Abstract
Caspase-6 activation occurs early in Alzheimer disease and sometimes precedes the clinical manifestation of the disease in aged individuals. The active Caspase-6 is localized in neuritic plaques, in neuropil threads, and in neurofibrillary tangles containing neurons that are not morphologically apoptotic in nature. To investigate the potential consequences of the activation of Caspase-6 in neurons, we conducted a proteomics analysis of Caspase-6-mediated cleavage of human neuronal proteins. Proteins from the cytosolic and membrane subcellular compartments were treated with recombinant active Caspase-6 and compared with undigested proteins by two-dimensional gel electrophoresis. LC/MS/MS analyses of the proteins that were cleaved identified 24 different potential protein substrates. Of these, 40% were cytoskeleton or cytoskeleton-associated proteins. We focused on the cytoskeleton proteins because these are critical for neuronal structure and function. Caspase-6 cleavage of α-Tubulin, α-Actinin-4, Spinophilin, and Drebrin was confirmed. At least one Caspase-6 cleavage site was identified for Drebrin, Spinophilin, and α-Tubulin. A neoepitope antiserum to α-Tubulin cleaved by Caspase-6 immunostained neurons, neurofibrillary tangles, neuropil threads, and neuritic plaques in Alzheimer disease and co-localized with active Caspase-6. These results imply that the early and neuritic activation of Caspase-6 in Alzheimer disease could disrupt the cytoskeleton network of neurons, resulting in impaired neuronal structure and function in the absence of cell death. This study provides novel insights into the pathophysiology of Alzheimer disease.
Caspases have been investigated in human neurodegenerative diseases based on the finding that the Caspase-3 (Csp3)1-null mice forego developmental neuronal cell death. Two studies have shown active Csp3 in granulovacuolar degeneration and in a few apoptotic neurons, but co-localization of active Csp3 with the hallmark pathological features of Alzheimer disease (AD) has not been reported (1, 2).
In contrast, Csp6 is highly activated in neuritic plaques, neuropil threads, and neurofibrillary tangles in the brain of AD individuals as demonstrated by immunohistochemistry with neoepitope antisera against active Csp6 and Tau cleaved by Csp6 (TauΔCsp6) (3, 4). Csp6 activation is observed during all stages of AD and in some mildly cognitively impaired and non-cognitively impaired aged individuals. The activation of Csp6 correlates with a lower cognitive score in normal aged individuals. Unexpectedly active Csp6 does not translocate to the nuclei in AD neurons (3). In contrast, nuclear translocation of Csp6 occurs in morphologically apoptotic neurons in human ischemia (3) and is responsible for the condensed chromatin in apoptotic cell cultures (5). Therefore, active Csp6 in AD neurons could indicate its implication in neurodegeneration rather than apoptosis. However, most of the identified substrates for Csp6 are nuclear (5–19). In the cytosol, Csp6 cleaves desmin, vimentin, and cytokeratin intermediate filament proteins; periplakin; Ufd2p; Nedd4; TRAF1; and focal adhesion kinase (20–27). Csp6 neuronal substrates are the microtubule-associated protein Tau (3), the amyloid precursor protein (28, 29), huntingtin (30), and presenilins 1 and 2 (31). Otherwise there are no other known neuronal substrates of Csp6. Several innovative approaches based on proteomics and mRNA display methods have been used recently to identify caspase substrates, but none have focused specifically on Csp6 or on neuronal protein substrates (32–35).
In this study, we exploited a simple proteomics approach to identify substrates of Csp6 in neurons. Human primary neuron cultures were fractionated into cytosolic and membrane fractions, and proteins were extracted and digested with recombinant active Csp6 (RCsp6). The proteins were submitted to two-dimensional gel electrophoresis, and those proteins digested by RCsp6 were sequenced by LC/MS/MS. Twenty-four different cleaved proteins of which 40% are cytoskeleton or cytoskeleton-associated proteins were identified. We confirmed α-Tubulin, Drebrin, Spinophilin, and α-Actinin-4 as Csp6 substrates. Furthermore a neoepitope antiserum raised against α-Tubulin cleaved by Csp6 showed specific immunoreactivity to the pathological hallmarks of AD. The results indicate that this proteomics approach is useful to identify previously unknown substrates of Csp6 and allow novel insight into Csp6-mediated defects in human neurons and AD.
EXPERIMENTAL PROCEDURES
Human Primary Neuron Cultures—
Human primary neurons were prepared from fetal cerebrum tissue according to the Canadian Institute of Health Research regulations and as approved by the McGill University Institutional Review Board (36).
Subcellular Fractionation and Digestion of Proteins with RCsp6—
Approximately 7.2 × 107 neurons were homogenized with a glass Dounce homogenizer (Kontes, Vineland, NJ) in ice-cold homogenization buffer containing protease inhibitors (8% sucrose, 1 mm EDTA, 20 mm Tricine, pH 7.8, 2 μg/ml chymostatin, 2 μg/ml pepstatin A, 2 μg/ml antipain HCl). Nuclei were removed by centrifugation at 385 × g for 5 min at 4 °C. The supernatant containing cytosolic and membrane proteins was centrifuged at 100,000 × g for 30 min at 4 °C. The membrane proteins were extracted with Stennicke's buffer (20 mm PIPES, 30 mm NaCl, 1 mm EDTA, 0.1% CHAPS, 10% sucrose, 10 mm DTT freshly added, pH 7.4) (37), and both fractions were stored at −80 °C. Protein concentrations were quantified by bicinchoninic acid (BCA) protein assay (Pierce). One hundred micrograms of total protein were digested with 270 ng of RCsp6 (BD Pharmingen) in Stennicke's buffer in a volume of 252 μl. Non-digested and digested proteins were incubated for 4 h at 37 °C, and proteins were precipitated with trichloroacetic acid.
Two-dimensional Gel Analysis—
One hundred micrograms of protein extracts, digested or not with RCsp6, were sent for two-dimensional (2D) gel analysis (38) to Kendrick Laboratories (Madison, WI). Fifty nanograms of tropomyosin was added to the samples as an internal isoelectric focusing standard. Proteins were separated based on their pI in a linear gradient of pH 3.5–10 (2% Ampholines, Amersham Biosciences) in glass tubes of 3.0-mm inner diameter. After separation in the first dimension for 20,000 V-h, the tube gels were equilibrated for 10 min in equilibration buffer (10% glycerol, 50 mm DTT, 2.3% SDS, 0.0625 m Tris, pH 6.8) and then sealed in agarose atop 1.0-mm-thick 10% acrylamide gels, and SDS-PAGE was carried out for 4 h at 12.5 mA/gel. Myosin, phosphorylase A, catalase, actin, carbonic anhydrase, and lysozyme were added as molecular weight markers in a well corresponding to the basic end of the gel. Proteins were silver-stained according to the method of O'Connell and Stults (39), and gels were subsequently dried between sheets of cellophane paper.
Selection of Spots from 2D Gels and LC/MS/MS—
The levels of 80% of the proteins chosen for analysis decreased by at least 50% (the rest decreased by at least 30%) in the digested samples versus the non-digested samples. The proteins were excised by Kendrick Laboratories and sent for analysis by LC/MS/MS to Protana Inc. (since acquired by Transition Therapeutics Inc., Ontario, Canada). Gel plugs were first washed in water and DTT followed by treatment with iodoacetamide. An in-gel digest with trypsin, which cuts C-terminal to a Lys or Arg residue, was done on the proteins, and the resulting peptides were extracted using acidic and basic conditions. All of these above steps were performed robotically (ProGest digestion robot, Genomic Solutions, Ann Arbor, MI). The peptides were analyzed by an LTQ-FT mass spectrometer (Thermo Finnigan, Waltham, MA) operated in data-dependent mode after first being separated based on polarity by reverse-phase chromatography on C18 resin. However, the ions were not transferred into the ICR cell because of poor transfer efficiency and the low levels of the samples, thus resulting in LTQ mass spectrometry. All peptides above a specified intensity were subjected to tandem mass spectroscopy fragmentation.
Analysis of MS/MS Peptide Data—
The search engine Mascot version 2.1 (Matrix Science, Boston, MA) was used to create the peak list and to compare the raw data files obtained with the entire National Center for Biotechnology Information (Bethesda, MD) non-redundant mammalian database files (NCBInr). The number of sequences and residues, the taxonomy, and the time stamp of each search are provided in the supplemental protein analysis. The searches were performed on a Mascot Daemon attached to an in-house 13-node cluster version of Mascot, and the search parameters were set at a parent ion tolerance of 1.5 Da; a fragment ion tolerance of 0.4 Da; one possible missed tryptic cut; and search for 1+, 2+, and 3+ ions with the machine type ESI trap. All results were validated manually for verifiability. The expectation scores and peptide scores correspond to the p values and the Mowse scores, respectively, as provided by Matrix Science. All peptide sequences end with either Lys or Arg with the exception of those peptides generated that are the sequence of the last C-terminal amino acids of the protein. Fixed modification of carbamidomethyl (Cys) and variable modifications of N-acetyl (protein) and oxidation (Met) were considered.
Assignment of Peptide Data Set to a Specific Protein—
A protein was identified as significant if the Mowse score was ≥70 (40), if the expectation score of the peptide sequenced was less than 10−3, if more than one peptide sequenced had a significant expectation score of <10−3, and if the identified protein had a molecular weight close to the expected experimental molecular weight. The protein identified from one spot was the protein with the highest protein coverage and with the highest number of peptide matches. The alternate names of each identified protein taken from the NCBI database or the Human Protein Resource Database (HPRD) are provided in the supplemental protein analysis, and care was taken not to have redundancy in the list of proteins identified. When a non-human protein was selected as the significant match for the peptide set, we confirmed that all peptides also matched the human counterpart of this protein. A table of all accession numbers matching the identified human protein is also provided in the supplemental material. When the set of peptides matched several members of the protein family, we indicated the match to this family and not to a specific member. If some of the peptides matched only one specific member, this member was chosen rather than the whole family. Peptide subsets matching other family members are indicated in the supplemental protein analysis.
