Abstract
The intestinal fatty acid binding protein (IFABP) is composed of two β-sheets with a large hydrophobic cavity into which ligands bind. After incorporating eight 4-19F-phenylalanines into the protein, the acid state of both apo- and holo-IFABP (at pH 2.8 and 2.3) was characterized by means of 1H-NMR diffusion measurements, circular dichroism and 19F-NMR. Diffusion measurements show a moderately increased hydrodynamic radius while near and far-UV CD measurements suggest that the acid state has substantial secondary structure as well as persistent tertiary interactions. At pH 2.8, these tertiary interactions have been further characterized by 19F-NMR, and show an NOE cross peak between residues that are located on different β-strands. Side chain conformational heterogeneity on the millisecond time scale was captured by phase sensitive 19F-19F NOESY. At pH 2.3, native NMR peaks are mostly gone but the protein can still bind fatty acid to form the holo-protein. An exchange cross peak of one phenylalanine in the holo-protein is attributed to increased motional freedom of the fatty acid backbone caused by the slight opening of the binding pocket at pH 2.8. In the acid environment Phe128 and Phe17 show dramatic line broadening and chemical shift changes, reflecting greater degrees of motion around these residues. We propose that there is a separation of specific regions of the protein that gives rise to the larger radius of hydration. Temperature and urea unfolding studies indicate that persistent hydrophobic clusters are native-like and may account for the ability of ligand to bind and induce native-like structure, even at pH 2.3.
The acid state of a protein is one in which all the ionizable groups have been protonated but the protein may still retain some structure. It is believed to have the characteristics of a molten globule in that it is less stable, has a radius of gyration larger than the native protein and shows considerable conformational heterogeneity. It was proposed over a decade ago that the molten globule state may be a general intermediate in protein folding (1, 2) and thus it is of interest to investigate the properties of the acid state of IFABP, one of the most thoroughly studied β-sheet proteins.
The usual NMR techniques used to study molten globule state focus on the backbone properties such as chemical shift perturbations and backbone dynamics (3). Such studies cannot sort out the crucial side chain interactions that provide stability of the molten globule state. The importance of side chains in stabilizing the molten globule state is at least two-fold. First, the packing itself of side chains is the driving force for the topology of molten globule state and second, the interaction among side chains could bring distant segments of the protein within range of other interactions, e.g., hydrogen bonds. The backbone dynamics revealed by usual 1H-15H-13C-NMR methods for the molten globule state usually provides information about the main chain on the ps-ns time scale. A full understanding of motions of internal mobility and dynamics on slower time scales, however, is essential for understanding protein function. The present study characterizes the acid state of a β-sheet protein and examines the internal mobility of a hydrophobic cluster by focusing on the response of side chains under acidic conditions using 19F-NMR, a method which is rarely used to characterize the molten globule state (4). 19F-NMR is unique in the ability to characterize side chains due to its much greater dispersion in chemical shift than 1H-15N-13C-NMR, which allows conformational heterogeneity on μs-ms time scale to be more easily characterized.
In our studies using 19F-NMR we have examined side chain behavior of IFABP under a variety of conditions and time scales (5-8) since side chain interactions play crucial roles in the overall tertiary structure (5, 9) and are critical in understanding the structure of β-sheet proteins.
IFABP contains 8 phenylalanine residues that are distributed on different secondary structural elements (Figure 1). We have previously assigned the 19F-NMR resonances for each phenylalanine residue (5) and examined the NMR spectrum as a function of urea (6). IFABP is an excellent model system for studies of the acid state since it is mostly β-sheet and may show different properties compared to other proteins that contain either α-helical or mixtures of α-helical and β-sheet structures. Such studies of β-sheet proteins should be particularly useful because α-helical structures are stabilized by local hydrogen bonds whereas β-sheet structures are stabilized by inter-strand hydrogen bonds between residues that are not close in sequence.
Fig.1.
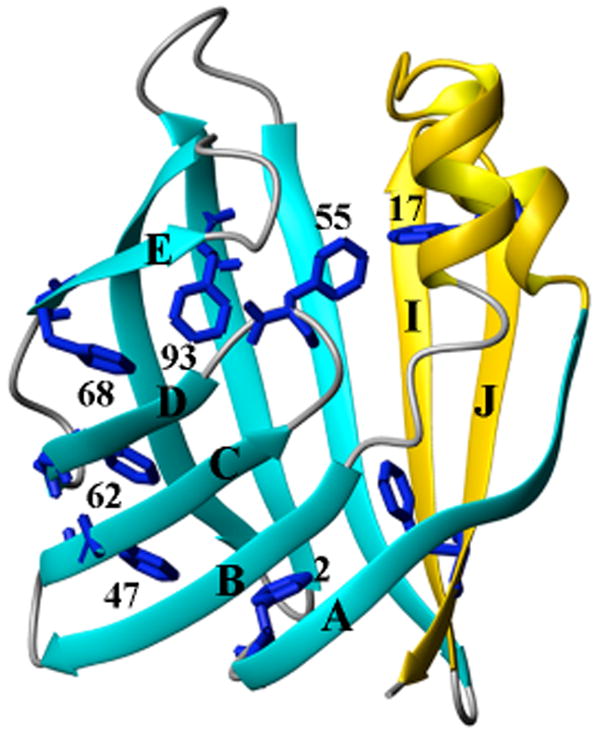
The crystal structure (PDB entry: 1IFB) of apo-IFABP (25). The 8 phenylalanines are shown in blue. The helical region and the last two β-strands are colored gold while the rest is colored cyan for β-strands and gray for turns. The diagram was prepared using MolMol (26)
Fatty acid binding to IFABP depends on the formation of a hydrophobic pocket, which is a network of hydrophobic side chains. Unlike the binding of ANS (1-anilino-8-napthalenesulphonate) to probe hydrophobic clusters (10) which usually provides relatively low resolution structural information and possible structural distortion, binding of its native ligand (fatty acid) to the acid state can give detailed information about the appearance of native-like topology using 19F-NMR.
