Abstract
Two-hybrid methods have augmented the classical genetic techniques biologists use to assign function to genes. Here, we describe construction of a two-bait interaction trap that uses yeast cells to register more complex protein relationships than those detected in existing two-hybrid systems. We show that such cells can identify bridge or connecting proteins and peptide aptamers that discriminate between closely related allelic variants. The protein relationships detected by these cells are analogous to classical genetic relationships, but lend themselves to systematic application to the products of entire genomes and combinatorial libraries. We show that, by performing logical operations on the phenotypic outputs of these complex cells and existing two-hybrid cells, we can make inferences about the topology and order of protein interactions. Finally, we show that cells that register such relationships can perform logical operations on protein inputs. Thus these cells will be useful for analysis of gene and allele function, and may also define a path for construction of biological computational devices.
Keywords: two-hybrid, interaction trap, two-bait, biological computation, functional genomics
Genetic analysis is a tool for understanding the regulatory networks that govern biological processes (1–7). The manipulations performed by geneticists (e.g., staging of temperature-sensitive mutants, construction and analysis of double mutants) define relationships among gene activities, which in turn reflect underlying biochemical mechanisms. For example, these relations may suggest that one gene product normally acts on another to carry out a process (epistasis), that two gene products physically interact (allele specific suppression) (1, 5), or that action of one gene product precedes that of another in time (dependency) (4).
Information obtained from classical genetic manipulations is typically of high quality, but can be difficult to acquire. The recent increase in the rate of identification of new coding sequences has stimulated interest in global systematic methods to understand gene function. These methods include DNA sequencing (8), analysis of expressed transcripts (see, for example, ref. 9), and two-hybrid methods, including the interaction trap, that we and others have developed to assay contact between two proteins (10–14). Such contact defines a physical relationship that frequently has functional significance.
In classical two-hybrid systems (10–13), transcription of reporter genes depends on an interaction between a DNA-bound “bait” protein and an activation-domain containing “prey” protein. The underlying molecular biology of these systems converts a single initial event input into billions of molecules that produce an output signal. The initial event, introduction of a plasmid encoding an interacting protein into a single cell, is first typically amplified by the growth of that cell into a colony of 107 cells. When protein interaction occurs, the amount of translated reporter protein from the reporter’s mRNA and the amount of substrate converted by reporter protein into signal vary linearly with the amounts of reporter mRNA. The detection of reporter phenotypes in these systems is subject to thresholds, above which a phenotype is scored (14); the end result is a nonlinear amplification of the input signal.
Here we construct cells that register more complex protein relationships, and view these relationships in symbolic-logical terms. We demonstrate that ability to use these cells to select proteins that satisfy complex relationships, particularly bridging and the two discrimination relationships, allows identification of proteins and peptide aptamers from library screens that can illuminate the function of genes and alleles, and can help distinguish among different models of protein topology and sequence of action in protein networks. Finally, we demonstrate that these complex cells can perform logical operations on input proteins, suggesting a path for constructing computational circuitry.
MATERIALS AND METHODS
The Two-Bait System.
In two-hybrid systems, we refer to the activation tagged protein as the prey (P) and the LexA-fusion protein as the bait (Ba) (12). Here, we constructed a two-bait two-hybrid system in which a plasmid, pCWX200, directed the synthesis of one bait (Ba1), whereas plasmids pEG202 and pJG4-5 (12) directed synthesis of the second bait (Ba2) and the prey. The Ba1/P interaction was measured by expression of TetOp-URA3 reporter constructs integrated into Saccharomyces cerevisiae (strains CWXY1 and CWXY2), and the Ba2/P interaction was measured by expression of LexOp-LacZ from pCWX24, an episomal LYS2 plasmid. (A detailed description of these constructions can be found at http://xanadu.mgh.harvard.edu.)
Assays for the And and Discrimination Relationships.
We cloned c-Raf1 (1–313) into the EcoRI/XhoI sites of pCWX200 and JG4-5. After amplifying RasA15A186, hSos1 (601–1,019) and Cdc25 (907–1,589) with RasA15A186 primers 5′-GCCTGAATTCATGACGGAATATAAGCTGG-3′ and 5′-CCCGAACTCGAGTCAGGAGAGCACTGCCTTGCAGC-3′, hSos1 (601–1,019) primers 5′-GCCTGAATTCAAAGCAGGAACTGTT-3′ and 5′-CCCGAACTCGAGCTATCGTGGTTCTATTTCTAG-3′, and Cdc25 catalytic domain (907–1,589) primers 5′-GCCTGAATTCATGTCTTCGGTCTCCTCAG-3′ and 5′-CCCGAACTCGAGTTATCGAAATAACCTAGAAGG-3′, we cloned the EcoRI/XhoI-ended PCR products of RasA15A186, hSos1 (601–1,019) and Cdc25 (907–1,589) into EcoRI/XhoI sites of pCWX200, pEG202, and pJG4-5. We transformed (15) CWXY2/pCWX24 with combinations of the above baits and preys, streaked transformants onto LHKW/Glu dropout plates (16) that contain 100 μg/ml 6-azauracil, an inhibitor of the URA3 gene product (17), for 12–48 hr at 30°C. We subsequently replicated the yeast streaks onto dropout plates LHKW/Glu + 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside (X-Gal), LHKW/Gal + raffinose (Raff) + X-Gal, LHKWU/Glu, and LHKWU/Gal + Raff and scored the results after 12–72 hr at 30°C.