In Vitro Translation and Site-directed Mutagenesis—
In vitro translation was conducted with the TnT system from Promega Corp. (Madison, WI) as described by the manufacturer. The human Drebrin cDNA was obtained from the American Type Culture Collection (Manassas, VA) and cloned in pET23b(+) (Novagen, EMD Bioscience Inc., San Diego, CA). The Spinophilin cDNA was kindly provided by Dr. G. La Mantia (Department of Structural and Functional Biology, University “Federico II,” Naples, Italy) in pcDNA3.1HisA. α-Actinin-4 (pCMV-XL4) cDNA was obtained from Origene (Rockville, MD). The wild type (WT), D431A, and D438A α-Tubulin cDNAs were a kind gift from Dr. Seamus J. Martin (Smurfit Institute, Dublin, Ireland). Mutations at specific sites were generated using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's protocol with the following primers: Spinophilin D411A, 5′-gccctggaggaggccgacgaagacgac-3′; Spinophilin D500A, 5′-gagctggagaaggcctccgagggcctg-3′; Spinophilin D551A, 5′-cctggtggaggtggctggaacaagtctgg-3′; Spinophilin D198A/D200A, 5′-ggacaagctggccgctgccgccgtgtccc-3′; Spinophilin D106A, 5′-gaacgagaacgtggcccacagcgccctgctg-3′; Spinophilin D125A, 5′-gtgagccgcttcgcctccaagcccgcg-3′; Spinophilin D198A, 5′-ggacaagctggccgctgacgccgtgtccc-3′; α-Actinin-4 D462A, 5′-ccttcgagagcgccctggctgcgca-3′; α-Actinin-4 D483A, 5′-gagctcaacgagctggcttactacgactcccac-3′; α-Actinin-4 D194A, 5′-ccacatcagctggaaggctggtcttgccttcaatg-3′; α-Actinin-4 D448A, 5′- cggccacactatcggccatcaaagccctcat-3′; α-Actinin-4 D487A, 5′-gagctggattactacgcctcccacaatgtcaac-3′; α-Actinin-4 D503A, 5′-gtgaccagtgggccgccctcggctc-3′; α-Actinin-4 D550A, 5′-gagagcgccatggaggccctccaggacatgttc-3′; α-Actinin-4 D579A, 5′-gtccaccctgcctgccgccgatagggagc-3′; α-Tubulin D33A, 5′-ggcatccagcccgctggccagatgcc-3′; and Drebrin D456A, 5′-accattgaaactgccactgccactgct-3′. All constructs were sent for DNA sequencing at the McGill University and Genome Quebec Innovation Centre.
Purified Proteins—
Tubulin was purified from bovine brains and was a kind gift from Dr. Hemant Paudel (Department of Neurology and Neurosurgery, McGill University). The β-actin was obtained from Cytoskeleton Inc. (Denver, CO).
Digestion of in Vitro Translated (IVT) or Purified Proteins with Caspases—
Proteins were digested for 4 h at 37 °C in Stennicke's buffer with RCsp6 (0.65 μg prepared from the pET23b cDNA construct generously provided by Dr. G. Salvesen (Burnham Institute, La Jolla, CA) or with RCsp3 (0.25 μg prepared from the pET21b cDNA construct generously provided by Dr. C. Clark, North Carolina State University) in a 20-μl final volume. IVT digested proteins were then separated by 8% (for Tubulin) or 10% (for all other proteins) SDS-PAGE. Gels were fixed in 50% methanol, 10% acetic acid and dried. Labeled proteins were visualized with Kodak BioMax MR film (Eastman Kodak Co.). Purified proteins were analyzed on Western blots.
Western Blot Analyses—
The rabbit neoepitope antiserum against Csp3-cleaved β-actin (Fractin) was kindly provided by Dr. Greg Cole (Department of Medicine, University of California, Los Angeles, CA) (41). Rabbit neoepitope antiserum to the p20 subunit of Csp1 was generated with the PGVVWFKD peptide through the services of Sigma-Genosys Proligo (The Woodlands, TX). Rabbit polyclonal antisera to Csp2 (Neomarker, Freemont, CA), Csp3 (Cell Signaling Technology, Inc., Danvers, MA), Csp4 (Medical & Biological Laboratories Co., Ltd., Nagoya, Japan), Csp5 (BioMol, Plymouth Meeting, PA), and Csp9 (BD Pharmingen) and monoclonal antibodies to Csp4 (MBL International Corp., Woburn, MA) and Csp8 (BD Pharmingen) were used for Western blots. Recombinant active caspases were a kind gift from Guy Salvesen (Burnham Institute, La Jolla, CA). Rabbit polyclonal antisera to α-Actinin-4 (1:500) and PARP (1:2000) were obtained from Alexis Biochemicals (Lausen, Switzerland) and Roche Applied Science, respectively. Rabbit antiserum to Spinophilin (0.1 μg/ml) was a kind gift from Dr. Patrick Allen (Department of Psychiatry, Yale University School of Medicine, New Haven, CT). Mouse monoclonal antibodies to clone C92F3A-5 cytosolic Hsp70 (cHsp70; 1:1000), clone MA3-028 mitochondrial Hsp70 (mtHsp70; 1:500), clone TU-16 α-Tubulin (1:400), clone AC-15 β-actin (1:3000), and clone M2F6 Drebrin (1:1000) were obtained from Stressgen Biotech (Victoria, British Columbia, Canada), Affinity Bioreagents Inc. (Golden, CO), AbCam (Cambridge, MA), Sigma, and MBL International Corp., respectively. The Western blots were prepared from polyacrylamide gel-separated proteins transferred to Immobilon-P PVDF membranes and probed with the antibodies at the dilutions indicated above or recommended by the manufacturer. Immunodetection was revealed with horseradish peroxidase-conjugated donkey anti-rabbit (GE Healthcare) or goat anti-mouse (Jackson ImmunoResearch Laboratories, West Grove, PA) secondary antibodies and ECL (GE Healthcare) development.
Caspase Activity Assays—
Caspase activity was assessed by in vitro fluorogenic assays using Ac-Val-Glu-Ile-Asp-7-amino-4-trifluoromethylcoumarin (Ac-VEID-AFC) for Csp6 or Csp8, Ac-DEVD-AFC for Csp3 or Csp7, Ac-YVAD-AFC for Csp1, Ac-LEHD-AFC for Csp9, and Ac-VDVAD-AFC for Csp2. The activity was measured using a Bio-Rad Fluoromark plate reader (excitation, 390 nm; emission, 538 nm) every 2 min for 1 h at 37 °C in Stennicke's buffer supplemented with 2 μg/ml pepstatin A, 2 μg/ml antipain HCl, 2 μg/ml chymostatin, 135 μm AFC-conjugated substrate, 1 μg of cytosolic neuronal proteins (±2.7 ng of RCsp6), and BSA to give 5 μg of total proteins in the reaction. Fluorescence units were converted to the amount of moles of AFC released based on a standard curve of 0–50 μm free AFC. Cleavage rates were calculated from the linear phase of the assay. Statistical evaluations were preformed with one-way analysis of variance and Tukey's posthoc test.
Immunohistochemistry—
The C-terminal six amino acids of α-Tubulin cleaved by Csp6, EEVGVD, were synthesized as a peptide with an N-terminal cysteine and conjugated with keyhole limpet hemocyanin. The peptide synthesis, conjugation, and rabbit serum production were done at Sigma-Genosys. Sera from two different rabbits (GN20621 and GN20622) were tested on Csp6-cleaved and full-length Tubulin by Western blotting. Human AD brain tissue was obtained from Dr. Catherine Bergeron (University of Toronto) and Dr. Steffen Albrecht (McGill University), and tissue sections were prepared and immunostained as described previously (3) with a 1:1000 dilution of the antiserum. The control human brain tissue sections from five individuals between the ages of 20 and 40 years were obtained from the Brain and Tissue Bank for Developmental Disorders (University of Maryland, College Park, MD). The serum was adsorbed with 50 μg of antigenic peptide in 1 ml of diluted antiserum overnight, and adsorbed serum was recovered by centrifugation for the immunostaining.
To perform co-immunostaining of active Csp6 with Tubulin cleaved by Csp6 (TubulinΔCsp6), we performed sequential immunostaining starting with the TubulinΔCsp6 antiserum (1:1000), which was developed with the Ventana DAB (diaminobenzidine) detection kit according to the manufacturer's instructions. This was then followed with the 10630 anti-active Csp6 antiserum at 1:1000 as described previously (3) and detected with the Ventana Enhanced Alkaline Phosphatase Red kit as recommended by the manufacturer. No immunostaining was obtained when primary antisera were not added (results not shown).
RESULTS
Characterization of Subcellular Fractions and RCsp6 Digestion—
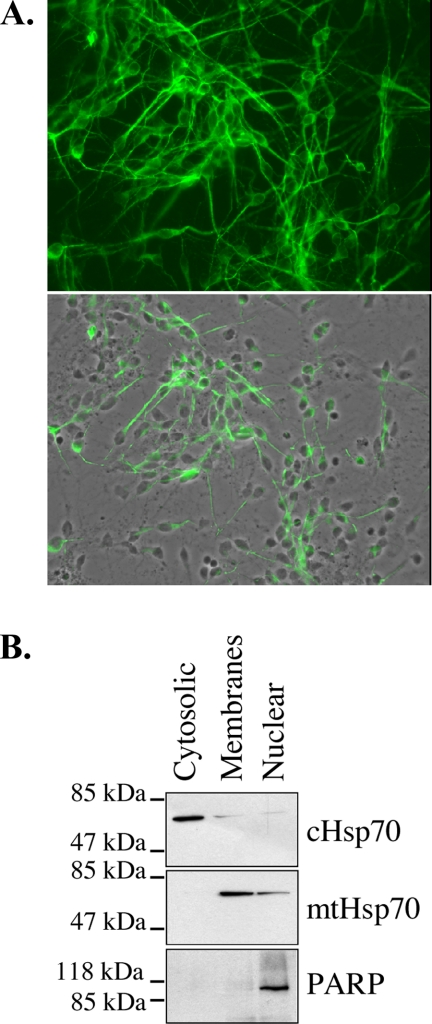
Primary cultures of human neurons produced extensive neuritic networks within 10 days of culture as shown by MAP2 immunofluorescence staining (Fig. 1A). Western blot analyses of the proteins extracted from the cytosolic, membrane, and nuclear subcellular fractions of human neurons with anti-cytosolic and mitochondrial Hsp70 and anti-PARP (nuclear marker) indicated that the cytosolic fraction is free of mitochondrial and nuclear proteins, the membrane fraction is slightly contaminated with cytosolic proteins, and the nuclear fraction contains a small amount of mitochondrial and cytosolic proteins (Fig. 1B). We did not attempt further purification because our interest was mostly in the cytosolic proteins.
Fig. 1.
Purification of subcellular fractions from primary human neurons. A, micrograph of MAP2 immunofluorescent staining (upper panel) and micrograph overlay of MAP2 fluorescent staining and phase contrast (lower panel) of the human neuronal cultures. B, Western blot analysis of primary human neuron subcellular fractions probed with mouse anti-cytosolic Hsp70 (cHsp70), mouse anti-mitochondrial Hsp70 (mtHsp70), and rabbit anti-PARP antibodies.
