly reduced relative to the entire data set (blue bars with stars). With the deleterious component fraction, those classified as cell process components were significantly increased (red bar with a star). The levels of membrane proteins, which are likely to misfold and aggregate when expressed in vitro, were not increased in the deleterious component fraction. These observations suggested that the GFP synthesis reaction is not affected by the presence of aggregating and sticky proteins and that the majority of the deleterious components do not inhibit the GFP synthesis reaction via stickiness of the modifiers. We focused on the individual functions of the modifiers with increased ratios relative to the entire data set and found for example that 40 Fe-S cluster-containing proteins (carrier proteins) were clustered close to and within the beneficial region (Fig. 3C). Although the basis of this observation is not yet clear, this result indicates that proteins belonging to this class stimulate GFP synthesis. It is also important to note that the differences in the ratios of functional classification of the modifiers from the entire data set indicate that the modifiers were not selected randomly but instead that there were reasons for their selection.
Distance of the Modifiers from the Minimal Protein Components in the Protein-Protein Interaction Network—
The PPI network showed physical contact between the proteins, but these were not necessarily functional. Therefore, we investigated whether the experimentally identified modifiers tend to have direct contact with the minimal components in the PPI network. At present, two sets of data are available for the E. coli PPI network (4, 5). In this study for further analysis, we used the data of Butland et al. (4) that consisted of 1255 proteins among the ∼4300 predicted ORFs (3); these data maintained the cellular stoichiometry when acquiring the data and included the interactions within large complexes. The details of the differences between the two PPI networks and the reasons for using that of Butland et al. (4) are described in supplemental Table 2. In this data set, the minimal protein components, of which 91% (83 of 93) were present in this network, were shown to interact directly with 423 proteins (see supplemental Table 3 for details regarding the number of proteins in the PPI network).
Within this PPI network, we first investigated the distance from each modifier to the closest minimal protein components where the distance between two proteins is defined as the minimum number of links between the two. The distance distribution shown in Fig. 4A indicated that ∼34% of the modifiers present in the PPI network were linked directly (i.e. showed physical contact) with the minimal protein components in the PPI network (□ and ▪), whereas the others were not. Furthermore beneficial (□) and deleterious (▪) components were found to have average distances of 1.69 and 1.76, respectively, to the closest minimal components, similar to the average distance from the entire protein complement (○) to the closest minimal components (1.73). In addition, no bias was found in the distance distribution of the modifiers compared with the entire protein, implying that modifiers were not clustering in close proximity to the minimal components in the PPI network. The protein synthesis reaction involves not only the protein components but also RNA and other low molecular weight substances. Therefore, functional modifiers do not necessarily have to interact with the minimal protein components; this in fact was the case. This is the first report of identification of the locations of functional modifiers of the protein translation system on the PPI network based on experimental results.
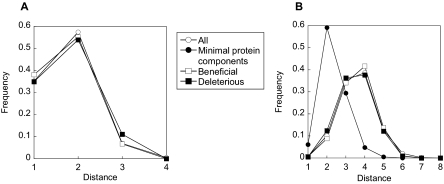
Fig. 4.
The protein synthesis system in the PPI network. A, the distribution of the distances from each protein to the nearest minimal components. B, the distribution of the distances between all protein pairs in the same group. The differences in the internal distance with the minimal components and the modifiers were confirmed by an unequal variance t test (p = 2.2 × 10−16).
Topological Relationship between Modifiers and the Minimal Protein Components in the Protein-Protein Interaction Network—
As described in the previous section, the modifiers did not cluster in close proximity to the minimal components in the PPI network. This information alone does not provide a topological relationship between modifiers and the minimal protein components in the PPI network. For example, it could be explained by the clustering of the minimal components and the modifiers separately but with the two clusters being distant from each other. Alternatively the minimal components may be highly clustered with each other, whereas the modifiers may be spread across the entire network. To obtain further insight into the topological relationships between modifiers and minimal protein components in the PPI network, we thus investigated whether the proteins in the same group (minimal components versus modifiers) were clustered in the PPI network. The distributions of the distances between all protein pairs in the same group are shown in Fig. 4B. The distance between two proteins is defined as the minimum number of links between the two. From the distance distribution, beneficial (□) and deleterious (▪) components were found to have average internal distances of 3.62 and 3.51, respectively, similar to that of the entire protein complement (○), which was found to be 3.60. On the other hand, minimal components (•) were found to have an average distance of 2.34. These results indicate that although the minimal components were highly clustered with each other modifiers were spread across the entire network as the distance distribution and mean distance between all protein pairs within beneficial and deleterious components were similar to those of the entire protein complement (Fig. 4B).