In addition to examining the side chain behavior of these residues we also use 1H-NMR diffusion experiment to characterize the change of hydrodynamic radii during acid denaturation. Unlike such techniques as small angle X-ray scattering, the diffusion experiment can be carried out under the very similar solution conditions as are the 19F-NMR experiments, making data interpretation more straightforward.
Materials and Methods
Chemicals
Ultrapure urea was obtained from United States Biochemical. 4-19F-Phe was obtained from ACROS Organics (New Jersey). All other chemicals were reagent grade.
Protein production and purification
Incorporation of 4-19F-labeled phenylalanine into IFABP and subsequent purification has been described elsewhere (5).
Sample preparation for 19F-NMR
Holo-form stock IFABP was prepared as described elsewhere (5). Samples with a final protein concentration of 200 μM were prepared by mixing 2.4 mM of the protein stock solution (containing 20 mM potassium phosphate, 0.25 mM EDTA, pH 7.3 (NMR buffer)) with 20 mM citric acid and NMR buffer at different ratios to give final pH values of 7.3, 6.3, 5.5, 4.8, 3.9 and 2.8. Samples at pH 8.2 and 2.3 were made by diluting 2.4 mM protein stock solution into 20 mM K2HPO4 or 100 mM citric acid, respectively.
Sample preparation for pulsed field gradient (PFG) diffusion 1H-NMR
The stock protein solution (2.4 mM in NMR buffer), 20 mM and 100 mM citric acid and NMR buffer in H2O was lyophilized and redissolved in D2O. This was repeated three times and the protein then dissolved in 99.99% D2O. The sample at pH 7.3 was prepared by diluting stock protein solution in D2O into NMR buffer in D2O to a final protein concentration of 1 mM. The samples at pH 2.8 and pH 2.3 were prepared by mixing the protein stock solution in D2O with 20 mM and 100 mM citric acid, respectively, to give a final concentration of 200 μM. The volume for all above samples is 250 _l. To each sample, 2.5 μl of 100 mM dioxane solution in D2O was added to act as an internal viscosity standard. Shigemi NMR tubes with 1.5 cm sample height were used.
Circular Dichroism (CD) spectra
CD spectra were recorded at 20 °C on a Jasco-715 spectropolarimeter. The protein concentration was kept at 87 μM for both far- and near-UV measurements. Samples at different pH values were prepared from a 1.8 mM stock solution of wild type IFABP by mixing 20 mM potassium phosphate buffer (pH 7.3, containing 0.25 mM EDTA) and 20 mM citric acid in different ratios. The sample at the lowest pH value (pH ∼2.3) was prepared by mixing 1.8 mM stock IFABP solution with 100 mM citric acid to give a final concentration of 87 μM. The pH value of each sample was confirmed after each measurement. A 0.02 cm path length cuvette was used for the far-UV measurement between 260 nm and 180 nm. All CD spectrum were corrected for solvent background. At different pH values, the same sample was used for near-UV study between 310 nm and 250 nm using a 1.0 cm path length cell. Other parameters were same as those in far-UV measurements. No differences in spectra were observed 12-24 hours after the spectra were first recorded.
19F-NMR spectroscopy
19F-NMR spectroscopy measurements are essentially the same as described elsewhere (5). The chemical exchange rate was determined by performing phase-sensitive 2D-NOESY experiments at different mixing times from 50 ms to 200 ms (11, 12). The 2D data were analyzed by Pipp program (13).
PFG diffusion 1H-NMR spectroscopy
PFG diffusion measurements were made as described for urea denaturation of IFABP (6). The length of the diffusion gradient was optimized for each sample. For samples at pH 7.3 and 2.8, the length of the diffusion gradient was set to 3 ms and to 4 ms for the sample at pH 2.3. All other experimental parameters and data fitting protocols are the same as described earlier (5).
Results
Probing secondary and tertiary structure of the acid state using CD studies
CD measurements have been made as a function of pH and are shown in Figure 2. In Figure 2A the far-UV CD shows no difference in secondary structure between pH 4.1 and 8.0 and only a slight change at pH 2.8. Even at pH 2.3, the protein still retains some secondary structure. In the near-UV CD (Figure 2B), the regions (250∼270 nm) that are attributed to phenylalanine show some moderate changes between pH 8 and 2.8 but a dramatic change at pH 2.3. Signals (270∼300 nm), which are attributable to tyrosine and tryptophan, are similar below pH 4.1. Those data indicate significant tertiary structure changes below pH 4.1, consistent with 19F-NMR data in which dramatic chemical shift changes occur below pH 3.9 (see below).
Fig.2.