Two-dimensional Analyses of RCsp6-treated Cytosolic and Membrane Proteins from Subcellular Fractionated Human Primary Neurons—
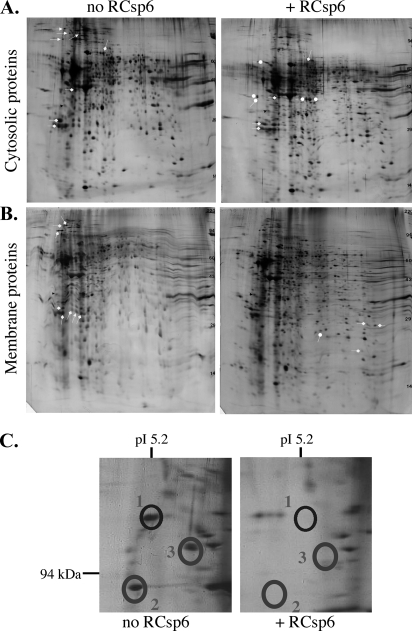
The two-dimensional polyacrylamide gels revealed a number of proteins that completely disappear, strongly decrease, or increase in the sample treated with RCsp6 (Fig. 2, A and B). We opted to focus on the proteins whose abundance decreased considerably with active Csp6 because these might represent the most vulnerable Csp6 substrates in the neurons. The protein levels in 80% of the spots analyzed decreased by at least 50%, and the rest decreased by 30% in intensity. A zoom of one of the sections shows the complete disappearance of three such abundant proteins in one area of the gel (Fig. 2C). Several other less abundant proteins disappeared and others appeared in the Csp6-treated sample. Therefore, we did not expect to identify all of the Csp6 protein substrates because many must be present at very low levels or are only weakly digested in this assay. We sent 72 spots to LC/MS/MS for sequencing.
Fig. 2.
Two-dimensional gel analysis of Csp6-cleaved proteins. Shown is a comparison of 2D gels: non-digested (no RCsp6) versus RCsp6 (+RCsp6)-digested proteins extracted from the cytosolic (A) and membrane (B) subcellular fractions. Spots that disappeared with RCsp6 are labeled with an arrow, spots that decreased with RCsp6 are labeled with a block arrow, and spots that increased with RCsp6 are indicated with a rounded arrow. C, high magnification of some of the spots that disappeared (encircled) shown in A.
Identification of Csp6-mediated Proteolytically Degraded Human Neuronal Proteins—
A protein is identified as significant if the Mowse score is ≥70 (40), if the expectation score of the peptide sequenced is <10−3, if more than one peptide sequenced has a significant expectation score of <10−3 (Table I), and if the identified protein has a molecular weight close to the expected experimental molecular weight (Table II). Some proteins, for example Drebrin, migrate at a much higher molecular mass (116 kDa) than the calculated molecular mass (72 kDa) due to the highly acidic nature of the protein. Accordingly we found the experimental molecular weight on the 2D gels to match the 116 kDa molecular mass reported previously (42).
Table I.
LC/MS/MS identification of human neuronal proteins altered by Csp6
Several scores for one protein indicate that this protein was identified in more than one spot. The number of non-redundant peptides with expectation score <10−3 (No. of peptides exp <10−3) are listed for each of these spots. % coverage indicates the coverage of peptides in the identified protein. Total no. of peptides includes additional peptides that did not reach an expectation score of <10−3.
| Protein | Mowse score | No. of peptides exp <10−3 | % coverage | Total no. of peptides |
|---|---|---|---|---|
| Cytoskeleton and cytoskeleton-associated | ||||
| Drebrin 1 isoform a | 252, 251, 512, 364 | 3, 4, 7, 5 | 11, 18, 29, 13 | 4, 7, 13, 6 |
| β-Actin | 184, 395 | 3, 5 | 17, 44 | 7, 15 |
| Spinophilin | 208 | 3 | 12 | 7 |
| α-Actinin-1 | 219, 212 | 4, 4 | 7, 10 | 4, 8 |
| α-Actinin-4 | 472 | 7 | 22 | 14 |
| Capping protein α | 124 | 2 | 27 | 3 |
| Ezrin | 177 | 3 | 20 | 12 |
| Cofilin I | 94 | 4 | 48 | 7 |
| Glial fibrillary acidic protein | 390 | 9 | 46 | 16 |
| α-Tubulin | 146 | 3 | 19 | 6 |
| Signaling | ||||
| 14-3-3ζ | 519, 153, 170, 300, 244, 285 | 8, 2, 3, 4, 4, 5 | 38, 24, 14, 28, 24, 28 | 11, 8, 3, 8, 8, 8 |
| 14-3-3ε | 694 | 9 | 74 | 17 |
| Inhibitor-2 of protein phosphatase 2A | 229, 177, 136 | 4, 4, 2 | 38, 31, 24 | 7, 5, 5 |
| Chaperones | ||||
| Hsp90α | 970 | 11 | 27 | 18 |
| Heat shock protein gp96 precursor | 296, 329 | 2, 7 | 8, 22 | 6, 17 |
| Valosin-containing protein | 1569 | 22 | 52 | 31 |
| Protein synthesis and conjugation | ||||
| Eukaryotic elongation factor 1γ | 223 | 5 | 21 | 9 |
| Metabolism | ||||
| Inorganic pyrophosphatase | 223 | 2 | 30 | 8 |
| Glyceraldehyde-3-phosphate dehydrogenase | 172, 221, 241, 178, 152 | 4, 5, 3, 4, 3 | 17, 25, 25, 19, 12 | 4, 6, 6, 5, 3 |
| Proteases | ||||
| Neurolysin (EC 3.4.24.16) | 125 | 2 | 4 | 3 |
| Prolyl endopeptidase (prolyl oligopeptidase, EC 3.4.21.26) | 185 | 2 | 14 | 7 |
| Membrane and lipid binding | ||||
| Rab GDP dissociation factor inhibitor α | 130 | 5 | 21 | 8 |
| Chain A, crystal structure of brain fatty acid-binding protein oleic acid | 303 | 5 | 77 | 8 |
| Annexin V | 508 | 11 | 66 | 18 |
Table II.
Comparison of calculated and experimental mass with that identified from the 2D gel analysis
The calculated mass was obtained based on the amino acid sequence in GenBank™. The experimental mass was taken from the results of data from the literature and company data sheets.
| Protein | Calculated mass | Experimental mass | Mass on 2D gel |
|---|---|---|---|
| kDa | kDa | kDa | |
| Cytoskeleton and cytoskeleton-associated | |||
| Drebrin 1 isoform a | 71.9 | 116/125 | 125/132/134/131 |
| β-Actin | 42 | 42 | 46/32 |
| Spinophilin | 89.6 | 120–140 | 131 |
| α-Actinin-1 | 103 | 107 | 123 |
| α-Actinin-4 | 105 | 100–115 | 120, 123 |
| Capping protein α | 33 | 34 | 35 |
| Ezrin | 69.4 | 81 | 100 |
| Cofilin I | 18.7 | 19–21 | 13.25 |
| Glial fibrillary acidic protein | 49.5 | 50 | 47 |
| α-Tubulin | 50 | 57 | 75 |
| Signaling | |||
| 14-3-3ζ | 31 | 30 | 26/26/34/32/32/31 |
| 14-3-3ε | 28 | 31 | 30 |
| Inhibitor-2 of protein phosphatase 2A | 31–33 | 30 | 48/48/46 |
| Chaperones | |||
| Hsp90α | 85.1 | 90 | 113/111 |
| Heat shock protein gp96 precursor | 92.7, 90.3 | 100 | 125/122 |
| Valosin-containing protein | 90 | 97 | 118 |
| Protein translation and conjugation | |||
| Eukaryotic elongation factor 1γ | 50.5 | 46 | 58 |
| Metabolism | |||
| Inorganic pyrophosphatase | 33.1 | 33 | 35 |
| Glyceraldehyde-3-phosphate dehydrogenase | 36.2 | 38 | 36, 38 |
| Proteases | |||
| Neurolysin (EC 3.4.24.16) | 81.3 | 80 | 96 |
| Prolyl endopeptidase (prolyl oligopeptidase, EC 3.4.21.26) | 81.6 | 79.6 | 96 |
| Membrane and lipid binding | |||
| Rab GDP dissociation factor inhibitor α | 51 | 50 | 78 |
| Chain A, crystal structure of brain fatty acid-binding protein oleic acid | 14.8 | 13–14 | 11 |
| Annexin V | 35.8 | 38 | 38 |
Of 72 spots analyzed, 24 different proteins from 41 spots were identified with high confidence (Table I) with scores ranging from 94 to 1569, number of peptides <10−3 ranging from 2 to 22, and the percentage of coverage ranging from 4 to 77%. Additional peptides with expectation scores >10−3 are indicated in the total peptide column of Table I. The duplicates are excluded from these numbers. The molecular weights of all of these proteins except inhibitor-2 of protein phosphatase 2A match almost exactly the expected experimental molecular weights if considering potential post-translational modifications (Table II). Three proteins, keratins, dermicidin precursor from skin, and bovine serum albumin, are excluded as contaminants from culture media serum, and 10 spots could not be identified with any significance.
Classification of Csp6-mediated Cleaved Proteins According to Function—
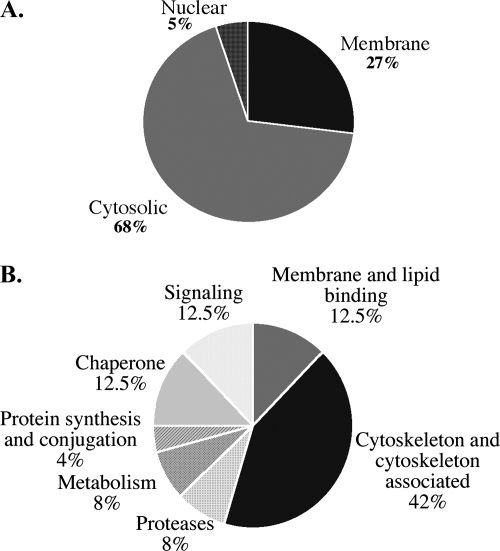
In total, 68% of the proteins identified were in the cytosolic fraction, 27% were in the membrane fraction, and 5% were in the nuclear fraction (Fig. 3A). Of the proteins identified, 42% are either cytoskeleton or cytoskeleton-associated proteins, 12.5% are signaling molecules, 12.5% are membrane and lipid-binding proteins, 12.5% are chaperones, 8% are proteins from metabolic pathways, 8% are proteases, and 4% are involved in protein synthesis and conjugation (Fig. 3B).
Fig. 3.
Schematic diagram of proteomics analysis results. A, pie chart showing the percentage of proteins identified as putative Csp6 substrates in the membrane, cytosolic, and nuclear subcellular fractions of primary cultures of human neurons. B, pie chart of the various functional categories of putative Csp6 substrates. The percentage of proteins in each category is indicated.