Assays for the And Operation on Protein Inputs.
We cloned into pCWX200 a EcoRI/XhoI-ended human GAP1 PCR product amplified with primers 5′-GCCTGAATTCATGAAGGGGTGGTATCACGGA-3′ and 5′-CCCGAACTCGAGCTACTTGACATCATTGGTTTTTG-3′. Similarly, we introduced EcoRI/XhoI-ended Cdc25 (907–1,589) into pCWX200. Subsequently, we transformed CWXY2 containing pEG202-RasA186 and pJG-c-Raf (1–313) with one of following: pCWX200, pCWX200-CDC25 (907–1,589), and pCWX200-GAP. We then replicated streaks of transformants pooled from >50 independent transformants onto LHKW/Glu + X-Gal and LHKW/Gal + Raff + X-Gal dropout plates and incubated them at 30°C for 2–3 days. Cells on the former showed no blue color, but on the latter showed blue color of varied intensity. We assayed β-galactosidase activity from liquid assays (14) in triplicate cultures grown from pooled colonies, each of which contained >50 independent transformants.
RESULTS
Preliminary Considerations: Logical Protein Relationships in Two-Hybrid Systems.
In classical (one-bait) two-hybrid systems, the output of the reporter gene depends on the interaction between the DNA-bound bait and activation-tagged prey. Genetic markers expressed by some reporters, for example URA3 (ref. 17 and this work), allow selection against reporter transcription and thus selection for lack of interaction. We can describe the relation between proteins in these systems in symbolic-logical terms.
In this view, contact between a bait (Ba1) and a prey (P) defines a variable (A1), here called the touching relationship. Because A1 is operationally defined by the reporter output, A1 can also to refer to the reporter output. There are four possible Boolean operations, or functions, on this relationship (18). Two of these are constant functions: F1 (A1) = 0 and F2 (A1) = 1; two are not: F3 (A1) = A1 and F4 (A1) = Not A1.
In one-bait systems, the phenotype dependent on the reporter output can register both the F3 and F4 operations on the touching relationship. Both operations have important biological correlates. Consider a one-bait system in which contact between Ba1 and P (for example, because these proteins heterodimerize) gives a positive output (blue color on X-Gal, growth on Ura− medium), whereas loss of that contact [for example by mutation, or disruption by a peptide aptamer (19)] gives a negative output [white color on X-Gal, growth on 5-fluoroorotic acid (5-FOA) medium]. We can think of such a cell growing on 5-FOA medium as a device that performs the logical Not operation on the touching relation, or, alternatively, as a cell that registers the state (Not A1). Although this formalism is not necessary for the analysis of data from one-bait systems, it provides a convenient way of describing data from more complex systems (see below).
Cells That Detect More Complex Protein Relationships.
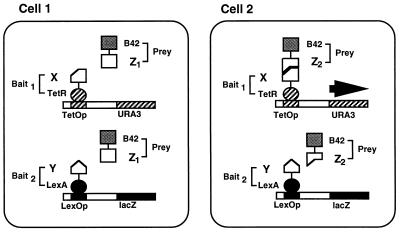
We constructed cells that allow simultaneous selection for and against two distinct protein–protein interactions (Fig. 1). This two-bait interaction trap utilizes the DNA binding moieties of LexA and TetR, the tetracycline repressor of bacterial transposon Tn10 (20). The LexA and TetR fusion bait proteins are expressed in a yeast cell that contains an integrated TetOp-URA3 reporter and an episomal LexAOp-lacZ reporter. The ADH1 promoter controls expression of baits in these cells, and the GAL1 promoter conditionally directs expression of activation-tagged protein (the prey) from a 2-μm plasmid.
Figure 1.
The two-bait interaction trap. Cells contain bait1, a protein fusion of Tet repressor and protein X, and bait2, a fusion of LexA and protein Y. In cell 1, a prey, a fusion of protein Z1 and transcriptional activator B42, does not interact with either X or Y. The reporters are not activated, the cell grows on 5-FOA, does not grow on Ura− medium, and is white on X-Gal. In cell 2, another prey, a fusion of Z2 and B42 interacts with X but not with Y, the TetOp-URA3 reporter is selectively expressed, and the cell grows on Ura− medium but is white on X-Gal.
Logical Operations on the Touching Relationship.