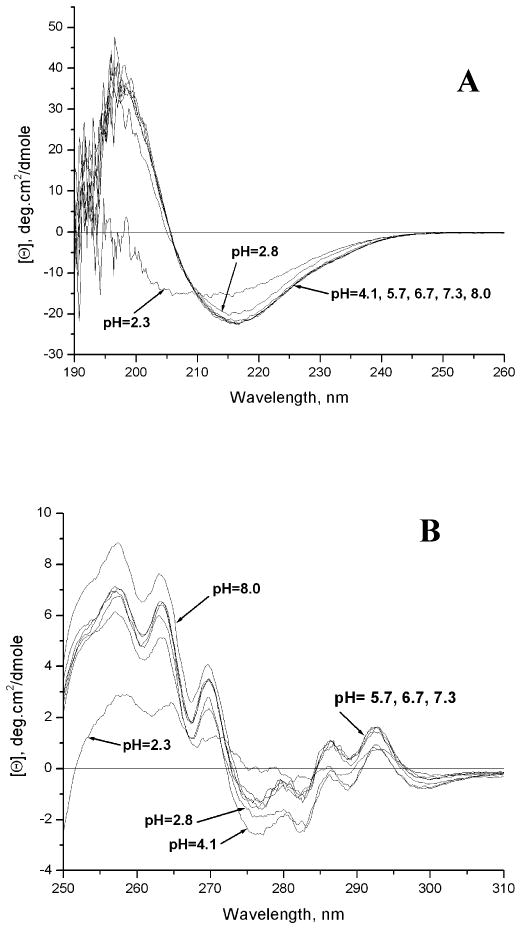
Far-UV (A) and near-UV (B) spectra of the IFABP at different pH values as described in the text.
19F-NMR Spectra of apo-IFABP As a Function pH
Figure 3A shows changes in the spectrum of apo-IFABP monitored by 1D-19F-NMR as a function of pH. IFABP remains structured at pH 2.8 as indicated by the dispersion of chemical shifts. There is extensive heterogeneity at pH 2.8 as ∼ 15 peaks appear for the eight phenylalanine residues. Below pH 3.9, at least two conformations in slow exchange become obvious as indicated by the appearance of minor peaks for Phe55, Phe62, Phe68 and Phe93, Phe128 (Figure 4). The peaks observed at pH 2.8 between −40 ppm and −41 ppm are not denatured peaks but multiple conformations of Phe2, Phe55 and Phe128 (Figure 4). At pH ∼2.3, the protein is mostly unfolded since most native peaks are either gone or substantially reduced. Even at pH 2.3, however, the unfolded IFABP is in slow equilibrium with a small amount of native-like structure as indicated by the resonances outside the denatured range (−40.3 ∼ -41.3 ppm). Figure 3B shows the dependence of chemical shift from pH 8 to 2.3 and indicate that, except for Phe2 (Figure 3B), the overall structure is barely perturbed from pH 8 to 5. At pH values lower than 5, however, the chemical shift changes are profound. The overall tendency of the chemical shift changes are similar, but not identical to, those observed with urea at concentrations below the urea denaturation midpoint as described elsewhere (6). The similarity in the chemical shift changes in low urea concentrations and pH values below 5 suggest that the two conditions yield similar structural changes.
Fig.3.
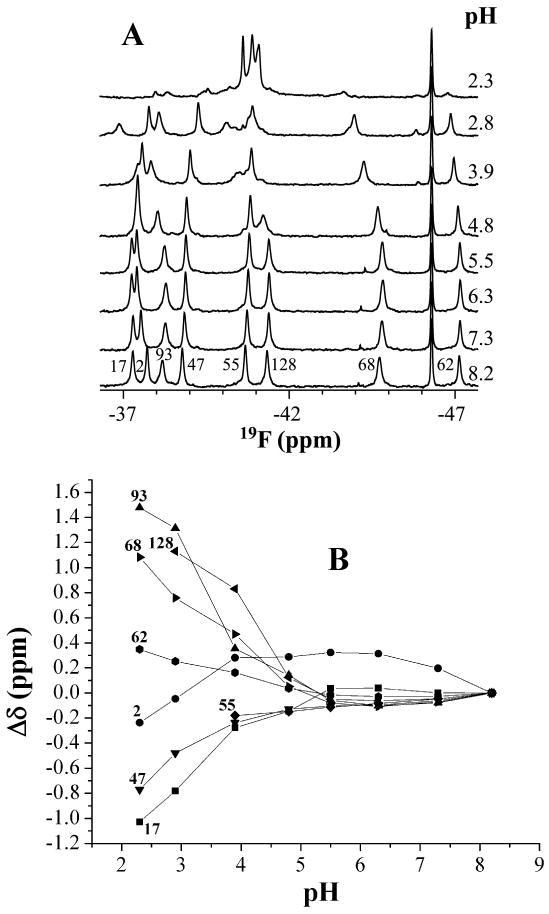
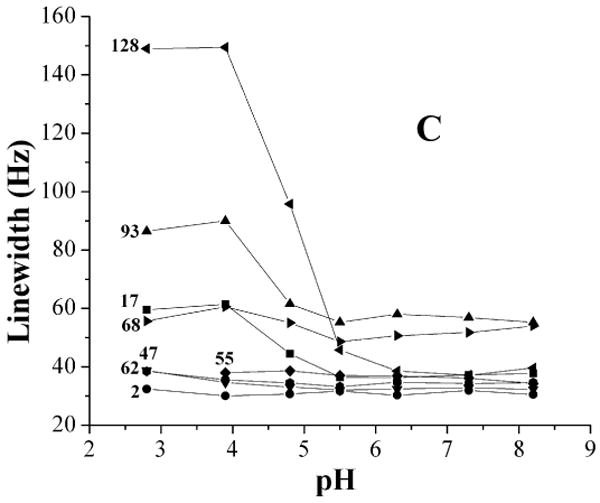
(A) 19F-NMR spectra of IFABP as a function of pH. The spectra were recorded at 20 °C with 200 μM protein. The samples with different pH were made as described in the text. Each spectrum was acquired with sixty-four scans and processed with 12 Hz exponential line broadening. The peak at -46.3 ppm is the reference peak of 6-19F-tyrptophan. (B) Chemical shift change of 19F-Phe-IFABP as a function of pH. At pH values above 3.9, an average chemical shift (broad peak) was used for Phe68 and Phe93 while the chemical shift of the major conformation was used below pH 3.9. (C) Line width change of 19F-Phe-IFABP as a function of pH.