Identification of Putative, Probable, and Confirmed Caspase Substrate Sites—
The identified protein sequences were examined for the presence of potential Csp6 substrate sites (Table III). Probable sites predicted based on combinatorial studies as (I/D/E/L/T/V)(D/E/Q)XD sequences (43, 44) are present in all except one of the identified proteins. However, when using a broader definition based on XEXD, VXXD, or demonstrated unusual Csp6 sites in the literature (Table IV), Csp6 cleavage sites were found in all of the proteins identified. The IETD (45), PEED and EEED (6), and VEVD (20) previously confirmed sites are bold in Table III when they are present in the identified proteins. The non-canonical Csp6 site, SWKD (Table IV), is present in Actinin-1 (172SWKD175) and -4 (191SWKD194). The presence of these putative Csp6 sites does not exclude the possibility that some of these proteins are also substrates of Csp2, -3, -7, and -8 because these caspases also cleave some of these sites (43).
Table III.
Probable and putative caspase substrate sites in proteins identified by LC/MS/MS
Probable Csp6 sites are identified as (I/D/E/L/T/V)(E/D/Q)XD based on combinatorial chemistry (43, 44). Because of the identification of unusual Csp6 substrate sites (see Table IV), we also added putative VXXD or XEXD sites. The accession numbers of the sequence used for the analyses are listed in the right-hand column. Bold indicates sites that were already defined as Caspase-6 substrate sites in other proteins.
| Protein | VXXD or XEXD | (I/D/E/L/T/V)(D/E/Q)XD | GenBank no. |
|---|---|---|---|
| Cytoskeleton and cytoskeleton-associated | |||
| Drebrin 1 isoform a | 86VGED89, 626PEID629 | 122EDID125, 453IETD456 | AAH00283 |
| β-Actin | 212VALD215, 360QEYD363 | AAH08633 | |
| Spinophilin | 386SEAD389, 615GEDD618 | 197LDAD200, 375EEVD378, 408LEED411, 409EEDD412, 411DDED414, 412DEDD415, 415DEED418, 437EEED440, 463EDYD466, 470EDVD473, 497LEKD500, 548VEVD551, 626TDED629, 790EEMD793 | NP_115984 |
| α-Actinin-1 | 15PEED18, 440FESD443, 462NELD465, 888GESD891 | 17EDWD20, 55IEED58, 481DQWD484, 688LEGD691 | AAP35871 |
| α-Actinin-4 | 34QEDD37, 459FESD462, 481NELD484, 907GESD910 | 36DDWD39, 74IDED77, 210IEYD213, 500DQWD503, 801VEND804, 840TDTD843 | NP_004915 |
| Capping protein α | 32VFND35, 111EEAD114 | AAH00144 | |
| Ezrin | 228YEKD231 | 389LEAD392 | NP_003370 |
| Cofilin I | 14VFND17 | NP_005498 | |
| Glial fibrillary acidic protein | 174QEAD177 | 139VERD142, 222VELD225 | AAH41765 |
| α-Tubulin | 66VFVD69, 303VKCD306, 324VPKD327, 435VGVD438 | 30IQPD33, 428LEKD331 | AAD33871 |
| Signaling | |||
| 14-3-3ζ | 276AELD279 | 307TQGD310 | AAH51814 |
| 14-3-3ε | 204AELD207 | BAA32538 | |
| Inhibitor-2 of protein phosphatase 2A | 207VIKD210, 40NEID43, 97GEED100, 232GEED235, 257GEED260, 269GEED272, 274GEDD 277 | 233EEDD236, 234EDDD237, 235DDDD238, 236DDDD239, 246EDID249, 250EEGD253, 259EDED262, 260DEDD263, 261EDDD264 | AAQ79833 |
| Chaperones | |||
| Hsp90α | 172VRTD175, 230VSDD233, 262VGSD265, 224KERD227, 650AEAK653 | 698IDED701, 699DEDD702, 719LEGD722, 729EEVD732 | NP_005339 |
| Heat shock protein gp96 precursor | 19VRAD22, 232VIAD235, 496VIED499, 596VKFD599, 784VGTD787, 223WESD226, 364KESD367, 581PEFD584, 798AEKD801 | 23DEVD26, 25VDVD28, 31VEED34, 259LELD262, 304EESD307, 350VEED353, 437VDSD440, 703EDED706, 704DEDD707, 749IDPD752, 774TEQD777, 780EEMD783 | NP_003290 |
| Valosin-containing protein | 70VLSD74, 176VAPD179, 201VGYD204, 407VGAD410, 447VTMD450, 666VAKD669, 365REVD368 | 166VETD169, 304DELD307, 392DDVD395, 577DELD580, 606TEMD609, 722VEED725, 723EEDD726, 798EDND801 | CAH70993 |
| Protein synthesis and conjugation | |||
| Eukaryotic elongation factor 1γ | 61FEGD64 | 102VDSD105, 261EEMD264 | AAH13918 |
| Metabolism | |||
| Inorganic pyrophosphatase | 276VPTD279, 150GETD153 | 162DDPD165, 278TDVD281 | AAP97214 |
| Glyceraldehyde-3-phosphate dehydrogenase | 163VIHD166, 282VSSD285, 78QERD81, 222PELD225 | CAA25833 | |
| Proteases | |||
| Neurolysin (EC 3.4.24.16) | 112VSSD115, 207NEDD210, 350FEYD353 | 123TEAD125, 234TDDD237, 520VETD523 | CAC27329 |
| Prolyl endopeptidase (prolyl oligopeptidase, EC 3.4.21.26) | 9VYRD12, 112VFLD115, 288GEYD291, 333HEKD336, 623PEAD626 | 32EDPD35 | BA A04661 |
| Membrane and lipid binding | |||
| Rab GDP dissociation factor inhibitor α | 151YEND154 | 3EEYD6, 413TEND416 | BAA08078 |
| Chain A, crystal structure of brain fatty acid-binding protein oleic acid | 84VSLD87, 44QEGD47 | 68EEFD71, 86LDGD89 | IFE3_A |
| Annexin V | 141VVGD144, 277SEID280, 317GEDD320 | 137LEDD140, 172VEQD175 | AAH01429 |
Table IV.
Unusual confirmed Csp6 substrate sites in various proteins
These sites were not predicted by combinatorial chemistry but were found to be specific for Csp6 in these proteins. FAK, focal adhesion kinase; TGEV, transmissible gastroenteritis coronavirus.
Confirmation of Csp6-mediated Cleavage in Some Proteins Identified by LC/MS/MS—
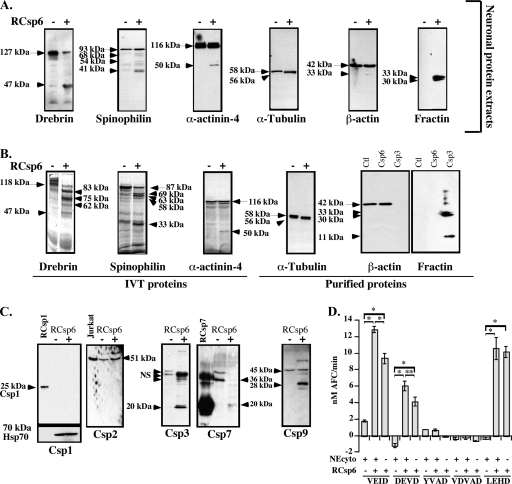
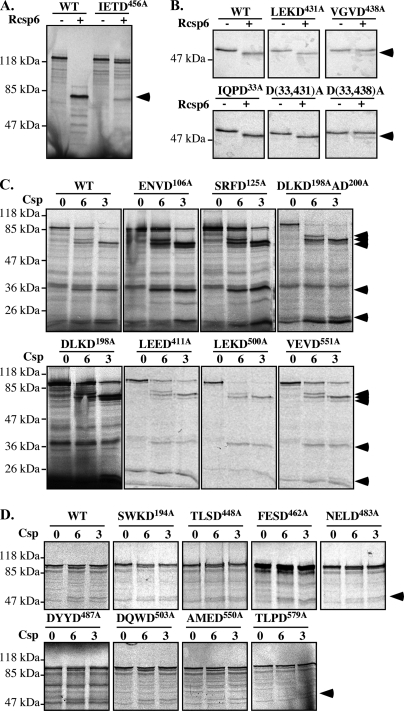
To confirm that the LC/MS/MS correctly identified the proteins, we further investigated Csp6-mediated cleavage of cytoskeleton and cytoskeleton-associated proteins. We showed that RCsp6 induces the cleavage of Drebrin, Spinophilin, α-Actinin-4, α-Tubulin, and β-actin in total protein extracts from primary cultures of human neurons (Fig. 4A).
Fig. 4.
Cleavage of specific proteins by Csp6. A and B, Western blot analyses or autoradiogram (for IVT Drebrin, Spinophilin, and α-Actinin-4) of neuronal protein extracts (A) or IVT or purified proteins (B) cleaved with RCsp6. The arrows indicate full-length proteins, and the arrowheads indicate the fragments. C, Western blot analysis of Csp1, Csp2, Csp3, Csp7, and Csp9 in 50 μg of neuronal cytosolic proteins treated without (−) or with (+) RCsp6. D, fluorogenic caspase activity assays of 1 μg of cytosolic neuronal protein extracts (NEcyto) treated with 2.7 ng of RCsp6 on YVAD-AFC for Csp1, VDVAD-AFC for Csp2, DEVD-AFC for Csp3 and Csp7, VEID-AFC for Csp6 and Csp8, and LEHD-AFC for Csp9. * indicates a p < 0.001 and ** indicates a p < 0.01 statistically significant difference between the indicated assays. NS, nonspecific. Error bars indicate standard deviation.