In two-bait cells, we can view the two touching relationships (and the output of the corresponding reporters) as Boolean variables, A1 and A2. There are 16 possible operations on these variables (18), four of which are registered by these cells (Table 1). We refer to these operations as And, Nor, and the two discrimination operations, logic state 1 (Ls1) and logic state 2 (Ls2). We imagined that And, Ls1, and Ls2 should represent particularly useful operations for determining protein function.
Table 1.
Boolean operations on the touching relationship detected in the two-bait interaction trap
| Value of | |||
|---|---|---|---|
| Variable | variable | ||
| A1 | 0 1 0 1 | ||
| A2 | 0 0 1 1 | Alternative name | Interpretation |
| Operation | Results of operation | ||
| F2 | 1 0 0 0 | Nor | No interactions |
| F9 | 0 0 0 1 | And, bridging, | P interacts with |
| A1 And A2 | Ba1 and Ba2 | ||
| F3 | 0 0 1 0 | Ls1, discrimination, | P interacts with |
| (Not A1) And A2 | Ba2 and not with Ba1 | ||
| F5 | 0 1 0 0 | Ls2, discrimination, | P interacts with |
| A1 And (Not A2) | Ba1 and not with Ba2 |
We denote the touching relationship between Bait1 (Ba1) and prey (P) as A1, measured by the output of the TetOp–URA3 reporter, and between Bait2 (Ba2) and P as A2, is measured by the output of the LexAop–lacZ reporter. The binary values of A1 and A2 refer both to values of the touching relationship and to the values (off and on states) of the reporter. The four subcolumns under “value of variables” denote the four different possible combinations of values of these two variables. The column labeled “operation” shows the four operations on these variables enabled in this system (18). The column labeled “alternative name” gives common names for the operations. The column labelled “interpretation” describes the state of interaction between the bait and prey proteins.
To test this idea, we used a set of derivatized interacting proteins described in Materials and Methods. Some of these are involved in Ras-Raf-dependent signal transduction: RasA186, RasA15A186, RasV12A186, GAP, hSos1 (residues 601–1,019), Cdc25 (residues 907–1,589), and c-Raf1 (residues 1–313); their sequence differs from that of the wild-type human proteins as noted. These proteins are here referred to as Ras, RasV12, RasA15, GAP, SOS, Cdc25, and c-Raf1. In addition, we used derivatives of human Max and Mxi1, which heterodimerize (21) as positive and negative controls for interaction.
The Harvey ras gene product, Ras, is a GTPase that exists in two conformations, a GDP-bound (inactive) and a GTP-bound (active) form. Because Ras can cycle between these conformations, it can act as a switch protein in signal transduction pathways that control cell proliferation (22). The Ras proteins used here all contained a Cys-186 → Ala mutation, which inactivates a farnesylation site and increases apparent nuclear concentration (A. Makris, personal communication; C.W.X. and R.B., unpublished data). Ras exists in a mixture of GDP- and GTP-bound forms, whereas RasA15 and RasV12 are respectively predominantly in the GDP- and GTP-bound forms. GAP, which stimulates Ras GTPase activity, binds to GTP-bound Ras. c-Raf1, a downstream target of Ras, also binds GTP-bound Ras. By contrast, hSos1 and Cdc25, both of which activate Ras, only bind GDP-bound Ras.
Table 2 summarizes experiments that show that two-bait cells can register the logical And. This relationship is fulfilled by proteins, such as bridging proteins in pathways, that can interact with both baits. In this experiment, CWXY2 carried B42-Ras as prey, TetR-c-Raf1 and LexA-hSos1 as the baits, and TetOp-URA3 and LexAOp-LacZ as the reporters. B42-Ras interacted with both TetR-c-Raf1 and LexA-hSos1. The cell was blue on X-Gal and grew on medium lacking uracil. This bridging relationship is expected from the fact that Ras interacts with both proteins, and suggests that proteins that activate both reporters can be selected from genome-wide screens.
Table 2.
Examples of cells that register the And and discrimination relationships (Ls1 and Ls2) on the touching relationships A1 and A2
| Baits | Prey | Reporter output
|
Logical relationship | |
|---|---|---|---|---|
| Ura- | X-Gal | |||
| 1 TetR-c-Raf1 | Ras | Growth | Blue | And |
| LexA-hSos1 | ||||
| 2 TetR-RasV12 | c-Raf1 | Growth | White | Ls2 |
| LexA-Max | ||||
| 3 TetR-RasV12 | Mxi1 | No growth | Blue | Ls1 |
| LexA-Max | ||||
| 4 TetR-RasA15 | c-Raf1 | No growth | Blue | Ls1 |
| LexA-RasV12 | ||||
| 5 TetR-RasA15 | Cdc25 | Growth | White | Ls2 |
| LexA-RasV12 | ||||
Cells were grown on raffinose medium, on which preys are expressed. Identities of the bait and prey proteins are given in the first two column headings. Reporter output phenotype is indicated by growth or blue color on the indicated medium. The logical relationship registered is given in the last column.