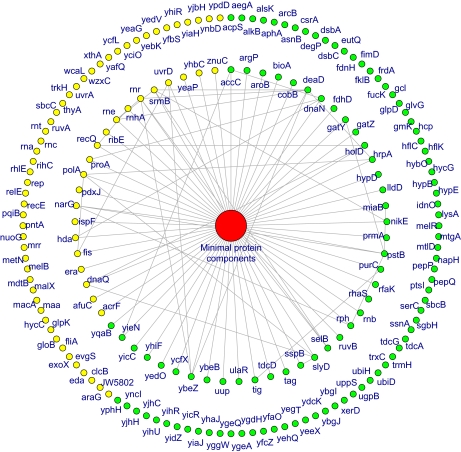
The minimal protein components were clustered, and modifiers were not in close proximity to the minimal protein components but were instead distributed across the entire network. From these observations (Fig. 4, A and B), a schematic of topology of the protein synthesis system in the PPI network was drawn (Fig. 5). The inner and outer circles of the modifiers are those that interact directly or indirectly with the minimum protein components, respectively.
Fig. 5.
Schematic illustration of the protein synthesis system in the PPI network. The minimal protein components were clustered in the PPI network. We located these at the center, and the modifiers are outside as they were distributed evenly across the entire network. The inner circle shows the modifiers that were found to interact directly with the minimal protein components in the PPI data. The outer circle shows the other modifiers for which direct interactions were not detected but were present in the PPI data. Green and yellow circles indicate the beneficial and deleterious protein components, respectively. Gene names are also shown.
PPI data have been suggested to include false positives (40, 41). It is also possible that the fraction of modifiers was underestimated because our assay was performed with one particular protein, GFP, and required the ORF products to be expressed in their functional form in vitro. These two considerations may affect the topology of the PPI network. However, as the distance distribution of modifiers was nearly identical to that of the entire protein complement (Fig. 4), the topology identified above will not be affected by the presence of errors in the network unless the locations of the false negatives or positives are highly biased to show a certain distance from the minimal components. Thus, we concluded that errors in the PPI network as well as the underestimation of the fraction of modifiers are unlikely to affect the identified topological structure of the protein synthesis system in the PPI network shown in Fig. 5.
Evolutionary Relationship between Minimal Components and the Modifiers of the Protein Synthesis System—
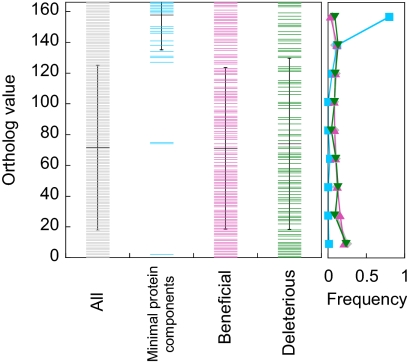
To obtain insight into the evolution of the protein translation system and its relationship with the topology in the PPI network described above, we investigated how each E. coli protein is conserved among bacterial species. As a measure, we used the Microbial Genome Database (22) and obtained the number of bacterial species among 166 included in the orthologous groups where each E. coli ORF is classified. Fig. 6 shows that the minimal protein components, highly clustered in the PPI network, were highly conserved among bacterial species. On the other hand, beneficial and deleterious components, distributed evenly across the entire PPI network, were not as conserved as the minimal components, indicating that most but not all of the modifiers appeared later than the minimal components. A possible interpretation of this observation in relation to the evolution of the protein synthesis system is discussed below.