To confirm direct cleavage of the identified proteins by Csp6, we treated IVT Drebrin, Spinophilin, and α-Actinin-4 with RCsp6. IVT Drebrin was cleaved by RCsp6 (Fig. 4B). In contrast to the Western blot results, we observed several [35S]methionine-labeled fragments generated from IVT Drebrin after Rcsp6 digestion. There is one fragment at 47 kDa that is likely the one detected with the antiserum in Fig. 4A. The other fragments would not be detected with the anti-Drebrin monoclonal antibody. IVT Spinophilin was cleaved by RCsp6 and generated several fragments similar to those observed in neuronal extracts. The IVT α-Actinin-4 was directly cleaved by RCsp6 and generated the 50-kDa fragment observed by Western blotting. Purified bovine Tubulin, which is over 95% identical to human Tubulin, generated a Csp6-cleaved product migrating ∼2 kDa below the full-length α-Tubulin. α-Tubulin was also identified independently by LC/MS/MS as one of five proteins interacting with RCsp6 on a CL-4B affinity chromatography column.2 However, Csp6 could not cleave purified β-actin despite excellent cleavage by Csp3, which was detected by the loss of the full-length β-actin and the appearance of the Fractin epitope at 33 kDa. The Fractin antiserum detects Csp3-cleaved β-actin (41). The 33-kDa fragment generated from β-actin in neuronal protein extracts was also immunoreactive to the Fractin antibody raised against Csp3-cleaved actin (Fig. 4, A and B). The additional 20-kDa band is the result of cleavage at LVVD11 (46). Therefore, the cleavage of β-actin detected by LC/MS/MS was the result of Csp3 and not of Csp6 activity. Interestingly β-actin did not have a probable Csp6 site as determined by combinatorial chemistry (Table III). In general, the cleavage of IVT or purified proteins was more efficient than the cleavage of proteins in neuronal extracts. This could be due to the association of protein complexes that restrict access of the substrate site to the caspase in native conditions.
Further evaluation showed that Csp3 is indeed activated in the RCsp6-treated neuronal extracts as shown by the appearance of the p20 active subunit (Fig. 4C). Furthermore there was increased DEVDase activity in RCsp6-treated neuronal cytosolic proteins suggesting the activation of Csp3 (Fig. 4D). In addition, we found activation of Csp7 by Csp6 because the proenzyme disappeared and the p20 subunit appeared in Csp6-treated neuronal cytosolic proteins (Fig. 4C). However, we exclude significant Csp6-mediated activation of Csp1, -2, and -9 in these protein extracts. The 25-kDa active subunit of Csp1 was not detected with the anti-active Csp1 antiserum in Csp6-untreated or -treated neuronal cytosolic proteins, and there was no increase in YVADase activity (Fig. 2, C and D). Csp2 was not cleaved, and there was no increase of VDVADase activity. We would expect VDVADase activity because Csp3 was activated and RCsp3 was able to cleave VDVAD-AFC substrate (supplemental Fig. 1). However, the amount of active Csp3 present in the assay was probably below the threshold required for its VDVADase activity. The Csp9 pro-arm was cleaved to give the 28-kDa fragment, but there was no additional increase in Csp9 activity on the LEHD-AFC substrate in Csp6-treated neuronal cytosolic proteins (Fig. 4D). Csp4, -5, and -8, although detected by Western blot in Jurkat cells, were undetectable in the cerebral human neuronal proteins and are thus unlikely to represent significant caspase activity in these protein extracts (results not shown and Ref. 28).
These results confirm that the LC/MS/MS identification of caspase-cleaved proteins is correct. Whether cleavage is direct or indirect through the activation of other caspases or proteases in all proteins identified has to be confirmed on an individual basis.
Identification of Csp6 Cleavage Sites Based on Predictions from Combinatorial Studies—
To determine whether the Csp6 cleavage sites predicted from combinatorial studies are present in some of the proteins identified (Table III), we performed site-directed mutagenesis in a few of the identified proteins. The aspartic (Asp) residue was mutated to alanine (Ala) residue to eliminate the caspase cleavage site. The IETD456 site of Drebrin was easily identified as a Csp6 cleavage site because the mutation abolished the production of the 83-kDa fragment of IVT Drebrin incubated with RCsp6 (Fig. 5A). Similarly the VGVD438 to VGVA mutation abolished Csp6 cleavage of α-Tubulin (Fig. 5B) as observed previously for Granzyme B cleavage (47). However, Csp6 generated a lower molecular weight fragment in the D438A mutant, indicating that an alternative site has been revealed with the mutation. Based on the size of the fragment and possible Csp6 sites in the protein, we mutated the IQPD33 site, but the fragment was still generated. Therefore, it possibly represents another secondary Csp6 site or a nonspecific cleavage resulting from poorly folded IVT proteins. Csp6 did not cleave the predicted α-Tubulin LEKD431 site. In Spinophilin (Fig. 5C), the DLKDAD200 sequence was identified as a Csp6 substrate site because the double Asp mutations eliminated the generation of a 35-kDa fragment in protein treated with RCsp6 and RCsp3. We further generated a mutation at Asp198 only, and this also eliminated cleavage indicating that both Csp6 and Csp3 cleave at the DLKD198 site. Furthermore based on epitope mapping and a 20-kDa doublet that was generated by Csp3 cleavage, we assessed the ENVD106 and SRFD125 as possible caspase sites. We found that the D106A mutation eliminated the bottom protein, whereas the D125A eliminated the top protein of the doublet. Both the D106A and D125A mutants also resulted in the appearance of an additional fragment around 30 kDa when treated with either Csp3 or Csp6. The appearance of this additional fragment is likely due to the unmasking of another site when the N-terminal sites are mutated. More work is needed to clarify this issue. Nevertheless we identified Asp198, Asp106, and Asp125 as caspase sites in Spinophilin. In addition, the possible Csp6 cleavage sites VEVD551, LEED411, LEKD500, and VEVD551 were excluded because mutagenesis did not prevent Csp6 cleavage of Spinophilin (Fig. 5C).
Fig. 5.
Identification of Csp6 cleavage sites. Shown are autoradiograms of IVT WT and mutant D456A Drebrin (A) and WT and mutant D431A, D438A, D33A, D33A/D431A (D(33,431)A), and D33A/D438A (D(33,438)A) α-Tubulin (B) before (−) and after (+) Csp6 cleavage. C and D, autoradiograms of IVT WT and mutant Spinophilin (C) and α-Actinin-4 (D) before (0) and after cleavage with Csp6 (6) and Csp3 (3). The arrowheads indicate cleaved fragments.
Similarly the SWKD194, TLSD448, FESD462, NELD484, DYYD487, DQWD503, AMED550, and TLDP579 sites were excluded as caspase sites from α-Actinin-4 (Fig. 5D). Interestingly the SWKD site identified as a Csp6 substrate site in NS5A (Table IV) and the AMED and TLDP sites were Csp3 sites in nucleolin and GRASP65, respectively (32, 48). Csp3 cleaved these mutant Spinophilin and α-Actinin-4 proteins, indicating that other sites on these proteins are responsible for caspase cleavage. These results indicate that the structure of the protein likely has a significant impact on the ability of caspases to bind to and cleave a substrate site and that the sequences from combinatorial studies are not always useful to identify caspase sites in proteins.
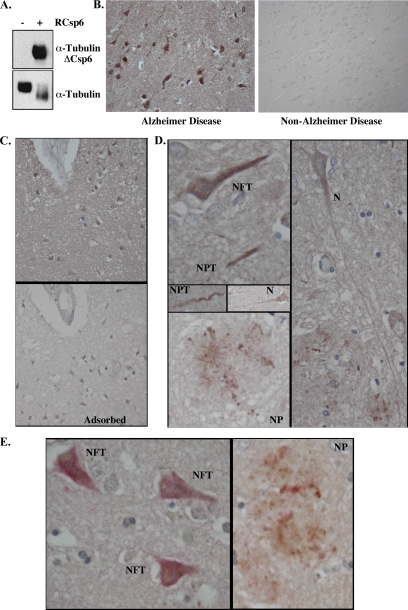
α-Tubulin Cleaved by Csp6 (TubulinΔCsp6) Is Detected in Pathological Hallmarks in AD—
To determine whether one of the identified Csp6 substrates is associated with Csp6 cleavage in AD tissues, we developed a neoepitope antiserum against Csp6-cleaved α-Tubulin (TubulinΔCsp6). The antiserum recognizes only the purified bovine Tubulin cleaved by Csp6, whereas the control total α-Tubulin antiserum recognizes both the full-length and cleaved proteins (Fig. 6A). The TubulinΔCsp6 antiserum did not prove to be useful on Western blots of total brain proteins. A likely reason is that caspase-cleaved proteins are often degraded rapidly, or too few cells are affected at any given time to be detected in total protein extracts. Therefore, we opted to conduct immunohistochemical analyses with this antiserum. We found that the TubulinΔCsp6 antiserum recognizes tangle-like formations and neurites in AD brain tissue sections but not in the non-AD control sections (Fig. 6B). Staining was adsorbed by the antigenic peptide (Fig. 6C). No immunoreactivity was observed with preimmune serum (not shown). Interestingly immunostaining was observed in the major pathological hallmarks of AD, neurofibrillary tangles (NFT), neuropil threads, and neuritic plaques (NP) (Fig. 6D), as observed with TauΔCsp6 and active Csp6 antisera (3, 4). Furthermore some immunopositive neurons did not seem to be affected by AD pathology or have an apoptotic morphology. The TubulinΔCsp6 immunoreactivity co-localized with anti-active Csp6-positive NFT and neuritic plaques (Fig. 6E). Therefore, the proteomics approach allowed identification of a novel Csp6 protein substrate that is generated in AD.
Fig. 6.
Identification α-Tubulin cleaved by Csp6 in AD brains. A, Western blot analysis of purified Tubulin without or with RCsp6 cleavage using Tubulin cleaved by Csp6 (anti-TubulinΔCsp6) and Tubulin antisera. B, micrographs of immunohistochemical detection of anti-TubulinΔCsp6 with diaminobenzidine in tissue sections of AD and non-AD temporal cortex. C, micrograph of immunohistochemical detection of anti-TubulinΔCsp6 in AD tissue sections showing the adsorption of the antiserum on the antigenic peptide. D, micrographs of α-TubulinΔCsp6 immunopositive NFT, neuropil threads (NPT), NP, and neurons (N) that otherwise look normal. E, micrograph of double immunostaining of TubulinΔCsp6 with diaminobenzidine (brown) and active Csp6 with fast red (pink) in NFT and NP.
DISCUSSION
In AD brains, Csp6 is strongly activated in neuropil threads, neuritic plaques, and neurofibrillary tangles in the absence of classical apoptotic features (3). Normally active Csp6 translocates to the nuclei in apoptotic cells (5). The absence of active Csp6 in the nuclei of neurons combined with its presence in neurofibrillary tangles, neuritic plaques, and neuropil threads suggests that Csp6 is involved in neurodegeneration in AD. To better understand the function of active Csp6 in the neurites of AD brains, we used a proteomics approach to identify Csp6-mediated proteolytic events in human neurons. We discovered 24 different proteins that were cleaved after the addition of Csp6 to neuronal protein extracts. These proteins were stringently selected, and all six proteins chosen from this list for further analyses (five shown here and one not shown) were confirmed by Western blotting to be cleaved in RCsp6-treated human neuronal protein extracts. We further showed using pure recombinant or IVT proteins that α-Tubulin, Drebrin, Spinophilin, and α-Actinin-4 are substrates of active Csp6.