Table 2 also shows that the two-bait system can register the discrimination relationships Ls1 or Ls2. These relationships are expected for proteins that interact with one bait but not another. In cells that contained TetR-RasV12 and LexA-Max baits, and expressed a B42-c-Raf1 prey, c-Raf1 interacted with TetR-RasV12 and activated the URA3 reporter, but did not interact with LexA-Max, and thus did not activate the lacZ reporter. In cells that contained TetR-RasV12 and LexA-Max baits, and that expressed a B42-Mxi1 prey, Mxi1 interacted with Max and activated the lacZ reporter, but did not interact with RasV12 to activate the URA3 reporter.
Finally, Table 2 shows that these cells can discriminate between allelic variants. In a cell that expressed TetR-RasA15, LexA-RasV12 and B42-c-Raf1, the Raf/RasV12 interaction only activated expression of the LexAop-LacZ reporter: the cells turned blue on X-Gal, but did not grow on medium lacking uracil. By contrast, in a cell that expressed the same TetR-RasA15 and LexA-RasV12 baits, but a different prey, B42-Cdc25, the Cdc25/RasA15 interaction activated the TetOp-URA3 reporter, and allowed the cell to grow on medium lacking uracil, but caused it to remain white on X-Gal. This result suggested that these cells could identify, from genomic or combinatorial libraries, proteins and peptides (19) that interact with allelic protein variants specific for disease states.
Identification of Peptide Aptamers That Discriminate Between Allelic Variants.
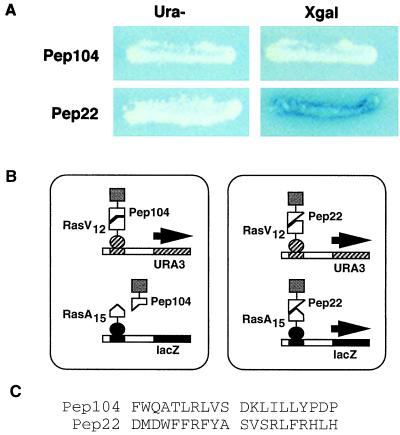
To test this idea, we used a two-bait cell that contained TetR-RasV12 and LexA-RasA15 to isolate members of a peptide aptamer library that interacted specifically with RasV12. We screened URA+ library transformants for lacZ− cells, which presumably contained aptamers that did not interact with LexA-RasA15. We then rescued plasmids encoding aptamers from these and lacZ+ cells and reconfirmed their phenotypes. Fig. 2 shows that these cells can easily detect discriminatory aptamers: Pep22 interacted with both RasV12 and RasA15, whereas, by contrast, Pep104 interacted only with RasV12. These results demonstrate the utility of this system in selection of specific peptide aptamers. For Pep22, the second bait increased the selectivity of the system by eliminating potential false positives that might arise from artifactual activation of a single reporter. For Pep104, the second bait allowed detection of aptamers specific for an allelic form of the protein active in signal transduction.
Figure 2.
Peptide aptamers that discriminate between Ras alleles. (A) Reporter phenotypes of cells on Ura− or X-Gal medium. These cells contain TetR-RasV12 and LexA-RasA15, and also peptide aptamer Pep104 or Pep22. The Pep104 containing cell grows on Ura− medium but is white on X-Gal medium. The Pep22 cell grows on Ura− medium and is blue on X-Gal. (B) Protein relationships in these two-bait cells. Pep104 interacts only with RasV12 but not with RasA15, and thus selectively activates the TetOp-URA3 reporter. Pep22 interacts with both RasV12 and RasA15, and thus activates both the TetOp-URA3 and LexOp-lacZ reporters. (C) Sequences of the variable (recognition) regions of peptide aptamers Pep104 and Pep22.
Logical Operations on Protein Inputs.
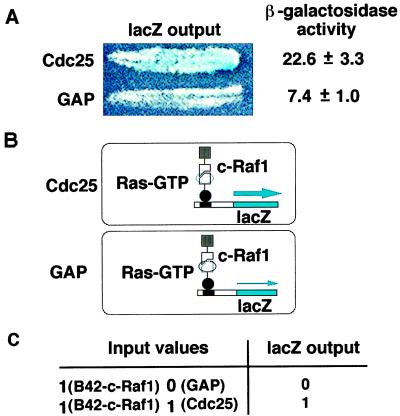
These cells can also perform logical operations on protein inputs, and register their result by changes in output. Fig. 3 shows a logical And operation on protein inputs. In a cell that expressed LexA-Ras, B42-c-Raf1, and TetR-GAP, the output of the LexAop-lacZ reporter was low (light blue on X-Gal medium; Fig. 3A) presumably because GAP drove most of the Ras into the GDP-bound, Raf nonbinding conformational state (Fig. 3B). By contrast, input of TetR-Cdc25 instead of TetR-GAP increased the Ras/c-Raf1 interaction, as shown by the intensity of blue color on X-Gal plates (Fig. 3A), presumably by changing Ras into the GTP-bound, Raf binding conformation (Fig. 3B). In this experiment, the cell had two inputs, one of which, B42-c-Raf1 (logical value 1) was constantly present, whereas the other was either TetR-GAP (logical value 0) or TetR-Cdc25 (logical value 1); and the cell’s output was either high (1) or low (0) (Fig. 3C). Thus, in these cells a LexA-Ras switch, whose conformation depended on the values of the inputs, controlled the output of the lacZ reporter.