Fig.4.
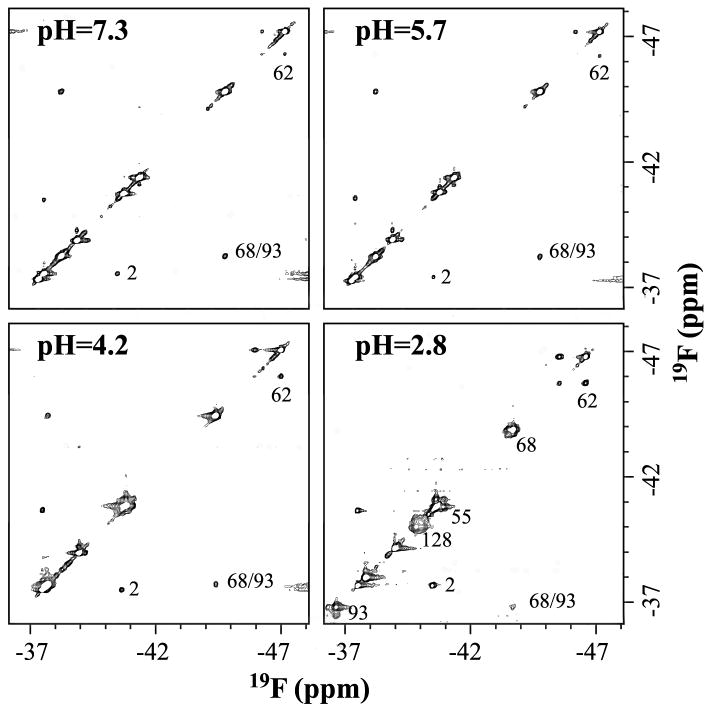
2D-NOESY spectra of 4-19F-Phe labeled IFABP at pH values of 7.3, 5.7, 4.2 and 2.8. The spectra were recorded at 20 °C with mixing time set to 250 ms and 100 points were used in F1 dimension. The spectra were reference to 6-19F-tryptophan at −46.3 ppm. The samples at different pH values were made as described in the text. The protein concentrations at pH values of 7.3, 5.7 and 4.2 are 1 mM and that at pH 2.8 is 200 μM.
Figure 3C shows the line width changes with pH. Phe128 (in the last β-strand) and Phe17 (in Helix 1) experience dramatic line broadening at pH below 4.8 and Phe68 and Phe93 (close to D-E turn) started to broaden at ∼ pH 3.9. It should be pointed out that all these line-broadenings are accompanied by the larger chemical shift changes (Fig. 3B). Other residues did not show significant line width changes in the pH range studied. These observations are consistent with previous studies (14, 15) showing that two regions appear to be involved in the early stage of urea folding, one is close to D-E turn, the other one is close to I-J turn and helical region (Figure 1).
Homonuclear 19F-NOE between 4-19F-Phe68 and 4-19F-Phe93 as a function of pH
Phe68 and Phe93 are part of a hydrophobic cluster and have been suggested to be involved in the early stages of folding. The distance between 4-19F-Phe68 and 4-19F-Phe93 was monitored at pH 7.3, 5.7, 4.2 and 2.8, respectively, by obtaining 2D-NOESY spectra (Figure 4). A Phe68/Phe93 cross peak was present at all pH values. The distance barely changes from pH 7.3 to pH 2.8 (Table 1). Essentially all of the native-like structure persists at those pH values as fully unfolded peaks at ∼-40.8 ppm barely grow (Figure 3A). The protein concentrations used at pH values 7.3, 5.7 and 4.2 were 1 mM while 200 μM sample was used at pH 2.8 due to aggregation with 1 mM protein.
Table 1. Relative distance change between 4-19F-Phe68 and 4-19F-Phe93 as a function of pHa.
The distance was determined by the relationship I ∝ 1/r6, where I is the intensity and r is the distance between atoms.
R0 is defined as the distance between the fluorine nuclei of Phe68 and Phe93 based on the crystal structure.
The distance is obtained with 200 μM protein sample at pH 2.8, while 1 mM protein was used for other pH values.
Chemical exchange rates determined by 19F-NMR for apo-IFAPB at pH 2.8
Conformational heterogeneity is a common property of both the molten globule state and acid state. Yet few methods can extract both dynamic (especially for slower time regime process) and thermodynamic (populations) parameters for those states. With 19F-NMR, conformational heterogeneity can be unambiguously characterized. As noted in the results for acid denaturation, the exchange phenomena among phenylalanine residues become more obvious at lower pH with a shift in the equilibrium between two different conformations. In the apo-form (Figure 4), not only is an NOE cross peak observed between Phe68 and Phe93, but also are chemical exchange peaks of Phe2, Phe55, Phe62, Phe68, Phe93 and Phe128. Their exchange rates (11) and population ratios are shown in Table 2. As shown in Table 2, six of the eight phenylalanines are in slow exchange in the apo-form, suggesting high mobility in the internal hydrophobic clusters. This appears to be contradictory to the intuitive view that hydrophobic core is “more rigid”. However, more studies are beginning to demonstrate that hydrophobic core can be very flexible with large amplitude motions (16) (Li & Frieden, unpublished results). The exchange rate of Phe2 and Phe128 are about half of the other phenylalanine residues that could be measured in the apo-protein. This indicates that the whole structure is still compact even at the ends of the protein.