Fig. 6.
Ortholog values of proteins involved in the protein synthesis reaction. Ortholog value (vertical axis) was defined as the number of prokaryotic bacterial species among 166 included in the orthologous groups (22) where each E. coli ORF was classified. Average and S.D. of the values of each protein group are also shown (black lines). The frequency distributions of the ortholog values for each protein group (all, minimal, beneficial, and deleterious components) are shown at the right of the figure.
DISCUSSION
The protein translation reaction, one of the most important regulators of cell behavior, can be operated by 91 protein components (55 ribosomal proteins + 36 factors), representing only 2.1% of the entire complement of ORFs in E. coli. Through comprehensive analysis, we showed experimentally that at least 12% of the E. coli genes affected the activity of the minimal protein synthesis system (Fig. 1C). By mapping the modifiers and the minimal components on the E. coli PPI network, we found that the functional modifiers are not necessarily linked physically with the minimal proteins but are instead spread across the entire PPI network (Fig. 4A). We also found that the minimal components are clustered in the network, whereas the modifiers tend to be distributed across the entire network (Figs. 4B and 5).
We showed that 12% of E. coli genes interact with the protein synthesis system composed of a minimal number of protein components. Our results shown in Fig. 1C are likely to be highly reliable for several reasons. First, measurement errors were smaller than the variety obtained by using different ORFs (Fig. 1, B and C). Second, ORF products that are expected to suppress the translation reaction were indeed found to be deleterious (Fig. 1C, inset). Third, ORF products that were found to be beneficial to the translation reaction through high throughput screening (Fig. 1C) were indeed found to show such effects as confirmed by adding the purified proteins (Fig. 2). Finally functional classification charts of the modifiers were different from those of the entire data set (Fig. 3). From these observations, although the 12% may include false positives, we concluded that a substantial fraction of the ORFs identified as interacting with the translation reaction are reliable. However, it should be noted that 12% is the minimum value as our assay was performed with one particular protein, GFP, and more importantly our assay requires the ORF products to be expressed in vitro and also in functional form. We used a tetracysteine tag (17, 18) to obtain qualitative data regarding whether the ORFs are expressed and found that more than 98% of ORF products gave detectable signals (supplemental Fig. 2A). Moreover as three arbitrarily chosen ORF products were expressed in the μm range in vitro (supplemental Fig. 2B), a substantial fraction is likely to be expressed to such an extent. However, it is also possible that proteins were not expressed functionally because of the lack of subunits with which to assemble or because of the inappropriate environment, resulting in false negatives. False negatives can also appear if the ORF products were not expressed at appropriate levels. For example, we identified HrpA as a beneficial modifier; however, if its expression level had been either significantly higher or lower, HrpA would have been assigned as a deleterious component or as one with no effect, respectively, on the GFP synthesis reaction (Fig. 2A). False positives and false negatives occur in our assay for example because of the above reasons. Nevertheless as we consider the fraction of false positives to be small, at least 12% of the genes, and very likely more, interact with the minimal protein synthesis system, although the underestimation is unlikely to affect the topology as described under “Results.” Here components interacting with the minimal protein synthesis system were defined as those that modified the protein synthesis activity without distinguishing how the activity is modified. Although there are number of possible ways to modify the activity of the system, we are not yet able to demonstrate how each modifier affected the reaction. Nevertheless we successfully determined the fraction of the entire ORF products that had an effect on the GFP synthesis reaction.
What does the location of the functional modifiers on the PPI network tell us? One possibility is that this is a reflection of the evolutionary process of the protein synthesis system. We investigated how each E. coli protein is conserved among bacterial species using the Microbial Genome Database (22), and we found that minimal components were highly conserved, whereas most of the beneficial and deleterious components were not conserved to such an extent (Fig. 6), indicating that most but not all appeared later than the minimal components. Thus, the identified locations of the modifiers relative to the minimal protein synthesis system in the PPI network (Fig. 4, A and B) suggest that such attachment has occurred not within the network composed of minimal components but mostly on the outer network. These observations further suggest that the protein synthesis system may have evolved by first establishing a system with minimal components and then adding components further outside rather than integrating them into the network composed of the minimal components.