Figure 3.
Cells that perform the logical And operation on input proteins. (A) Expression of the LexOp-lacZ reporter in cells containing TetR-Cdc25 (907–1,589) or GAP as measured by the intensity of blue color on X-Gal medium and by β-galactosidase activity in Miller units. (B) Inferred protein relationships in these cells. In the cell expressing TetR-Cdc25, Cdc25 loads LexA-Ras with GTP, increasing the amount of Ras that is in the GTP bound conformation (black outline) with which B42-c-Raf1 can interact to stimulate transcription of the lacZ reporter, and decreasing the amount of Ras in the GDP-bound conformation (grey outline) with which B42-c-Raf-1 cannot interact. In the cell expressing TetR-GAP, this switch is reversed. In this cell, TetR-GAP stimulates Ras GTPase activity, increasing the amount of Ras in the GDP-bound form (black outline), with which B42-c-Raf1 cannot interact, and decreasing the amount of Ras in the GTP-bound form (grey outline), thus resulting in lowered lacZ output. (C) Table depicting results of these operations on protein inputs. In the table, the 1’s in the first column indicate the presence of B42-Raf, the 0 and 1 in the second column denote, respectively, the presence of TetR-Gap and TetR-Cdc25, and the 0 and 1 in the third column denote respectively low and high outputs of β-galactosidase.
DISCUSSION
Construction of Cells with Independently Functional Interaction Reporters.
We constructed cells that allow simultaneous detection of two touching relationships (Fig. 1). These cells utilize the DNA binding moieties of LexA and TetR, the Tn10 tetracycline repressor. Fusion proteins containing these moieties are expressed as baits in cells that also contain TetOp-URA3 and LexAop-lacZ reporters.
The inclusion of TetR baits and TetOp-URA3 reporters significantly facilitates conventional applications of the interaction trap (12). The phenotype dependent on the TetOp-URA3 reporter is more sensitive than that of previously described lacZ and LEU2 reporters (12, 14), which should facilitate detection of even weaker interactions (C.W.X. and R.B., unpublished data). In addition, both the URA3 (17) and LacZ reporters can be quantitatively assayed. Moreover, the sensitivity of the URA3 reporter can be down-regulated in two ways. Expression of the URA3 reporter can be titrated by 6-azauracil, an inhibitor of the URA3 gene product (17, 23, 24). Reporter activity can also be reduced by tetracycline and its derivatives, which disrupt binding of the TetR bait to Tet operators (C.W.X. and R.B., unpublished data). Both kinds of inhibitors thus diminish the sensitivity of the URA3 reporter, allowing its use with baits that activate transcription, and, along with lacZ, to estimate interaction affinities. Moreover, the URA3 reporter allows the use of 5-FOA (25) to select against its gene expression. In this case tetracycline and 6-azauracil can alter the threshold amount of transcription selected against, facilitating the selection of peptide aptamers that break specific protein interactions (C.W.X. and R.B., unpublished data).
Protein Relationships in Two-Bait Systems.
As Tables 1 and 2 show, the two-bait cells register four logical relations, Nor, And, Ls1, and Ls2. Three are particularly important. The And relationship [A1 (between Ba1 and P) And A2 (between (Ba2 and P)] is found for preys that contact both baits (connecting proteins). Cells that register the And relationship will speed the identification of such proteins, which will help connect pathways not previously known to be related and help elaborate protein networks.
Ls1 and Ls2, the discrimination relationships, are more complex: for example, the Ls1 relationship [Not A1 (between Ba1 and P) And A2 (between Ba2 and P)] involves two operations on two interactions. These relationships have numerous biological correlates, one of which (see below) corresponds to the detection of peptide aptamers that interact differently with allelic variants. The use of these relationships to survey the products of combinatorial libraries and genomes will allow selection of proteins that interact specifically with proteins encoded by disease-state alleles, or that interact specifically with proteins that differ from wild-type due to differential splicing or posttranslational modification.
Use of Two-Bait Cells to Identify Allele-Specific Peptide Aptamers.
To demonstrate the utility of these cells in isolating interacting molecules, we used them to select a peptide aptamer that interacts specifically with an allelic form of Ras that causes cell proliferation (Fig. 2). That is, we isolated a peptide aptamer that satisfies the Ls1 discrimination relation and interacts with an active allelic form of Ras but not with an inactive allelic form. The ability to select proteins that satisfy this relationship may open the way to select molecules that specifically suppress the function of activated Ras alleles, as well as to select reagents to probe the function of allelic variants of other genes.
