Abstract
The exocyst is a protein complex required for the late stages of secretion in yeast. Unlike the SNAREs (SNAP receptors), important secretory proteins that are broadly distributed on the target membrane, the exocyst is specifically located at sites of vesicle fusion. We have isolated cDNAs encoding the rexo70, rsec5, and rsec15 subunits of the mammalian complex. The amino acid sequences encoded by these genes are between 21% and 24% identical to their yeast homologs. All three genes are broadly expressed and multiple transcripts are observed for rexo70 and rsec15. Characterization of cDNAs encoding the 84-kDa subunit of the mammalian complex revealed a novel protein. mAbs were generated to the mammalian rsec6 subunit of the exocyst complex. rsec6 immunoreactivity is found in a punctate distribution at terminals of PC12 cell processes at or near sites of granule exocytosis.
Intracellular vesicle trafficking pathways underlie organization of membrane compartments and secretion of extracellular messengers. The fidelity of membrane fusion is defined by a set of vesicle and target membrane proteins [SNAREs (SNAP receptors)] that form specific sets of complexes insuring fusion of the appropriate membranes. The best understood membrane fusion step is vesicle exocytosis, a component of which is mediated by the formation of a “core complex” consisting of the vesicle protein vesicle-associated membrane protein (VAMP, also called synaptobrevin) (Snc1p and Snc2p in yeast) and the plasma membrane proteins SNAP-25 (synaptosome-associated protein of 25 kDa) (Sec9p in yeast) and syntaxin1 (Sso1p and Sso2p in yeast). This SNARE complex is regulated by three families of soluble proteins: nsec1 (Sec1p in yeast); α, β, and γ-SNAP (γ-soluble NSF-attachment protein) (Sec17p in yeast); and the ATPase NSF (N-ethylmaleimide-sensitive factor) (Sec18p in yeast) (1–5). This relatively simple machinery is reiterated, through the use related SNAREs and SEC1 genes, at multiple steps within the secretory pathway. The precise role of the low molecular weight GTPase Rab proteins, also present at each stage of the secretory pathway, is less well understood.
This basic membrane recognition and fusion machinery is regulated to fit the needs of individual trafficking steps, specific differentiated cells, and particular physiological states. For example, exocytosis in the nerve terminal is tightly coupled to calcium concentration (6). Additionally, polarized cells such as neurons and epithelial cells must strictly govern the sites of fusion of vesicles with subregions of the target membrane defined by particular SNARE proteins. One set of proteins important in this process in yeast are the components of the exocyst complex (7). Six of these genes were defined as secretory mutants in yeast (SEC3, -5, -6, -8, -10, and -15) (8), and a seventh was characterized as a component of the complex (exo70) (9). These seven yeast genes are found in a stoichiometric complex with a molecular mass of 743 kDa. Whereas Sso1p, Sso2p, and Sec9p are present all along the plasma membrane, the exocyst complex exhibits a restricted localization at the distal end of the budding cell, the site of active vesicle fusion (7, 10). These data suggest that the exocyst complex is important in defining the site of vesicle fusion, possibly linking the machinery responsible for bud site selection and polarization to the secretory apparatus.
Mammalian homologs of the sec6 and sec8 subunits of the exocyst complex were characterized and shown to be constituents of a multisubunit 17S complex (11). Antibodies raised against these subunits were used to follow immunoreactivity through purification of the complex. Like its yeast counterpart, the mammalian exocyst complex is comprised of eight protein bands with a combined molecular mass of 734 kDa (12). Amino acid sequences from the eight protein bands were determined allowing the additional characterization of cDNAs encoding mammalian sec10 (9, 13). Database searches with the sequence information available from these three genes revealed no significant homologies to other characterized mammalian proteins. The mammalian complex is present in soluble and plasma membrane associated forms and is expressed throughout the nervous system and nonneural tissues as well. Although the complex is found in axons, immunocytochemical studies using antibodies directed against rsec8 did not reveal a highly restricted localization pattern as in yeast.
To more fully understand the structure and function of the mammalian exocyst complex, we have characterized three additional subunits from rat brain, those encoding rexo70, rsec5, and rsec15. These genes are expressed in all tissues examined. A fourth gene corresponding to the 84-kDa subunit is not similar to any previously characterized yeast subunit. mAbs raised against the rsec6 subunit of the complex reveal punctate and peripheral immunoreactivity in PC12 cells close to the sites of exocytosis.
MATERIALS AND METHODS
cDNA Characterization.
rexo70. A search of the human expressed sequence tag (EST) cDNA database from the Institute for Genomic Research (14) with amino acid sequences derived from the 79-kDa subunit of the mammalian exocyst complex revealed two overlapping ESTs (THC84610 and THC94542).
rsec15.
An EST clone (AA046774) was identified from a database search with amino acid sequences derived from the 96-kDa subunit.
rsec5.
PCR primers were generated based on an EST clone (AA161356) identified by searching cDNA database using amino acid sequences from the 102-kDa subunit.
exo84.