Table 2. Population ratio and chemical exchange rates of the two conformations at pH 2.8 for both apo- and holo-IFABP. The ratio was determined by measuring area under each peak.
| Residue
number |
Apo-IFABP | Holo-IFABP | ||
|---|---|---|---|---|
| Ratioa | k (s-1) | Ratioa | k (s-1) | |
| 62 | 22/78 | 20.4±1.1b | 22/78 | 15.4±0.4 |
| 68 | 28/72 | 15.8±2.6 | ||
| 93 | 22/78 | 21.5±1.6 | 32/68 | 9.7±1.4 |
| 2 | 73/27 | 7.6±1.5 | 70/30 | 9.8±0.9 |
| 17 | 92/8 | 0.95±0.03 | ||
| 128 | 72/28 | 8.7±2.0 | ||
| 55 | 64/36 | 19.5±4.5 | ||
Downfield population/upfield population
Deviations from the average of 5 determinations
Chemical exchange rates determined by 19F-NMR for holo-IFAPB at pH 2.8
Similar to the apo-form, in the holo-form at pH 2.8 (Supplementary Material, Figure 1), Phe2, Phe62, Phe68 and Phe93 show exchange cross peaks. But the chemical shifts of the two conformations of Phe68 are too close to be able to correctly measure the intensities of diagonal and cross peaks in the holo-form. Therefore, the exchange rate of Phe68 of holo-form at pH 2.8 is not included in Table 2. In addition, both Phe62 and Phe68 have an extra cross peak with a very minor conformation, upfield of the major conformation. However, we cannot determine whether they are minor conformations in the holo-form or minor apo-form IFABP as their populations are negligible.
There is an exchange cross peak for Phe17 at pH 2.8 in the holo-form but not in the apo-form (Supplementary Material, Figure 1). We observed the exchange cross peak of Phe17 in the holo-form at pH 7.3 (5) with the population of the other conformation very minor (<1%). At pH 2.8, however, the other conformation of Phe17 increased to about 8%. Interestingly, the exchange rate of Phe17 is almost a magnitude slower than those of other residues (Table 2), those being on the same order of magnitude. The exchange of other residues reflects the overall conformational interconversion and may be closely related, while the exchange of Phe17 could be dictated by the backbone conformation of the fatty acid, since Phe17 has extensive contacts with the hydrocarbon tail (17). At low pH, the binding pocket might be slightly opened, giving more motional freedom to the tail of fatty acid and thus to Phe17 as it maintains extensive contact with the backbone of fatty acid.
PFG diffusion NMR studies
The apparent hydrodynamic radii of IFABP at different pH values are listed in Table 3. The radii are 17.2 Å and 23.7 Å at pH 2.8 and 2.3, respectively, which represent 11.0% and 52.8 % increases in hydrodynamic radius or 36.7% and 256.7% increase in hydrodynamic volume, respectively, compared to the native state (15.5 Å). Yet, both are smaller than that of fully denatured state (36.7 Å) in 7 M urea (6), suggesting that the acid state has the characteristics of a molten globule.
Table 3. hydrodynamic radii of IFABP at different pH values.
| pH | pH = 7.3 | pH = 2.8 | pH = 2.3 |
| Rh (Å) | 15.5 ± 0.4a | 17.2 ± 0.3 | 23.7 ± 0.5 |
Deviations from the average of 5 determinations
Probing the structure perturbation of binding pocket at low pH
Fig 5 shows the spectra of holo-IFABP (containing oleate) and apo-IFABP at low pH values. Apo-IFABP is substantially unfolded at pH 2.3 with only small amount of native-like structure existing (Figure 5). Holo-IFABP at pH 2.3 is more structured and its spectrum is superimposable with that at pH 2.8 except that a greater percentage of denatured peaks exists around −41 ppm. Not only are there dramatic changes in the chemical shifts of the holo form compared to the apo form it is clear that oleate stabilizes the protein at pH 2.3. At this pH, the NMR spectrum for holo-IFAPB is similar to that when oleate is added to apo-IFAPB (Figure 5).
Fig. 5.
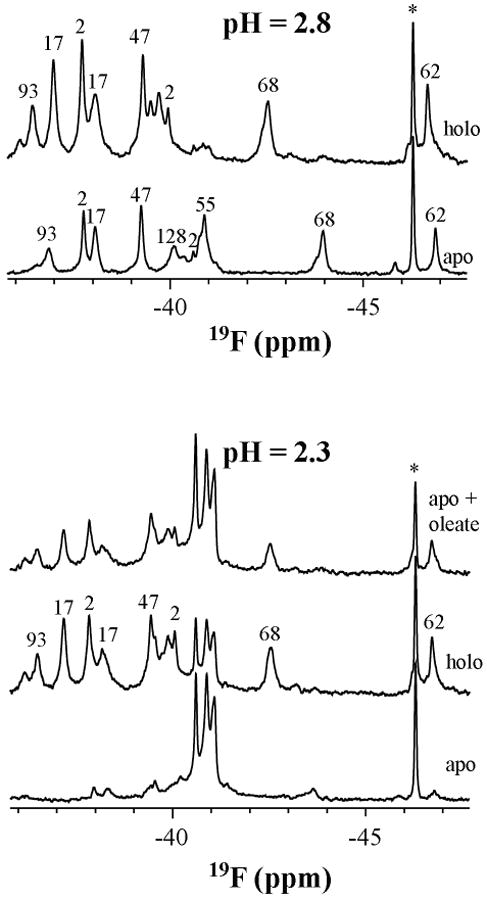
Spectra of apo-, and holo-IFABP and the stabilization of apo-IFABP in the presence of oleate at pH 2.3. The samples were made by mixing 2.6 mM 19F-Phe-IFABP (both apo and holo) in NMR buffer with 20 mM and 100 mM citric acid to give protein concentration of 200 uM and pH of 2.8 and 2.3, respectively. The sample for the upper spectrum at pH 2.3 was made by mixing the apo-IFABP with oleate heptane solution. The peak marked with star (*) is the reference 6-19F-tryptophan (−46.3 ppm relative to TFA).