Analysis of Higher-Order Protein Relationships.
The protein relationships that can be inferred from two-bait cells are not always identical to those revealed by one-bait cells. For example, if both Ba1 and Ba2 oligomerize to form a surface that interacts with P, then neither the Ba1/P nor Ba2/P interaction will be detected in one-bait cells. Such differences in touching relationships are useful, since combining data from one- and two-bait cells allows the experimenter to make inferences about the topology, temporal sequence, and posttranslational modification dependence of the protein interactions.
Table 3 shows inferences about physical interactions among three proteins, X, Y, and Z: (i) from possible outputs of a two-bait cell that detects touching relationships A1 (between X and Z) and A2 (between Y and Z), and (ii) from outputs of three one-bait cells that detect A1 (between X and Z), A2 (between Y and Z), and A3 (between X and Y). We anticipate that experiments suggested by such linkages of one- and two-bait data will prove useful in ordering function of proteins in pathways and complexes.
Table 3.
Analysis of the interaction of three proteins by operations on reporter outputs in two- and one-bait cells
| Reporter output
|
Physical interpretation (one-bait and two bait data combined) | Physical interpretation (one-bait data only) | ||||
|---|---|---|---|---|---|---|
| One-bait cells
|
Two-bait cells
|
|||||
| A1 | A2 | A3 | A1 | A2 | ||
| Ba(X) | Ba(Y) | Ba(X) | Ba1(X) | Ba2(Y) | ||
| P(Z) | P(Z) | P(Y) | P(Z) | P(Z) | ||
| 0 | 0 | 0 | 0 | 0 | X, Y, Z do not interact | X, Y, Z do not interact |
| 0 | 0 | 0 | 0 | 1 | X, Y, Z associate weakly, modified Y and/or Z interact | X, Y, Z do not interact |
| 0 | 0 | 0 | 1 | 0 | X, Y, Z associate weakly, modified X and/or Z interact | X, Y, Z do not interact |
| 0 | 0 | 0 | 1 | 1 | X, Y, Z form trimer, requires X, Y, and Z | X, Y, Z do not interact |
| 0 | 0 | 1 | 0 | 0 | X, Y form dimer | X, Y form dimer |
| 0 | 0 | 1 | 0 | 1 | X modifies Y, modified Y binds Z | X, Y form dimer |
| 0 | 0 | 1 | 1 | 0 | Y modifies X, modified X binds Z | X, Y form dimer |
| 0 | 0 | 1 | 1 | 1 | X, Y form dimer, Z binds X/Y dimer to form X/Y/Z trimer | X, Y form dimer |
| 0 | 1 | 0 | 0 | 0 | X breaks Y/Z dimer | Y, Z form dimer |
| 0 | 1 | 0 | 0 | 1 | Y, Z form dimer, Z discriminates between X and Y | Y, Z form dimer |
| 0 | 1 | 0 | 1 | 0 | Y modifies Z, modified Z binds X | Y, Z form dimer |
| 0 | 1 | 0 | 1 | 1 | Y, Z form dimer, X binds to Y/Z dimer to form X/Y/Z trimer | Y, Z form dimer |
| 1 | 0 | 0 | 0 | 0 | Y breaks X/Z dimer | X, Z form dimer |
| 1 | 0 | 0 | 0 | 1 | X modifies Z, modified Z binds Y | X, Z form dimer |
| 1 | 0 | 0 | 1 | 0 | X, Z form dimer, Z discriminates between X and Y | X, Z form dimer |
| 1 | 0 | 0 | 1 | 1 | X, Z form dimer, Y binds to X/Z dimer to form X/Y/Z trimer | X, Z form dimer |
| 0 | 1 | 1 | 0 | 0 | X, Y and Y, Z form dimers, X/Y dimer precludes Y binding Z | Y, Z and Y, X form dimers |
| 0 | 1 | 1 | 0 | 1 | X, Y and Y, Z form dimers, Y/Z dimer precludes X binding Y | Y, Z and Y, X form dimers |
| 0 | 1 | 1 | 1 | 0 | Y modifies X, modified X binds Z | Y, Z and Y, X form dimers |
| 0 | 1 | 1 | 1 | 1 | X/Y and Y/Z dimers interact through Y to form X/Y/Z trimer | Y, Z and Y, X form dimers |
| 1 | 0 | 1 | 0 | 0 | X, Y and X, Z form dimers, X/Y dimer precludes X binding Z | X, Z and X, Y form dimers |
| 1 | 0 | 1 | 0 | 1 | X modifies Y, modified Y binds Z | X, Z and X, Y form dimers |
| 1 | 0 | 1 | 1 | 0 | X, Y and X, Z form dimers, X/Z dimer precludes X binding Y | X, Z and X, Y form dimers |
| 1 | 0 | 1 | 1 | 1 | X/Z and X/Y dimers interact through X to form X/Y/Z trimer | X, Z and X, Y form dimers |
| 1 | 1 | 0 | 0 | 0 | X, Z and Y, Z form dimers, dimers inactivate one another | X, Z and Y, Z form dimers |
| 1 | 1 | 0 | 0 | 1 | X, Z and Y, Z form dimers, Y/Z dimer precludes X binding Z | X, Z and Y, Z form dimers |
| 1 | 1 | 0 | 1 | 0 | X, Z and Y, Z form dimers, X/Z dimer precludes Y binding Z | X, Z and Y, Z form dimers |
| 1 | 1 | 0 | 1 | 1 | X, Z and Y, Z dimers form | X, Z and Y, Z form dimers |
| 1 | 1 | 1 | 0 | 0 | X, Y forms dimer, X/Y dimer inactivates Z | X, Y and X, Z and Y, Z form dimers, X/Y/Z trimer may form (16) |
| 1 | 1 | 1 | 0 | 1 | X, Y and X, Z and Y, Z form dimers, Y modifies X, modified X binds Z poorly | X, Y, and X, Z, and Y, Z form dimers, X/Y/Z trimer may form (16) |