Previously it was reported that modification of the center of the network, which is comprised of proteins with a high degree of connectivity (i.e. a number of interacting proteins), tends to alter the network properties markedly, often resulting in collapse of the system (42, 43). Although still controversial (44, 45), theoretical studies have suggested a negative correlation between evolutionary rate and connectivity (46, 47). There seems to be difficulty in accumulating mutations in proteins with higher connectivity as these proteins are often important for maintenance of the system (42). Minimal protein components were reported to exhibit high degrees of connectivity (4). Therefore, difficultly in modification of the network composed of minimal components can be expected, and this may be the reason why we found the modifiers to be attached to the outside of the network composed of the minimal components rather than integrated within the center of the network.
Additivity of the effects of the beneficial components on the GFP synthesis reaction, shown in Fig. 2C, may also be important for evolution of the protein synthesis system. If the effects are additive, addition of a beneficial modifier to the system is unlikely to suppress the effects of those attached previously. Thus, additivity will allow a gradual increase in function of the system by incorporation of modifiers. Otherwise the system would need to conduct a combinatory search for sets of components capable of acting together to have a beneficial effect. Therefore, after its establishment with the minimal components, the protein synthesis system may have evolved gradually by incorporation of modifiers without damaging the network constructed by the minimal components.
In summary, we have obtained a data set of the effects of 4194 E. coli ORF products on the protein synthesis reaction, mapped the components involved in the reaction on the PPI network, and discussed the possible scenario of the protein translation system in the PPI network. The comprehensive data obtained in this study will be useful in future studies on individual proteins as well as functional genomics and systems biology. Although we are still at the starting point of investigating the precise roles of the modifier proteins on the reaction, combining the experimental data with the PPI network may provide insight into how these proteins interact with the translation machinery. Moreover our data may be useful not only for technologies utilizing in vitro translation systems (19, 48, 49) but also for designing in vitro translation systems with significantly improved performance than those available at present and also for protein production in vivo, such as by coexpressing beneficial components together with a protein of interest.
Supplementary Material
Acknowledgments
We thank Naoko Miki for technical assistance and Drs. H. Matsuda, C. Furusawa, I. Urabe (Osaka University), and Y. Shimizu (University of Tokyo) for helpful discussions. This research was partially conducted in Open Laboratories for Advanced Bioscience and Biotechnology, Osaka University.
Footnotes
Published, MCP Papers in Press, May 2, 2008, DOI 10.1074/mcp.M800051-MCP200
The abbreviations used are: PPI, protein-protein interaction; GFP, green fluorescent protein; ASKA library, a complete set of E. coli K12 ORF archive; PURE system, protein synthesis using recombinant elements; a.u., arbitrary unit.