Two other experiments used to probe structural perturbation at low pH are urea denaturation and temperature studies of apo-IFABP at pH 2.8. As is typical of other molten globule states, the protein at low pH is much less stable than the native protein (Supplementary Material, Figure 2) with a urea denaturation mid-point of about 0.9 M compared to 4.8 M for native protein (6). Interestingly, the chemical shift induced by urea are similar to those induced by urea at pH 7.3 (6).
The spectra of apo- IFABP at pH 2.8 were recorded at 3 °C, 7.5 °C, 11.5 °C, 15.5°C, 20 °C, 24 °C, 28 °C, 32 °C and 36 °C (Supplementary Material, Figure 3). Generally, the resonances of Phe62, Phe68 and Phe93 show the largest temperature coefficient (Δδ/ΔT) in both the native (5) and acid state. Phe2 shows negative temperature correlation in both native and acid state, but its temperature coefficient is more negative in the acid state. Combined with the observation that the distance between Phe68 and Phe93 maintained up to pH 2.8, these data collectively suggest that the acid state at pH 2.8 and native state are closely related in all regions involving hydrophobic clusters.
Discussion
Protonation of all the ionizable groups of a protein will certainly lead to loss of salt bridge interactions and indeed to repulsion of surface charges with a corresponding loss of stability. Dyson and Wright (18) postulate that the equilibrium unfolded state of apomyoglobin at pH 2.3 may serve as a model for events that might occur at the earliest stages of folding with regard to local hydrophobic collapse and sampling of native-like secondary structure. Dabora et al. had earlier examined the acid state of ribonuclease H and postulated that the acid form had the characteristics of a molten globule (19). In either case, characterization of the acid form of different proteins is therefore of interest. Both apomyoglobin and ribonuclease H contain considerable α-helical content. If the idea that the acid state approximates a molten globule, examination of a protein that is primarily β-sheet may give different information especially since the former studies examined backbone properties while the present study focuses on side chain behavior.
Nature of the acid state
Below pH 5, all the 19F-labeled phenylalanines exhibit apparent chemical shift changes with Phe128, Phe17, Phe68 and Phe93 showing line broadening as well. At least six out of the eight phenylalanines are in slow exchange. In addition, the near UV-CD spectra show changes to side chains of tyrosines and tryptophans while the far UV-CD indicates that secondary structure is retained as low as pH 2.8. Even at pH 2.3, some β-structure remains. These data indicate that, with decreasing pH, side chain changes precede backbone changes with increased internal mobility despite the fact that these side chains are buried in the hydrophobic clusters.
Table 3 shows that the hydrodynamic radius of IFABP increases as the pH is lowered. The structural units, however, as measured by far UV-CD, stay intact. The 19F-NMR data presented here may explain this increase. The line broadening and chemical shift changes of Phe17 and Phe128 as the pH is lowered indicate both increased mobility and solvent exposure. These results would be consistent with separation of two subsections (shown in gold and cyan in Figure 1) as a consequence of more solvent entering the interior cavity. Also consistent with this proposal are the increased motion of Phe17 in the holo-form and the observation that the distance between Phe68 and Phe93, which are located in the same subsection of the protein, does not change with decreasing pH. Further justification for this proposal comes from the results of Hodsdon and Frieden (14) who examined the HSQC spectra as a function of urea concentration. One observation made there was a concerted motion of the C, D and E strands and that this subsection of the protein was particularly susceptible to intermediate urea concentrations.
Implications from oleate binding at low pH
As stated in the Results, the acid state at pH 2.8 is closely related to the native state in terms of hydrophobic clusters, as suggested by their chemical shift dispersion, side chain response to temperature and urea denaturation and almost unchanged distance between Phe68 and Phe93. Furthermore, the ability of IFABP to bind oleate at low pH provides a straightforward method to monitor the existence of native-like topology. Lowering the pH of holo-IFABP to 2.8 results in a shift of many of the peaks compared to either the apo-protein at this pH (Figure 5) or the holo-protein at pH 7.3 (Supplementary Material, Figure 4), suggesting that the binding pocket remains intact.
Although fluorine chemical shifts in proteins are difficult to interpret in term of specific structure perturbation, they are more meaningful when the structure is known or when structures are related. We have shown elsewhere that in the native state the binding of oleate caused dramatic downfield chemical shift changes for almost all 19F-phenylalanines (except Phe2) in IFABP (5) without any apparent structural changes (17) due to exclusion of some of the ordered water molecules which impose shielding effect on side chains in the pocket. Binding of oleate at pH 2.8 (Figure 5) also causes downfield chemical shift changes, but to a lesser extent, for all 19F-labeled phenylalanines except Phe47. Among those residues, Phe68 shifted the most (∼ 1.7 ppm). In the native state, binding of oleate also causes Phe68 shifted the most (∼4.64 ppm) (5). It should be noted that in the native state, Phe68 has more surrounding water molecules than other phenylalnines. We can postulate that the binding pocket at pH 2.8 retains more or less ordered water molecules and that binding of oleate is also accompanied by the exclusion of some of ordered water molecules, resulting in deshielding effect on the side chains surrounding the oleate.