We confirmed the cleavage of α-Tubulin in AD by immunohistochemistry with a neoepitope antiserum to the cleaved proteins. The α-TubulinΔCsp6 immunostained neurofibrillary tangles, neuropil threads, and neuritic plaques, and the immunostaining was co-localized with active Csp6 immunoreactivity. These results show that, in addition to Tau, α-Tubulin is a substrate of active Csp6 in vivo. We identified the site of cleavage at VGVD438. Cleavage at this site releases the 13 C-terminal residues, SVEGEGEEEGEEY, rich in acidic amino acids that interact with many microtubule-associated proteins such as MAP2 and dynein (49, 50). The axonal microtubule-associated protein Tau also interacts with α-Tubulin in this area at amino acids 430–441 (51). The cleavage of the functional α-Tubulin C-terminal domain indicates that Csp6 could seriously alter the state of microtubules in neurons. Removal of this C-terminal tail with the subtilisin protease, which cleaves at VDSV440, two amino acids downstream of the Csp6 site, results in aberrant polymerization of the microtubules (52, 53). Interestingly the cytotoxic T-lymphocyte serine protease Granzyme B has been found recently to also cleave the C terminus of α-Tubulin at Asp438 within a canonical Granzyme B cleavage site, VGVDSV440. The Granzyme B-truncated α-Tubulin increases the rate of microtubule polymerization (47). It is therefore likely that Csp6 will have effects on microtubule polymerization similar to those shown with Granzyme B and subtilisin. We already have shown that the axonal microtubule-associated protein Tau is cleaved by Csp6 and that Tau cleaved by Csp6 is highly abundant in AD pathology (3). Therefore, cleavage of α-Tubulin and Tau protein would affect microtubule integrity in both the axons and dendrites of neurons. Together these results suggest that the activity of Csp6 could alter microtubule function in the cellular cytoskeleton.
The other three Csp6 protein substrates, Drebrin, Spinophilin, and α-Actinin-4, are important proteins of postsynaptic densities (54–56). Interestingly these are localized to the dendritic spines, whose structure is mainly regulated by the actin cytoskeleton as dendritic spines lack intermediate filaments and microtubules. Drebrin is an actin-interacting protein that is highly concentrated in the dendritic spines of excitatory synapses in mature neurons of the central nervous system (42, 54, 57) and may be necessary for spine morphogenesis because it also regulates the distribution of other postsynaptic density proteins like PSD-95 (58). Antisense knockdown of Drebrin in rats results in cognitive problems (59). In AD, Drebrin immunoreactivity is decreased significantly (60–62). Similarly Spinophilin is an actin-binding protein of postsynaptic densities that regulates dendritic spine morphology and function (56, 63–65). Mice null for Spinophilin have learning problems (66). Furthermore Spinophilin interacts with protein phosphatase 1, a highly abundant protein of dendritic spines that regulates ionic conductance and synaptic plasticity (56). To our knowledge, there has been no study of Spinophilin in neurodegenerative diseases, but alteration of this protein by proteolytic cleavage in dendritic spines could result in a disruption of neuronal function because Spinophilin-null mice show reduced long term depression (65). The Actinins are thought to regulate the receptors at the synapses. α-Actinin-4 interacts with calmodulin kinase II and Densin 180 at the postsynaptic densities (67). These interactions are important in mediating neuronal function at the synapse. α-Actinin is a component of Hirano bodies, which increase in aging and AD, and Hirano bodies have also been shown to contain the Fractin epitope but no active Csp3 (68, 69). Therefore, these results allow the hypothesis that cleavage of Drebrin, Spinophilin, and α-Actinin-4 by caspases in AD or ischemia may alter the synaptic function of neurons or may be responsible for the loss of synapses in AD. Because the loss of synapses in AD is the pathological defect that best correlates with the progressive dementia of AD (70) and Csp6 activity is detected preclinically, our novel findings of Csp6 cleavage of these important synaptic proteins provide a tantalizing lead into the potential cause of the synaptic loss in AD.
We confirmed Csp6-mediated proteolytic cleavage of some proteins chosen from the list. However, identification of the site of cleavage predicted by combinatorial chemistry was not always possible (43, 44). We rapidly identified IETD456 and VGVD438 in Drebrin and α-Tubulin, respectively. We identified DLKD198 as a Csp3 or Csp6 site and the ENVD106 and SRFD125 sites as Csp3 cleavage sites in Spinophilin. However, in Spinophilin and α-Actinin-4, several potential and one unconventional site were excluded. Some of these, like VEVD551 in Spinophilin and SWKD194, AMED550, and TLPD579 in α-Actinin-4, have been confirmed previously as Csp6 or Csp3 cleavage sites in cytokeratin 18 and NS5A (20, 32, 48, 71) yet were not cleaved by Csp3 or Csp6 in our assays. The possibility that these sites are hidden in the protein is unlikely because a cursory look at the published partial structure of some of these proteins indicates that some of the predicted cleavage sites are exposed on the protein surface (data not shown). Therefore, at least in some instances, it is likely that the sequence surrounding the sites is most important in allowing caspase cleavage.
This proteomics approach successfully identified novel Csp6 protein substrates in human neurons and provides new leads into understanding the role of Csp6 in AD. In our study, we chose to focus on proteins whose levels were reduced by ∼50% in Csp6-treated protein extracts. Several other proteins could have been identified. However, there are some disadvantages to this approach. Some proteins were likely missed because of poor separation by two-dimensional gel analysis, low abundance in neurons, or restricted proteolysis. This could explain why we did not detect already known Csp6-cleaved proteins. Caspase cleavage of proteins in vitro may also unmask sites that would otherwise be protected by protein-protein or protein-membrane interactions in physiological conditions. Therefore, proteins identified will need to be studied on an individual basis to confirm Csp6 cleavage in physiological and pathological conditions. The main problem is that the addition of active Csp6 in neuronal protein extracts activates other caspases, and therefore, as seen with the selective Csp3 cleavage of β-actin, each candidate substrate will have to be investigated to assign protein cleavage to a specific caspase. Alternative proteomics approaches to identify caspase protein substrates, such as N-terminal tagging of cleaved proteins or mRNA display, suffer from the same problem because adding active caspase to a heterogeneous mixture of proteins is likely to activate other proteases in a cascade-dependent manner (32, 33, 35). The main advantage to this method relative to others is its relative simplicity. In addition, performing the study on proteins from subcellular fractions of human primary neurons avoids possible artifacts of compartmentalization.
In summary, we discovered a number of novel caspase protein substrates that could be very important in the regulation of neuronal structure and function. The Csp6 cleavage of three proteins regulating the microfilament networks in postsynaptic densities is interesting with respect to neurodegeneration. The confirmation of α-TubulinΔCsp6 in AD validates the approach. Because the active Csp6 is found at all stages of AD, in mild cognitive impairment, and in some aged individuals with lower cognitive abilities (4), our results suggest that the disruption of these proteins either directly or indirectly by active Csp6 could result in the early cognitive deficits of AD. The results suggest that local activation of Csp6 could have a significant impact on neuronal function in the absence of cell death.
Supplementary Material
Acknowledgments
We gratefully acknowledge the Birth Defects Research Laboratory at the University of Washington, Seattle, WA for providing conceptus tissue for research (National Institutes of Health Grant HD 000836) and Eveline Clair for helping with the Western blots. We thank Dr. Greg Cole (University of California, Los Angeles, CA) for the Fractin antibody, Dr. Patrick Allen (Yale University School of Medicine, New Haven, CT) for the Spinophilin antibody, Dr. Hemant Paudel (McGill University, Montreal, Quebec, Canada) for the purified Tubulin, Dr. Girolama La Mantia (University “Federico II,” Naples, Italy) for the Spinophilin construct, Dr. Guy Salvesen (Burnham Institute, La Jolla, CA) for the Caspase-6 prokaryotic construct and purified caspases, Dr. Clay Clark (North Carolina State University) for the Caspase-3 construct, and Dr. Seamus Martin (Smurfit Institute, Dublin, Ireland) for the α-Tubulin D431A, D438A, and WT constructs. We thank Dr. Catherine Bergeron (University of Toronto, Toronto, Canada) for the Alzheimer brain sections and Martine Bourdeau, Jocelyne Jacques, and Dr. Steffen Albrecht from the Department of Pathology at the Jewish General Hospital (Montreal, Quebec, Canada) for help in establishing and interpreting the immunohistochemistry.
Footnotes
Published, MCP Papers in Press, May 16, 2008, DOI 10.1074/mcp.M800007-MCP200
The abbreviations used are: Csp, Caspase; AD, Alzheimer disease; MEM, minimal essential medium; BCS, bovine calf serum; PARP, poly(ADP-ribose) polymerase; 2D, two-dimensional; RCsp, recombinant active Csp; Tricine, N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine; PIPES, 1,4-piperazinediethanesulfonic acid; WT, wild type; IVT, in vitro translated; AFC, 7-amino-4-trifluoromethylcoumarin; NFT, neurofibrillary tangles; NP, neuritic plaques.