This research was supported in part by “Special Coordination Funds for Promoting Science and Technology: Yuragi Project, Noda Institute for Scientific Research” and the “Global COE (Centers of Excellence) Program” of the Ministry of Education, Culture, Sports, Science, and Technology, Japan. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Nierhaus, K. H., and Wilson, D. N. ( 2004) Protein Synthesis and Ribosome Structure: Translating the Genome, Wiley-VCH, Weinheim, Germany
- 2.Shimizu, Y., Inoue, A., Tomari, Y., Suzuki, T., Yokogawa, T., Nishikawa, K., and Ueda, T. ( 2001) Cell-free translation reconstituted with purified components. Nat. Biotechnol. 19, 751–755 [DOI] [PubMed] [Google Scholar]
- 3.Riley, M., Abe, T., Arnaud, M. B., Berlyn, M. K., Blattner, F. R., Chaudhuri, R. R., Glasner, J. D., Horiuchi, T., Keseler, I. M., Kosuge, T., Mori, H., Perna, N. T., Plunkett, G., III, Rudd, K. E., Serres, M. H., Thomas, G. H., Thomson, N. R., Wishart, D., and Wanner, B. L. ( 2006) Escherichia coli K-12: a cooperatively developed annotation snapshot—2005. Nucleic Acids Res. 34, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butland, G., Peregrin-Alvarez, J. M., Li, J., Yang, W., Yang, X., Canadien, V., Starostine, A., Richards, D., Beattie, B., Krogan, N., Davey, M., Parkinson, J., Greenblatt, J., and Emili, A. ( 2005) Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature 433, 531–537 [DOI] [PubMed] [Google Scholar]
- 5.Arifuzzaman, M., Maeda, M., Itoh, A., Nishikata, K., Takita, C., Saito, R., Ara, T., Nakahigashi, K., Huang, H. C., Hirai, A., Tsuzuki, K., Nakamura, S., Altaf-Ul-Amin, M., Oshima, T., Baba, T., Yamamoto, N., Kawamura, T., Ioka-Nakamichi, T., Kitagawa, M., Tomita, M., Kanaya, S., Wada, C., and Mori, H. ( 2006) Large-scale identification of protein-protein interaction of Escherichia coli K-12. Genome Res. 16, 686–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maier, T., Ferbitz, L., Deuerling, E., and Ban, N. ( 2005) A cradle for new proteins: trigger factor at the ribosome. Curr. Opin. Struct. Biol. 15, 204–212 [DOI] [PubMed] [Google Scholar]
- 7.Qin, Y., Polacek, N., Vesper, O., Staub, E., Einfeldt, E., Wilson, D. N., and Nierhaus, K. H. ( 2006) The highly conserved LepA is a ribosomal elongation factor that back-translocates the ribosome. Cell 127, 721–733 [DOI] [PubMed] [Google Scholar]
- 8.Uetz, P., Giot, L., Cagney, G., Mansfield, T. A., Judson, R. S., Knight, J. R., Lockshon, D., Narayan, V., Srinivasan, M., Pochart, P., Qureshi-Emili, A., Li, Y., Godwin, B., Conover, D., Kalbfleisch, T., Vijayadamodar, G., Yang, M., Johnston, M., Fields, S., and Rothberg, J. M. ( 2000) A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403, 623–627 [DOI] [PubMed] [Google Scholar]
- 9.Ito, T., Chiba, T., Ozawa, R., Yoshida, M., Hattori, M., and Sakaki, Y. ( 2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. U. S. A. 98, 4569–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giot, L., Bader, J. S., Brouwer, C., Chaudhuri, A., Kuang, B., Li, Y., Hao, Y. L., Ooi, C. E., Godwin, B., Vitols, E., Vijayadamodar, G., Pochart, P., Machineni, H., Welsh, M., Kong, Y., Zerhusen, B., Malcolm, R., Varrone, Z., Collis, A., Minto, M., Burgess, S., McDaniel, L., Stimpson, E., Spriggs, F., Williams, J., Neurath, K., Ioime, N., Agee, M., Voss, E., Furtak, K., Renzulli, R., Aanensen, N., Carrolla, S., Bickelhaupt, E., Lazovatsky, Y., DaSilva, A., Zhong, J., Stanyon, C. A., Finley, R. L., Jr., White, K. P., Braverman, M., Jarvie, T., Gold, S., Leach, M., Knight, J., Shimkets, R. A., McKenna, M. P., Chant, J., and Rothberg, J. M. ( 2003) A protein interaction map of Drosophila melanogaster. Science 302, 1727–1736 [DOI] [PubMed] [Google Scholar]