At pH 2.3, the addition of oleate to the apo-protein yields a spectrum similar to that observed on lowering the pH of the holo-protein (Figure 5). The persistent native-like structure at pH 2.3 in the apo-form probably binds to oleate and thus shifts the equilibrium between the native-like structure and unfolded state toward the native-like state. Oleate may also induce the formation of binding pocket directly from the unfolded state. Whatever the reason is, the pocket is formed through oleate interaction with multiple native-like subsets of side chain clusters that persist at pH 2.3.
Possible role of a molten globule-like state in transporting fatty acid
The molten globule state has been suggested to be important for biological functions in vivo including, for example, delivery of retinol by retinol binding protein to target cells (20). This hypothesis can be extended to the function of molten globule state of IFABP. It has been demonstrated recently that electrostatic interactions are a major determinant of the collisional mechanism of fatty acid transfer between IFABP and membranes (21). The high density of negatively charged groups at the membrane surface could create a strong electrostatic potential leading to local acidic pH (22-24). The local acidic pH could facilitate transfer of uncharged fatty acid with the positively charged protein interacting with negatively charged membrane, resulting in subsequent release of the ligand at a physiologically reasonable rate. We note that the chemical exchange rate of Phe17, which is thought to be controlled by oleate as discussed above, is on the same order of magnitude as the fatty acid transfer rate determined between IFABP and mimic membrane (21). The loosely packed and slow breathing of the hydrophobic pocket at low pH, as demonstrated by their exchange rate, could be responsible for ligand exchange. More evidence however, would be needed to establish convincing relationship between the molten globule state and the transfer of ligand.
In conclusion, 19F-NMR, combined with other techniques, has enabled us to characterize the acid unfolded molten globule like state of a β-sheet protein through the side chain analysis. The secondary structure of molten globule state is closely related to its native state, i.e., the acid state contains most of the β-structure for β-sheet protein (IFABP) while helical proteins such as apo Mb (18) retain helices. Thus, there is no common structure for the acid form of a protein. Rather, the structure is similar to that of the native protein. In the acid unfolded state of IFABP, side chain changes occur before any backbone changes. The same could be true for helical proteins based on side chain analysis. In contrast to general definition of molten globule state that it lacks fixed tertiary packing interactions, the acid unfolded state of IFABP contains subsets of native-like hydrophobic clusters, which could be intermediates resulting from hydrophobic collapse.
Supplementary Material
Supporting Information Available
Figure 1. 2-D NOESY spectrum at pH 2.8
Figure 2. 19F-NMR spectrum at pH 2.8 as a function of urea concentration.
Figure 3. 19F-NMR spectrum at pH 2.8 as a function of temperature.
Figure 4. 19F-NMR spectrum of holo-IFABP as a function of pH.
This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
We would like to thank Dr. Andre D'Avignon for discussions about pulse field gradient experiments and Mr. Robert Horton for technical assistance. We also thank Dr. Linda Kurz for critical reading of the manuscript and insightful suggestions.
The abbreviations used are
- IFABP
intestinal fatty acid binding protein
- CD
circular dichroism
- PFG
pulsed field gradient
- NOESY
Nuclear Overhauser Effect Spectroscopy
- apo
ligand free
- holo
ligand bound
Footnotes
Supported by NIH Grant DK13332.
References
- 1.Kuwajima K. The molten globule state as a clue for understanding the folding and cooperativity of globular-protein structure. Proteins. 1989;6:87–103. doi: 10.1002/prot.340060202. [DOI] [PubMed] [Google Scholar]
- 2.Ptitsyn OB, Pain RH, Semisotnov GV, Zerovnik E, Razgulyaev OI. Evidence for a molten globule state as a general intermediate in protein folding. FEBS Lett. 1990;262:20–24. doi: 10.1016/0014-5793(90)80143-7. [DOI] [PubMed] [Google Scholar]
- 3.Eliezer D, Yao J, Dyson HJ, Wright PE. Structural and dynamic characterization of partially folded states of apomyoglobin and implications for protein folding. Nat Struct Biol. 1998;5:148–155. doi: 10.1038/nsb0298-148. [DOI] [PubMed] [Google Scholar]
- 4.Bai P, Luo L, Peng Z. Side chain accessibility and dynamics in the molten globule state of alpha-lactalbumin: a (19)F-NMR study. Biochemistry. 2000;39:372–80. doi: 10.1021/bi992056f. [DOI] [PubMed] [Google Scholar]
- 5.Li H, Frieden C. NMR studies of 4-(19)f-phenylalanine-labeled intestinal Fatty Acid binding protein: evidence for conformational heterogeneity in the native state. Biochemistry. 2005;44:2369–2377. doi: 10.1021/bi047600l. [DOI] [PubMed] [Google Scholar]