| 1 | 1 | 1 | 1 | 0 | X, Y and X, Z and Y, Z form dimers, X modifies Y, modified Y binds Z poorly | X, Y, and X, Z and Y, Z, form dimers, X/Y/Z trimer may form (16) |
| 1 | 1 | 1 | 1 | 1 | X, Y and X, Z and Y, Z form dimers, X/Y/Z trimer may form | X, Y and X, Z and Y, Z form dimers, X/Y/Z trimer may form (16) |
“Reporter output” shows the state of the touching relationships registered by outputs of the reporters in three one-bait cells and a single two-bait cell. In the one-bait cells, the reporter output shows the touching relationships A1, A2, and A3 for different combinations of proteins X, Y, and Z. Here, X and Z are fused to an activation domain to form preys [P(X) or P(Z)], and Y and X are fused to a DNA binding domain to form baits [Ba(X), Ba(Y)]. In a two-bait cell, the outputs of the reporters show the state of the touching relationship for proteins X, Y and Z where they are fused with one of two DNA binding domains [Ba1(X) or Ba2(Y)] and an activation domain [P(Z)]. “Physical interpretation” shows one possible biological interpretation of this set of reporter outputs for possible combinations of one- and two-bait data, or for one-bait data alone. Although all patterns here may not have biological correlates, many have been observed, for example, the interaction of Bait1 and prey depends on the presence of Bait2 (P. King, personal communication).
Application to the Analysis of Gene Function.
This two-bait system thus extends the utility of yeast interaction technology for determining gene function. It can aid the identification of proteins and peptide aptamers (this work) that distinguish between allelic variants of proteins. In addition, linkage of data from two-bait cells (likely to result from individual experiments) and from one-bait cells [perhaps obtained from genome-wide surveys (26, 27)] will allow detailed analysis of protein function and contact topology in pathways and complexes.
Toward Protein-and-Transcription-Based Logical Circuitry.
The cells used in this work illustrate another point. In these experiments, we used a protein, Ras, that cycles between two conformational states, and used an activation tagged protein, Raf, that binds Ras in one of these states. We showed that the state of the Ras switch, and its output measured by transcription, varied depending on whether the input protein was GAP or Cdc25. In these experiments the cells were acting as logical And gates, in which one input, c-Raf1, was held constant (logical value 1), the other was either Gap (logical value 0) or Cdc25 (logical value 1), and the phenotypes caused by expression of the reporters constitute the outputs.
In these cells, to change the inputs, we employed different DNA constructions that expressed interacting proteins; to measure outputs, we employed a human observer. Construction of wholly biological logic circuitry will require replacing these inputs and outputs with extracellular signals such as secreted peptide pheromones or light. Such logical devices might not be very fast: although the switch, Ras, can cycle in milliseconds, and a number of signal transduction pathways can provide inputs within minutes (28), reporter output may require minutes to hours to be detectable. However, because we know so much about how to perform the required construction work near the DNA, it is likely that gene expression will remain a useful output. Construction of such circuits will be facilitated by the rapid increase in the number of natural and artificially selected protein domains with useful allosteric and targeting properties (19). It is thus possible that transcription-based technologies will provide an early route to biological computation.
Acknowledgments
We thank R. Baumeister, B. Cohen, J. Colicelli, G. M. Cooper, J. A. Cooper, G. Fink, R. Finley, I. Ha, S. Hanes, A. Makris, F. McCormick, and E. Porfiri for plasmids; and L. Lok, L. Nyuyen, P. Silver, C. Kaiser, M. Franklin, C. Kenyon, L. Buck, and members of the lab for useful discussions. C.W.X. (formerly Chanxing Xu) was supported by the Massachusetts General Hospital fund for medical discovery and the Human Frontier Science Program, A.R.M. by an American Cancer Society postdoctoral fellowship, and R.B. by an American Cancer Society Faculty Research Award. This work was funded by the National Human Genome Research Institute.