- 11.Li, S., Armstrong, C. M., Bertin, N., Ge, H., Milstein, S., Boxem, M., Vidalain, P. O., Han, J. D., Chesneau, A., Hao, T., Goldberg, D. S., Li, N., Martinez, M., Rual, J. F., Lamesch, P., Xu, L., Tewari, M., Wong, S. L., Zhang, L. V., Berriz, G. F., Jacotot, L., Vaglio, P., Reboul, J., Hirozane-Kishikawa, T., Li, Q., Gabel, H. W., Elewa, A., Baumgartner, B., Rose, D. J., Yu, H., Bosak, S., Sequerra, R., Fraser, A., Mango, S. E., Saxton, W. M., Strome, S., Van Den Heuvel, S., Piano, F., Vandenhaute, J., Sardet, C., Gerstein, M., Doucette-Stamm, L., Gunsalus, K. C., Harper, J. W., Cusick, M. E., Roth, F. P., Hill, D. E., and Vidal, M. ( 2004) A map of the interactome network of the metazoan C. elegans. Science 303, 540–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rual, J. F., Venkatesan, K., Hao, T., Hirozane-Kishikawa, T., Dricot, A., Li, N., Berriz, G. F., Gibbons, F. D., Dreze, M., Ayivi-Guedehoussou, N., Klitgord, N., Simon, C., Boxem, M., Milstein, S., Rosenberg, J., Goldberg, D. S., Zhang, L. V., Wong, S. L., Franklin, G., Li, S., Albala, J. S., Lim, J., Fraughton, C., Llamosas, E., Cevik, S., Bex, C., Lamesch, P., Sikorski, R. S., Vandenhaute, J., Zoghbi, H. Y., Smolyar, A., Bosak, S., Sequerra, R., Doucette-Stamm, L., Cusick, M. E., Hill, D. E., Roth, F. P., and Vidal, M. ( 2005) Towards a proteome-scale map of the human protein-protein interaction network. Nature 437, 1173–1178 [DOI] [PubMed] [Google Scholar]
- 13.Stelzl, U., Worm, U., Lalowski, M., Haenig, C., Brembeck, F. H., Goehler, H., Stroedicke, M., Zenkner, M., Schoenherr, A., Koeppen, S., Timm, J., Mintzlaff, S., Abraham, C., Bock, N., Kietzmann, S., Goedde, A., Toksoz, E., Droege, A., Krobitsch, S., Korn, B., Birchmeier, W., Lehrach, H., and Wanker, E. E. ( 2005) A human protein-protein interaction network: a resource for annotating the proteome. Cell 122, 957–968 [DOI] [PubMed] [Google Scholar]
- 14.Kitagawa, M., Ara, T., Arifuzzaman, M., Ioka-Nakamichi, T., Inamoto, E., Toyonaga, H., and Mori, H. ( 2005) Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 12, 291–299 [DOI] [PubMed] [Google Scholar]
- 15.Yokoyama, S. ( 2003) Protein expression systems for structural genomics and proteomics. Curr. Opin. Chem. Biol. 7, 39–43 [DOI] [PubMed] [Google Scholar]
- 16.Endo, Y., and Sawasaki, T. ( 2006) Cell-free expression systems for eukaryotic protein production. Curr. Opin. Biotechnol. 17, 373–380 [DOI] [PubMed] [Google Scholar]
- 17.Adams, S. R., Campbell, R. E., Gross, L. A., Martin, B. R., Walkup, G. K., Yao, Y., Llopis, J., and Tsien, R. Y. ( 2002) New biarsenical ligands and tetracysteine motifs for protein labeling in vitro and in vivo: synthesis and biological applications. J. Am. Chem. Soc. 124, 6063–6076 [DOI] [PubMed] [Google Scholar]
- 18.Martin, B. R., Giepmans, B. N., Adams, S. R., and Tsien, R. Y. ( 2005) Mammalian cell-based optimization of the biarsenical-binding tetracysteine motif for improved fluorescence and affinity. Nat. Biotechnol. 23, 1308–1314 [DOI] [PubMed] [Google Scholar]
- 19.Sunami, T., Sato, K., Matsuura, T., Tsukada, K., Urabe, I., and Yomo, T. ( 2006) Femtoliter compartment in liposomes for in vitro selection of proteins. Anal. Biochem. 357, 128–136 [DOI] [PubMed] [Google Scholar]