- 6.Li H, Frieden C. Phenylalanine side chain behavior of the intestinal fatty acid-binding protein: the effect of urea on backbone and side chain stability. J Biol Chem. 2005;280:38556–38561. doi: 10.1074/jbc.M505435200. [DOI] [PubMed] [Google Scholar]
- 7.Ropson IJ, Frieden C. Dynamic NMR spectral analysis and protein folding: identification of a highly populated folding intermediate of rat intestinal fatty acid-binding protein by 19F NMR. Proc Natl Acad Sci U S A. 1992;89:7222–7226. doi: 10.1073/pnas.89.15.7222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ropson IJ, Frieden C. Intermediates in the folding of rat intestinal fatty-acid binding-protein (ifabp) that lack secondary structure. FASEB J. 1993;7:A1287–A1287. [Google Scholar]
- 9.Clark PL, Liu ZP, Rizo J, Gierasch LM. Cavity formation before stable hydrogen bonding in the folding of a beta-clam protein. Nat Struct Biol. 1997;4:883–886. doi: 10.1038/nsb1197-883. [DOI] [PubMed] [Google Scholar]
- 10.Arighi CN, Rossi JP, Delfino JM. Temperature-induced conformational transition of intestinal fatty acid binding protein enhancing ligand binding: a functional, spectroscopic, and molecular modeling study. Biochemistry. 1998;37:16802–16814. doi: 10.1021/bi981827x. [DOI] [PubMed] [Google Scholar]
- 11.Jeener J, Meier BH, Bachmann PL, Ernst RR. Investigation of exchange processes by two-dimensional NMR spectroscopy. J Chem Phys. 1979;71:4546–4553. [Google Scholar]
- 12.Perrin CL, Dwyer TJ. Application of two-dimensional NMR to kinetics of chemical exchange. Chem Rev. 1990;90:935–967. [Google Scholar]
- 13.Garrett DS, Powers R, Gronenborn AM, Clore GM. A common sense approach to peak picking two-, three- and four-dimensional spectra using automatic computer analysis of contour diagrams. J Magnet Reson. 1991;95:214–220. doi: 10.1016/j.jmr.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 14.Hodsdon ME, Frieden C. Intestinal fatty acid binding protein: the folding mechanism as determined by NMR studies. Biochemistry. 2001;40:732–742. doi: 10.1021/bi001518i. [DOI] [PubMed] [Google Scholar]
- 15.Kim K, Frieden C. Turn scanning by site-directed mutagenesis: application to the protein folding problem using the intestinal fatty acid binding protein. Protein Sci. 1998;7:1821–1828. doi: 10.1002/pro.5560070818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skalicky JJ, Mills JL, Sharma S, Szyperski T. Aromatic ring-flipping in supercooled water: implications for NMR-based structural biology of proteins. J Am Chem Soc. 2001;123:388–397. doi: 10.1021/ja003220l. [DOI] [PubMed] [Google Scholar]
- 17.Sacchettini JC, Gordon JI, Banaszak LJ. Crystal structure of rat intestinal fatty-acid-binding protein. Refinement and analysis of the Escherichia coli-derived protein with bound palmitate. J Mol Biol. 1989;208:327–339. doi: 10.1016/0022-2836(89)90392-6. [DOI] [PubMed] [Google Scholar]
- 18.Dyson HJ, Wright PE. Insights into the structure and dynamics of unfolded proteins from nuclear magnetic resonance. Adv Protein Chem. 2002;62:311–340. doi: 10.1016/s0065-3233(02)62012-1. [DOI] [PubMed] [Google Scholar]
- 19.Dabora JM, Pelton JG, Marqusee S. Structure of the acid state of Escherichia coli ribonuclease HI. Biochemistry. 1996;35:11951–11958. doi: 10.1021/bi9611671. [DOI] [PubMed] [Google Scholar]
- 20.Bychkova VE, Berni R, Rossi GL, Kutyshenko VP, Ptitsyn OB. Retinol-binding protein is in the molten globule state at low pH. Biochemistry. 1992;31:7566–7571. doi: 10.1021/bi00148a018. [DOI] [PubMed] [Google Scholar]
- 21.Corsico B, Franchini GR, Hsu KT, Storch J. Fatty acid transfer from intestinal fatty acid binding protein to membranes: electrostatic and hydrophobic interactions. J Lipid Res. 2005;46:1765–1772. doi: 10.1194/jlr.M500140-JLR200. [DOI] [PubMed] [Google Scholar]
- 22.Prats M, Teissie J, Tocanne JF. Lateral proton conduction at lipid–water interfaces and its implications for the chemiosmotic-coupling hypothesis. Nature. 1986;322:756–758. [Google Scholar]
- 23.Stegmann T, Doms RW, Helenius A. Protein-mediated membrane fusion. Annu Rev Biophys Biophys Chem. 1989;18:187–211. doi: 10.1146/annurev.bb.18.060189.001155. [DOI] [PubMed] [Google Scholar]
- 24.van der Goot FG, Gonzalez-Manas JM, Lakey JH, Pattus F. A ‘molten-globule’ membrane-insertion intermediate of the pore-forming domain of colicin A. Nature. 1991;354:408–410. doi: 10.1038/354408a0. [DOI] [PubMed] [Google Scholar]
- 25.Sacchettini JC, Scapin G, Gopaul D, Gordon JI. Refinement of the structure of escherichia-coli-derived rat intestinal fatty-acid binding-protein with bound oleate to 1.75-angstrom resolution - correlation with the structures of the apoprotein and the protein with bound palmitate. J Biol Chem. 1992;267:23534–23545. [PubMed] [Google Scholar]
- 26.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Available
Figure 1. 2-D NOESY spectrum at pH 2.8
Figure 2. 19F-NMR spectrum at pH 2.8 as a function of urea concentration.
Figure 3. 19F-NMR spectrum at pH 2.8 as a function of temperature.
Figure 4. 19F-NMR spectrum of holo-IFABP as a function of pH.
This material is available free of charge via the Internet at http://pubs.acs.org.