G. Klaiman and A. C. LeBlanc, unpublished results.
This work was supported by Canadian Institutes of Health Research Grant MOP15118 and by Fonds de la recherche en santé du Québec (to A. LB). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Stadelmann, C., Deckwerth, T., Srinivasan, A., Bancher, C., Bruck, W., Jellinger, K., and Lassmann, H. ( 1999) Activation of caspase-3 in single neurons and autophagic granules of granulovacuolar degeneration in Alzheimer's disease. Am. J. Pathol. 155, 1459–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Selznick, L., Holtzman, D., Han, B., Gokden, M., Srinivasan, A., Johnson, E., and Roth, K. ( 1999) In situ immunodetection of neuronal caspase-3 activation in Alzheimer's disease. J. Neuropathol. Exp. Neurol. 58, 1020–1026 [DOI] [PubMed] [Google Scholar]
- 3.Guo, H., Albrecht, S., Bourdeau, M., Petzke, T., Bergeron, C., and LeBlanc, A. C. ( 2004) Active Caspase-6 and Caspase-6 cleaved Tau in neuropil threads, neuritic plaques and neurofibrillary tangles of Alzheimer's disease. Am. J. Pathol. 165, 523–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albrecht, S., Bourdeau, M., Bennett, D., Mufson, E. J., Bhattacharjee, M., and LeBlanc, A. C. ( 2007) Activation of caspase-6 in aging and mild cognitive impairment. Am. J. Pathol. 170, 1200–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruchaud, S., Korfali, N., Villa, P., Kottke, T. J., Dingwall, C., Kaufmann, S. H., and Earnshaw, W. C. ( 2002) Caspase-6 gene disruption reveals a requirement for lamin A cleavage in apoptotic chromatin condensation. EMBO J. 21, 1967–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Samejima, K., Svingen, P. A., Basi, G. S., Kottke, T., Mesner, P. W., Jr., Stewart, L., Durrieu, F., Poirier, G. G., Alnemri, E. S., Champoux, J. J., Kaufmann, S. H., and Earnshaw, W. C. ( 1999) Caspase-mediated cleavage of DNA topoisomerase I at unconventional sites during apoptosis. J. Biol. Chem. 274, 4335–4340 [DOI] [PubMed] [Google Scholar]
- 7.Rouaux, C., Jokic, N., Mbebi, C., Boutillier, S., Loeffler, J. P., and Boutillier, A. L. ( 2003) Critical loss of CBP/p300 histone acetylase activity by caspase-6 during neurodegeneration. EMBO J. 22, 6537–6549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nyormoi, O., Wang, Z., Doan, D., Ruiz, M., McConkey, D., and Bar-Eli, M. ( 2001) Transcription factor AP-2α is preferentially cleaved by caspase 6 and degraded by proteasome during tumor necrosis factor α-induced apoptosis in breast cancer cells. Mol. Cell. Biol. 21, 4856–4867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buendia, B., Santa-Maria, A., and Courvalin, J. C. ( 1999) Caspase-dependent proteolysis of integral and peripheral proteins of nuclear membranes and nuclear pore complex proteins during apoptosis. J. Cell Sci. 112, 1743–1753 [DOI] [PubMed] [Google Scholar]
- 10.Cross, T., Griffiths, G., Deacon, E., Sallis, R., Gough, M., Watters, D., and Lord, J. M. ( 2000) PKC-δ is an apoptotic lamin kinase. Oncogene 19, 2331–2337 [DOI] [PubMed] [Google Scholar]
- 11.Chiarini, A., Whitfield, J. F., Armato, U., and Dal Pra, I. ( 2002) Protein kinase C-β II Is an apoptotic lamin kinase in polyomavirus-transformed, etoposide-treated pyF111 rat fibroblasts. J. Biol. Chem. 277, 18827–18839 [DOI] [PubMed] [Google Scholar]
- 12.Eymin, B., Sordet, O., Droin, N., Munsch, B., Haugg, M., Van de Craen, M., Vandenabeele, P., and Solary, E. ( 1999) Caspase-induced proteolysis of the cyclin-dependent kinase inhibitor p27Kip1 mediates its anti-apoptotic activity. Oncogene 18, 4839–4847 [DOI] [PubMed] [Google Scholar]
- 13.Columbaro, M., Mattioli, E., Lattanzi, G., Rutigliano, C., Ognibene, A., Maraldi, N. M., and Squarzoni, S. ( 2001) Staurosporine treatment and serum starvation promote the cleavage of emerin in cultured mouse myoblasts: involvement of a caspase-dependent mechanism. FEBS Lett. 509, 423–429 [DOI] [PubMed] [Google Scholar]
- 14.Galande, S., Dickinson, L. A., Mian, I. S., Sikorska, M., and Kohwi-Shigematsu, T. ( 2001) SATB1 cleavage by caspase 6 disrupts PDZ domain-mediated dimerization, causing detachment from chromatin early in T-cell apoptosis. Mol. Cell. Biol. 21, 5591–5604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gotzmann, J., Meissner, M., and Gerner, C. ( 2000) The fate of the nuclear matrix-associated-region-binding protein SATB1 during apoptosis. Cell Death Differ. 7, 425–438 [DOI] [PubMed] [Google Scholar]
- 16.Hirata, H., Takahashi, A., Kobayashi, S., Yonehara, S., Sawai, H., Okazaki, T., Yamamoto, K., and Sasada, M. ( 1998) Caspases are activated in a branched protease cascade and control distinct downstream processes in Fas-induced apoptosis. J. Exp. Med. 187, 587–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lagace, T. A., Miller, J. R., and Ridgway, N. D. ( 2002) Caspase processing and nuclear export of CTP:phosphocholine cytidylyltransferase α during farnesol-induced apoptosis. Mol. Cell. Biol. 22, 4851–4862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Slee, E. A., Adrain, C., and Martin, S. J. ( 2001) Executioner caspase-3, -6, and -7 perform distinct, non-redundant roles during the demolition phase of apoptosis. J. Biol. Chem. 276, 7320–7326 [DOI] [PubMed] [Google Scholar]
- 19.Fernandes-Alnemri, T., Litwack, G., and Alnemri, E. S. ( 1995) Mch2, a new member of the apoptotic Ced-3/Ice cysteine protease gene family. Cancer Res. 55, 2737–2742 [PubMed] [Google Scholar]
- 20.Caulin, C., Salvesen, G. S., and Oshima, R. G. ( 1997) Caspase cleavage of keratin 18 and reorganization of intermediate filaments during epithelial cell apoptosis. J. Cell Biol. 138, 1379–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Byun, Y., Chen, F., Chang, R., Trivedi, M., Green, K. J., and Cryns, V. L. ( 2001) Caspase cleavage of vimentin disrupts intermediate filaments and promotes apoptosis. Cell Death Differ. 8, 443–450 [DOI] [PubMed] [Google Scholar]
- 22.Chen, F., Chang, R., Trivedi, M., Capetanaki, Y., and Cryns, V. L. ( 2003) Caspase proteolysis of desmin produces a dominant-negative inhibitor of intermediate filaments and promotes apoptosis. J. Biol. Chem. 278, 6848–6853 [DOI] [PubMed] [Google Scholar]
- 23.Gervais, F. G., Thornberry, N. A., Ruffolo, S. C., Nicholson, D. W., and Roy, S. ( 1998) Caspases cleave focal adhesion kinase during apoptosis to generate a FRNK-like polypeptide. J. Biol. Chem. 273, 17102–17108 [DOI] [PubMed] [Google Scholar]
- 24.Kalinin, A. E., Aho, M., Uitto, J., and Aho, S. ( 2005) Breaking the connection: caspase 6 disconnects intermediate filament-binding domain of periplakin from its actin-binding N-terminal region. J. Investig. Dermatol. 124, 46–55 [DOI] [PubMed] [Google Scholar]
- 25.Mahoney, J. A., Odin, J. A., White, S. M., Shaffer, D., Koff, A., Casciola-Rosen, L., and Rosen, A. ( 2002) The human homologue of the yeast polyubiquitination factor Ufd2p is cleaved by caspase 6 and granzyme B during apoptosis. Biochem. J. 361, 587–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harvey, K. F., Harvey, N. L., Michael, J. M., Parasivam, G., Waterhouse, N., Alnemri, E. S., Watters, D., and Kumar, S. ( 1998) Caspase-mediated cleavage of the ubiquitin-protein ligase Nedd4 during apoptosis. J. Biol. Chem. 273, 13524–13530 [DOI] [PubMed] [Google Scholar]
- 27.Leo, E., Deveraux, Q. L., Buchholtz, C., Welsh, K., Matsuzawa, S., Stennicke, H. R., Salvesen, G. S., and Reed, J. C. ( 2001) TRAF1 is a substrate of caspases activated during tumor necrosis factor receptor-α-induced apoptosis. J. Biol. Chem. 276, 8087–8093 [DOI] [PubMed] [Google Scholar]
- 28.LeBlanc, A. C., Liu, H., Goodyer, C., Bergeron, C., and Hammond, J. ( 1999) Caspase-6 role in apoptosis of human neurons, amyloidogenesis and Alzheimer's disease. J. Biol. Chem. 274, 23426–23436 [DOI] [PubMed] [Google Scholar]
- 29.Pellegrini, L., Passer, B., Tabaton, M., Ganjei, K., and D'Adamio, L. ( 1999) Alternative, non-secretase processing of Alzheimer's β-amyloid precursor protein during apoptosis by caspase-6 and -8. J. Biol. Chem. 274, 21011–21016 [DOI] [PubMed] [Google Scholar]
- 30.Wellington, C. L., Singaraja, R., Ellerby, L., Savill, J., Roy, S., Leavitt, B., Cattaneo, E., Hackam, A., Sharp, A., Thornberry, N., Nicholson, D. W., Bredesen, D. E., and Hayden, M. R. ( 2000) Inhibiting caspase cleavage of huntingtin reduces toxicity and aggregate formation in neuronal and nonneuronal cells. J. Biol. Chem. 275, 19831–19838 [DOI] [PubMed] [Google Scholar]
- 31.van de Craen, M., de Jonghe, C., van den Brande, I., Declercq, W., van Gassen, G., van Criekinge, W., Vanderhoeven, I., Fiers, W., van Broeckhoven, C., Hendriks, L., and Vandenabeele, P. ( 1999) Identification of caspases that cleave presenilin-1 and presenilin-2. Five presenilin-1 (PS1) mutations do not alter the sensitivity of PS1 to caspases. FEBS Lett. 445, 149–154 [DOI] [PubMed] [Google Scholar]
- 32.Ju, W., Valencia, C. A., Pang, H., Ke, Y., Gao, W., Dong, B., and Liu, R. ( 2007) Proteome-wide identification of family member-specific natural substrate repertoire of caspases. Proc. Natl. Acad. Sci. U. S. A. 104, 14294–14299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Enoksson, M., Li, J., Ivancic, M. M., Timmer, J. C., Wildfang, E., Eroshkin, A., Salvesen, G. S., and Tao, W. A. ( 2007) Identification of proteolytic cleavage sites by quantitative proteomics. J. Proteome Res. 6, 2850–2858 [DOI] [PubMed] [Google Scholar]
- 34.Taylor, R. C., Brumatti, G., Ito, S., Hengartner, M. O., Derry, W. B., and Martin, S. J. ( 2007) Establishing a blueprint for CED-3-dependent killing through identification of multiple substrates for this protease. J. Biol. Chem. 282, 15011–15021 [DOI] [PubMed] [Google Scholar]