- 20.Ito, Y., Suzuki, M., and Husimi, Y. ( 1999) A novel mutant of green fluorescent protein with enhanced sensitivity for microanalysis at 488 nm excitation. Biochem. Biophys. Res. Commun. 264, 556–560 [DOI] [PubMed] [Google Scholar]
- 21.Mathews, D. E., and Durbin, R. D. ( 1990) Tagetitoxin inhibits RNA synthesis directed by RNA polymerases from chloroplasts and Escherichia coli. J. Biol. Chem. 265, 493–498 [PubMed] [Google Scholar]
- 22.Uchiyama, I. ( 2007) MBGD: a platform for microbial comparative genomics based on the automated construction of orthologous groups. Nucleic Acids Res. 35, D343–D346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blattner, F. R., Plunkett, G., III, Bloch, C. A., Perna, N. T., Burland, V., Riley, M., Collado-Vides, J., Glasner, J. D., Rode, C. K., Mayhew, G. F., Gregor, J., Davis, N. W., Kirkpatrick, H. A., Goeden, M. A., Rose, D. J., Mau, B., and Shao, Y. ( 1997) The complete genome sequence of Escherichia coli K-12. Science 277, 1453–1474 [DOI] [PubMed] [Google Scholar]
- 24.Moriya, H., Kasai, H., and Isono, K. ( 1995) Cloning and characterization of the hrpA gene in the terC region of Escherichia coli that is highly similar to the DEAH family RNA helicase genes of Saccharomyces cerevisiae. Nucleic Acids Res. 23, 595–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang, X., Zhu, L., and Deutscher, M. P. ( 1998) Oligoribonuclease is encoded by a highly conserved gene in the 3′–5′ exonuclease superfamily. J. Bacteriol. 180, 2779–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miranda-Vizuete, A., Damdimopoulos, A. E., Gustafsson, J., and Spyrou, G. ( 1997) Cloning, expression, and characterization of a novel Escherichia coli thioredoxin. J. Biol. Chem. 272, 30841–30847 [DOI] [PubMed] [Google Scholar]
- 27.Stoller, G., Rucknagel, K. P., Nierhaus, K. H., Schmid, F. X., Fischer, G., and Rahfeld, J. U. ( 1995) A ribosome-associated peptidyl-prolyl cis/trans isomerase identified as the trigger factor. EMBO J. 14, 4939–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hottenrott, S., Schumann, T., Pluckthun, A., Fischer, G., and Rahfeld, J. U. ( 1997) The Escherichia coli SlyD is a metal ion-regulated peptidyl-prolyl cis/trans-isomerase. J. Biol. Chem. 272, 15697–15701 [DOI] [PubMed] [Google Scholar]
- 29.Chen, C. M., Ye, Q. Z., Zhu, Z. M., Wanner, B. L., and Walsh, C. T. ( 1990) Molecular biology of carbon-phosphorus bond cleavage. Cloning and sequencing of the phn (psiD) genes involved in alkylphosphonate uptake and C-P lyase activity in Escherichia coli B. J. Biol. Chem. 265, 4461–4471 [PubMed] [Google Scholar]
- 30.Koo, J. T., Choe, J., and Moseley, S. L. ( 2004) HrpA, a DEAH-box RNA helicase, is involved in mRNA processing of a fimbrial operon in Escherichia coli. Mol. Microbiol. 52, 1813–1826 [DOI] [PubMed] [Google Scholar]
- 31.Farabaugh, P. J. ( 1978) Sequence of the lacI gene. Nature 274, 765–769 [DOI] [PubMed] [Google Scholar]
- 32.Masuda, Y., Miyakawa, K., Nishimura, Y., and Ohtsubo, E. ( 1993) chpA and chpB, Escherichia coli chromosomal homologs of the pem locus responsible for stable maintenance of plasmid R100. J. Bacteriol. 175, 6850–6856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gotfredsen, M., and Gerdes, K. ( 1998) The Escherichia coli relBE genes belong to a new toxin-antitoxin gene family. Mol. Microbiol. 29, 1065–1076 [DOI] [PubMed] [Google Scholar]
- 34.Grady, R., and Hayes, F. ( 2003) Axe-Txe, a broad-spectrum proteic toxin-antitoxin system specified by a multidrug-resistant, clinical isolate of Enterococcus faecium. Mol. Microbiol. 47, 1419–1432 [DOI] [PubMed] [Google Scholar]