Note Added in Proof
While a previous version of this manuscript was under review, Jiang and Carlson (29) described a two-hybrid system that employs LexA and Gal4 baits.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: X-Gal, 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside; P, prey; Ba, bait; 5-FOA, 5-fluoroorotic acid.
To whom reprint requests should be addressed. e-mail: brent@ochre.mgh.harvard.edu.
References
- 1.Hartman P E, Roth J R. Adv Genet. 1973;17:1–105. doi: 10.1016/s0065-2660(08)60170-4. [DOI] [PubMed] [Google Scholar]
- 2.Botstein D, Maurer R. Annu Rev Genet. 1982;16:61–83. doi: 10.1146/annurev.ge.16.120182.000425. [DOI] [PubMed] [Google Scholar]
- 3.Herskowitz I. Nature (London) 1987;329:219–222. doi: 10.1038/329219a0. [DOI] [PubMed] [Google Scholar]
- 4.Hereford L M, Hartwell L H. J Mol Biol. 1974;84:445–461. doi: 10.1016/0022-2836(74)90451-3. [DOI] [PubMed] [Google Scholar]
- 5.Jarvik J, Bostein D. Proc Natl Acad Sci USA. 1973;70:2046–2050. doi: 10.1073/pnas.70.7.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hengartner M O, Horvitz H R. Curr Opin Gen Dev. 1994;4:581–586. doi: 10.1016/0959-437x(94)90076-f. [DOI] [PubMed] [Google Scholar]
- 7.Horvitz H R, Shaham S, Hengartner M O. Cold Spring Harbor Symp Quant Biol. 1994;59:377–385. doi: 10.1101/sqb.1994.059.01.042. [DOI] [PubMed] [Google Scholar]
- 8.Adams, M. D., Kerlavage, A. R., Fleischmann, R. D., Fuldner, R. A., Bult, C. J., et al. (1995) Nature (London) 337 (Suppl.), 3–17. [PubMed]
- 9.Lockhart D J, Dong H L, Byrne M C, Follettie M T, Gallo M V, Chee M S, Mittmann M, Wang C W, Kobayashi M, Horton H, Brown E L. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 10.Fields S, Song O. Nature (London) 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 11.Chien C T, Bartel P L, Sternglanz R, Fields S. Proc Natl Acad Sci USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gyuris J, Golemis E, Chertkov H, Brent R. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 13.Durfee T, Becherer K, Chen P L, Yeh S H, Yang Y, Kilburn A E, Lee W H, Elledge S J. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 14.Estojak J, Brent R, Golemis E A. Mol Cell Biol. 1995;15:5820–5829. doi: 10.1128/mcb.15.10.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gietz R D, Schiestl R H, Willems A R, Woods R A. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 16.Finley R L, Jr, Brent R. Proc Natl Acad Sci USA. 1994;91:12980–12984. doi: 10.1073/pnas.91.26.12980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Douarin B, Pierrat B, vom Baur E, Chambon P, Losson R. Nucleic Acids Res. 1995;23:876–878. doi: 10.1093/nar/23.5.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schneeweiss W G. Boolean Functions: With Engineering Applications and Computer Programs. Berlin: Springer; 1989. [Google Scholar]
- 19.Colas P, Cohen B, Jessen T, Grishina I, McCoy J, Brent R. Nature (London) 1996;380:548–550. doi: 10.1038/380548a0. [DOI] [PubMed] [Google Scholar]
- 20.Gossen M, Bujard H. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zervos A S, Gyuris J, Brent R. Cell. 1993;79:388. [PubMed] [Google Scholar]
- 22.Boguski M S, McCormick F. Nature (London) 1993;366:643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- 23.Pierrat B, Heery D M, Lemoine Y, Losson R. Gene. 1992;119:237–245. doi: 10.1016/0378-1119(92)90277-v. [DOI] [PubMed] [Google Scholar]
- 24.Shostak K, Christopherson R I, Jones M E. Anal Biochem. 1990;191:365–369. doi: 10.1016/0003-2697(90)90233-y. [DOI] [PubMed] [Google Scholar]
- 25.Boeke J D, LaCroute F, Fink G R. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 26.Bartel P L, Roecklein J A, SenGupta D, Fields S. Nat Genet. 1996;12:72–77. doi: 10.1038/ng0196-72. [DOI] [PubMed] [Google Scholar]
- 27.Finley R, Brent R. In: Two-Hybrid Systems: A Practical Approach. Bartel P, Fields S, editors. New York: Oxford Univ. Press; 1996. , in press. [Google Scholar]
- 28.Bray D. Nature (London) 1995;376:307–312. doi: 10.1038/376307a0. [DOI] [PubMed] [Google Scholar]
- 29.Jiang R, Carlson M. Genes Dev. 1996;10:3105–3115. doi: 10.1101/gad.10.24.3105. [DOI] [PubMed] [Google Scholar]