- 35.Van Damme, P., Martens, L., Van Damme, J., Hugelier, K., Staes, A., Vandekerckhove, J., and Gevaert, K. ( 2005) Caspase-specific and nonspecific in vivo protein processing during Fas-induced apoptosis. Nat. Methods 2, 771–777 [DOI] [PubMed] [Google Scholar]
- 36.LeBlanc, A. ( 1995) Increased production of 4 kDa amyloid β peptide in serum deprived human primary neuron cultures: possible involvement of apoptosis. J. Neurosci. 15, 7837–7846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stennicke, H. R., and Salvesen, G. S. ( 1997) Biochemical characteristics of caspases-3, -6, -7, and -8. J. Biol. Chem. 272, 25719–25723 [DOI] [PubMed] [Google Scholar]
- 38.O'Farrell, P. Z., Goodman, H. M., and O'Farrell, P. H. ( 1977) High resolution two-dimensional electrophoresis of basic as well as acidic proteins. Cell 12, 1133–1141 [DOI] [PubMed] [Google Scholar]
- 39.O'Connell, K. L., and Stults, J. T. ( 1997) Identification of mouse liver proteins on two-dimensional electrophoresis gels by matrix-assisted laser desorption/ionization mass spectrometry of in situ enzymatic digests. Electrophoresis 18, 349–359 [DOI] [PubMed] [Google Scholar]
- 40.Perkins, D. N., Pappin, D. J., Creasy, D. M., and Cottrell, J. S. ( 1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 41.Yang, F., Sun, X., Beech, W., Teter, B., Wu, S., Sigel, J., Frautschy, S., and Cole, G. ( 1998) Antibody to caspase-cleaved actin detects apoptosis in differentiated neuroblastoma and neurons and plaque microglia in Alzheimer's disease. Am. J. Pathol. 152, 379–389 [PMC free article] [PubMed] [Google Scholar]
- 42.Aoki, C., Sekino, Y., Hanamura, K., Fujisawa, S., Mahadomrongkul, V., Ren, Y., and Shirao, T. ( 2005) Drebrin A is a postsynaptic protein that localizes in vivo to the submembranous surface of dendritic sites forming excitatory synapses. J. Comp. Neurol. 483, 383–402 [DOI] [PubMed] [Google Scholar]
- 43.Thornberry, N. A., Rano, T. A., Peterson, E. P., Rasper, D. M., Timkey, T., Garcia-Calvo, M., Houtzager, V. M., Nordstrom, P. A., Roy, S., Vaillancourt, J. P., Chapman, K. T., and Nicholson, D. W. ( 1997) A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J. Biol. Chem. 272, 17907–17911 [DOI] [PubMed] [Google Scholar]
- 44.Talanian, R. V., Quinlan, C., Trautz, S., Hackett, M. C., Mankovich, J. A., Banach, D., Ghayur, T., Brady, K. D., and Wong, W. W. ( 1997) Substrate specificities of caspase family proteases. J. Biol. Chem. 272, 9677–9682 [DOI] [PubMed] [Google Scholar]
- 45.Srinivasula, S. M., Fernandes-Alnemri, T., Zangrilli, J., Robertson, N., Armstrong, R. C., Wang, L., Trapani, J. A., Tomaselli, K. J., Litwack, G., and Alnemri, E. S. ( 1996) The Ced-3/interleukin 1β converting enzyme-like homolog Mch6 and the lamin-cleaving enzyme Mch2α are substrates for the apoptotic mediator CPP32. J. Biol. Chem. 271, 27099–27106 [DOI] [PubMed] [Google Scholar]
- 46.Kayalar, C., Ord, T., Testa, M. P., Zhong, L. T., and Bredesen, D. E. ( 1996) Cleavage of actin by interleukin 1 β-converting enzyme to reverse DNase I inhibition. Proc. Natl. Acad. Sci. U. S. A. 93, 2234–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adrain, C., Duriez, P. J., Brumatti, G., Delivani, P., and Martin, S. J. ( 2006) The cytotoxic lymphocyte protease, granzyme B, targets the cytoskeleton and perturbs microtubule polymerization dynamics. J. Biol. Chem. 281, 8118–8125 [DOI] [PubMed] [Google Scholar]
- 48.Lane, J. D., Lucocq, J., Pryde, J., Barr, F. A., Woodman, P. G., Allan, V. J., and Lowe, M. ( 2002) Caspase-mediated cleavage of the stacking protein GRASP65 is required for Golgi fragmentation during apoptosis. J. Cell Biol. 156, 495–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paschal, B. M., Obar, R. A., and Vallee, R. B. ( 1989) Interaction of brain cytoplasmic dynein and MAP2 with a common sequence at the C terminus of tubulin. Nature 342, 569–572 [DOI] [PubMed] [Google Scholar]
- 50.Serrano, L., Avila, J., and Maccioni, R. B. ( 1984) Controlled proteolysis of tubulin by subtilisin: localization of the site for MAP2 interaction. Biochemistry 23, 4675–4681 [DOI] [PubMed] [Google Scholar]
- 51.Maccioni, R. B., Vera, J. C., Dominguez, J., and Avila, J. ( 1989) A discrete repeated sequence defines a tubulin binding domain on microtubule-associated protein tau. Arch. Biochem. Biophys. 275, 568–579 [DOI] [PubMed] [Google Scholar]
- 52.Sackett, D. L., Bhattacharyya, B., and Wolff, J. ( 1985) Tubulin subunit carboxyl termini determine polymerization efficiency. J. Biol. Chem. 260, 43–45 [PubMed] [Google Scholar]
- 53.Bhattacharyya, B., Sackett, D. L., and Wolff, J. ( 1985) Tubulin, hybrid dimers, and tubulin S. Stepwise charge reduction and polymerization. J. Biol. Chem. 260, 10208–10216 [PubMed] [Google Scholar]
- 54.Shirao, T., and Sekino, Y. ( 2001) Clustering and anchoring mechanisms of molecular constituents of postsynaptic scaffolds in dendritic spines. Neurosci. Res. 40, 1–7 [DOI] [PubMed] [Google Scholar]
- 55.Otey, C. A., and Carpen, O. ( 2004) α-Actinin revisited: a fresh look at an old player. Cell Motil. Cytoskelet. 58, 104–111 [DOI] [PubMed] [Google Scholar]
- 56.Allen, P. B., Ouimet, C. C., and Greengard, P. ( 1997) Spinophilin, a novel protein phosphatase 1 binding protein localized to dendritic spines. Proc. Natl. Acad. Sci. U. S. A. 94, 9956–9961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Allison, D. W., Chervin, A. S., Gelfand, V. I., and Craig, A. M. ( 2000) Postsynaptic scaffolds of excitatory and inhibitory synapses in hippocampal neurons: maintenance of core components independent of actin filaments and microtubules. J. Neurosci. 20, 4545–4554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takahashi, H., Sekino, Y., Tanaka, S., Mizui, T., Kishi, S., and Shirao, T. ( 2003) Drebrin-dependent actin clustering in dendritic filopodia governs synaptic targeting of postsynaptic density-95 and dendritic spine morphogenesis. J. Neurosci. 23, 6586–6595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kobayashi, R., Sekino, Y., Shirao, T., Tanaka, S., Ogura, T., Inada, K., and Saji, M. ( 2004) Antisense knockdown of drebrin A, a dendritic spine protein, causes stronger preference, impaired pre-pulse inhibition, and an increased sensitivity to psychostimulant. Neurosci. Res. 49, 205–217 [DOI] [PubMed] [Google Scholar]
- 60.Shim, K. S., and Lubec, G. ( 2002) Drebrin, a dendritic spine protein, is manifold decreased in brains of patients with Alzheimer's disease and Down syndrome. Neurosci. Lett. 324, 209–212 [DOI] [PubMed] [Google Scholar]
- 61.Hatanpaa, K., Isaacs, K. R., Shirao, T., Brady, D. R., and Rapoport, S. I. ( 1999) Loss of proteins regulating synaptic plasticity in normal aging of the human brain and in Alzheimer disease. J. Neuropathol. Exp. Neurol. 58, 637–643 [DOI] [PubMed] [Google Scholar]
- 62.Harigaya, Y., Shoji, M., Shirao, T., and Hirai, S. ( 1996) Disappearance of actin-binding protein, drebrin, from hippocampal synapses in Alzheimer's disease. J. Neurosci. Res. 43, 87–92 [DOI] [PubMed] [Google Scholar]
- 63.Satoh, A., Nakanishi, H., Obaishi, H., Wada, M., Takahashi, K., Satoh, K., Hirao, K., Nishioka, H., Hata, Y., Mizoguchi, A., and Takai, Y. ( 1998) Neurabin-II/spinophilin. An actin filament-binding protein with one PDZ domain localized at cadherin-based cell-cell adhesion sites. J. Biol. Chem. 273, 3470–3475 [DOI] [PubMed] [Google Scholar]
- 64.Hsieh-Wilson, L. C., Allen, P. B., Watanabe, T., Nairn, A. C., and Greengard, P. ( 1999) Characterization of the neuronal targeting protein spinophilin and its interactions with protein phosphatase-1. Biochemistry 38, 4365–4373 [DOI] [PubMed] [Google Scholar]
- 65.Feng, J., Yan, Z., Ferreira, A., Tomizawa, K., Liauw, J. A., Zhuo, M., Allen, P. B., Ouimet, C. C., and Greengard, P. ( 2000) Spinophilin regulates the formation and function of dendritic spines. Proc. Natl. Acad. Sci. U. S. A. 97, 9287–9292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stafstrom-Davis, C. A., Ouimet, C. C., Feng, J., Allen, P. B., Greengard, P., and Houpt, T. A. ( 2001) Impaired conditioned taste aversion learning in spinophilin knockout mice. Learn. Mem. 8, 272–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Walikonis, R. S., Oguni, A., Khorosheva, E. M., Jeng, C. J., Asuncion, F. J., and Kennedy, M. B. ( 2001) Densin-180 forms a ternary complex with the α-subunit of Ca2+/calmodulin-dependent protein kinase II and α-actinin. J. Neurosci. 21, 423–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Galloway, P. G., Perry, G., and Gambetti, P. ( 1987) Hirano body filaments contain actin and actin-associated proteins. J. Neuropathol. Exp. Neurol. 46, 185–199 [DOI] [PubMed] [Google Scholar]
- 69.Rossiter, J. P., Anderson, L. L., Yang, F., and Cole, G. M. ( 2000) Caspase-cleaved actin (fractin) immunolabelling of Hirano bodies. Neuropathol. Appl. Neurobiol. 26, 342–346 [DOI] [PubMed] [Google Scholar]
- 70.Scheff, S. W., and Price, D. A. ( 2003) Synaptic pathology in Alzheimer's disease: a review of ultrastructural studies. Neurobiol. Aging 24, 1029–1046 [DOI] [PubMed] [Google Scholar]
- 71.Kalamvoki, M., Georgopoulou, U., and Mavromara, P. ( 2006) The NS5A protein of the hepatitis C virus genotype 1a is cleaved by caspases to produce C-terminal-truncated forms of the protein that reside mainly in the cytosol. J. Biol. Chem. 281, 13449–13462 [DOI] [PubMed] [Google Scholar]
- 72.Eleouet, J. F., Slee, E. A., Saurini, F., Castagne, N., Poncet, D., Garrido, C., Solary, E., and Martin, S. J. ( 2000) The viral nucleocapsid protein of transmissible gastroenteritis coronavirus (TGEV) is cleaved by caspase-6 and -7 during TGEV-induced apoptosis. J. Virol. 74, 3975–3983 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.