- 35.Huang, S., and Deutscher, M. P. ( 1992) Sequence and transcriptional analysis of the Escherichia coli rnt gene encoding RNase T. J. Biol. Chem. 267, 25609–25613 [PubMed] [Google Scholar]
- 36.Joyce, C. M., Kelley, W. S., and Grindley, N. D. ( 1982) Nucleotide sequence of the Escherichia coli polA gene and primary structure of DNA polymerase I. J. Biol. Chem. 257, 1958–1964 [PubMed] [Google Scholar]
- 37.Richter, G., Ritz, H., Katzenmeier, G., Volk, R., Kohnle, A., Lottspeich, F., Allendorf, D., and Bacher, A. ( 1993) Biosynthesis of riboflavin: cloning, sequencing, mapping, and expression of the gene coding for GTP cyclohydrolase II in Escherichia coli. J. Bacteriol. 175, 4045–4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coles, M., Djuranovic, S., Soding, J., Frickey, T., Koretke, K., Truffault, V., Martin, J., and Lupas, A. N. ( 2005) AbrB-like transcription factors assume a swapped hairpin fold that is evolutionarily related to double-psi β barrels. Structure (Lond.) 13, 919–928 [DOI] [PubMed] [Google Scholar]
- 39.Schmidt, O., Schuenemann, V. J., Hand, N. J., Silhavy, T. J., Martin, J., Lupas, A. N., and Djuranovic, S. ( 2007) prlF and yhaV encode a new toxin-antitoxin system in Escherichia coli. J. Mol. Biol. 372, 894–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bader, G. D., and Hogue, C. W. ( 2002) Analyzing yeast protein-protein interaction data obtained from different sources. Nat. Biotechnol. 20, 991–997 [DOI] [PubMed] [Google Scholar]
- 41.von Mering, C., Krause, R., Snel, B., Cornell, M., Oliver, S. G., Fields, S., and Bork, P. ( 2002) Comparative assessment of large-scale data sets of protein-protein interactions. Nature 417, 399–403 [DOI] [PubMed] [Google Scholar]
- 42.Jeong, H., Mason, S. P., Barabasi, A. L., and Oltvai, Z. N. ( 2001) Lethality and centrality in protein networks. Nature 411, 41–42 [DOI] [PubMed] [Google Scholar]
- 43.Albert, R., Jeong, H., and Barabasi, A. L. ( 2000) Error and attack tolerance of complex networks. Nature 406, 378–382 [DOI] [PubMed] [Google Scholar]
- 44.Batada, N. N., Hurst, L. D., and Tyers, M. ( 2006) Evolutionary and physiological importance of hub proteins. PLoS Comput. Biol. 2, e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jordan, I. K., Wolf, Y. I., and Koonin, E. V. ( 2003) No simple dependence between protein evolution rate and the number of protein-protein interactions: only the most prolific interactors tend to evolve slowly. BMC Evol. Biol. 3, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim, P. M., Lu, L. J., Xia, Y., and Gerstein, M. B. ( 2006) Relating three-dimensional structures to protein networks provides evolutionary insights. Science 314, 1938–1941 [DOI] [PubMed] [Google Scholar]
- 47.Fraser, H. B., Hirsh, A. E., Steinmetz, L. M., Scharfe, C., and Feldman, M. W. ( 2002) Evolutionary rate in the protein interaction network. Science 296, 750–752 [DOI] [PubMed] [Google Scholar]
- 48.Matsuura, T., Yanagida, H., Ushioda, J., Urabe, I., and Yomo, T. ( 2007) Nascent chain, mRNA, and ribosome complexes generated by a pure translation system. Biochem. Biophys. Res. Commun. 352, 372–377 [DOI] [PubMed] [Google Scholar]
- 49.Yanagida, H., Matsuura, T., and Yomo, T. ( 2008) Compensatory evolution of a WW domain variant lacking the strictly conserved Trp residue. J. Mol. Evol. 66, 61–71 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.