EST clone AA354462 served as a starting point for isolating overlapping clones that define an ORF. Overlapping sets of clones were isolated and characterized by using standard molecular biology techniques.
Northern Blot Analysis.
The mRNA blots of different rat tissues were purchased from CLONTECH and used according to manufacturer’s instructions.
mAb Production and Regional Western Blot Analysis.
mAbs against rsec6 were obtained from the corresponding hybridoma cell lines generated by fusion of NS-1 mouse myeloma cells with spleen cells from BALB/c mice immunized with His-tagged rsec6 fusion protein (15). For regional Western blot analysis, rat tissues were prepared as described (12). rsec6 protein bands were visualized by enhanced chemiluminescence (Amersham).
Fluorescence Microscopy.
Confocal differential interference contrast and immunofluorescence micrographs were obtained with a Zeiss Axiovert Microscope equipped with a laser scanning apparatus constructed by Stephen J Smith, Noam Ziv, and Tim Ryan (Stanford University, Stanford, CA). Optical sections were processed using Adobe photoshop software.
RESULTS
Mammalian rexo70, rsec15, rsec5, and exo84.
Characterization of rexo70, rsec15, and rsec5 from rat brain allows assignment of a total of six among the eight mammalian constituents of the exocyst complex to their yeast counterparts. These assignments are based on amino acid sequence similarities and include the proteins sec5, -6, -8, -10, -15, and exo70 (Figs. 1 and 2). In addition, several C. elegans homologs are available in the database and are included for comparison (Fig. 2).
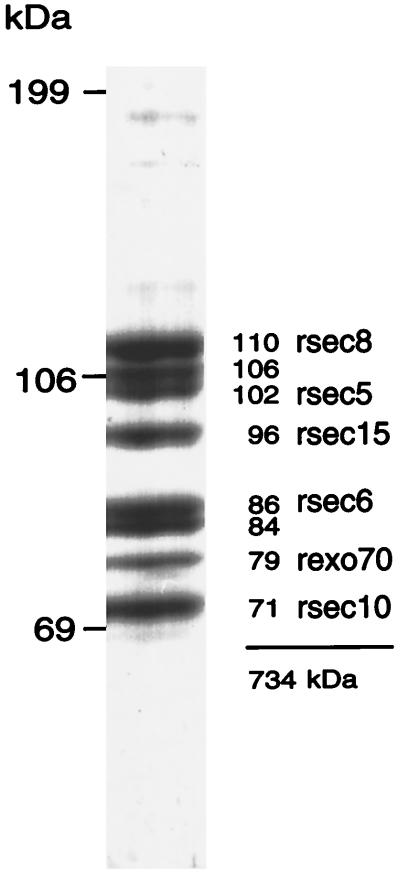
Figure 1.
Subunits of the mammalian exocyst complex. The purified mammalian exocyst complex is comprised of eight bands ranging in size from 71 to 110 kDa. Previous studies defined three subunits as rsec10, rsec6, and rsec8. In this report we characterize rexo70, rsec15, and rsec5 and a set of cDNAs that predicts a protein containing peptides derived from the 84-kDa band.
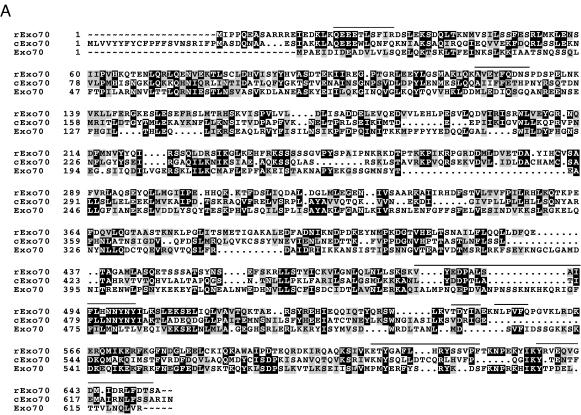
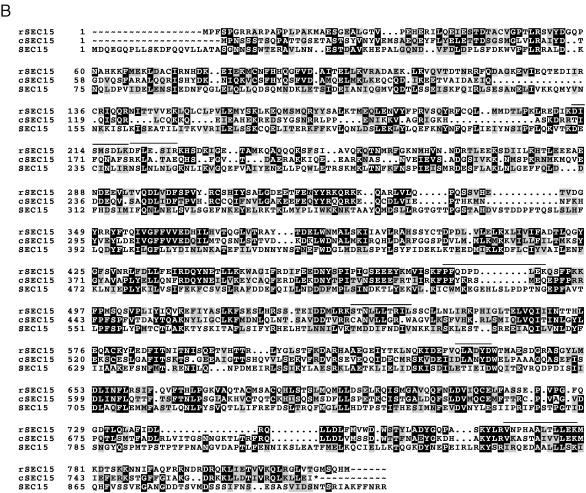
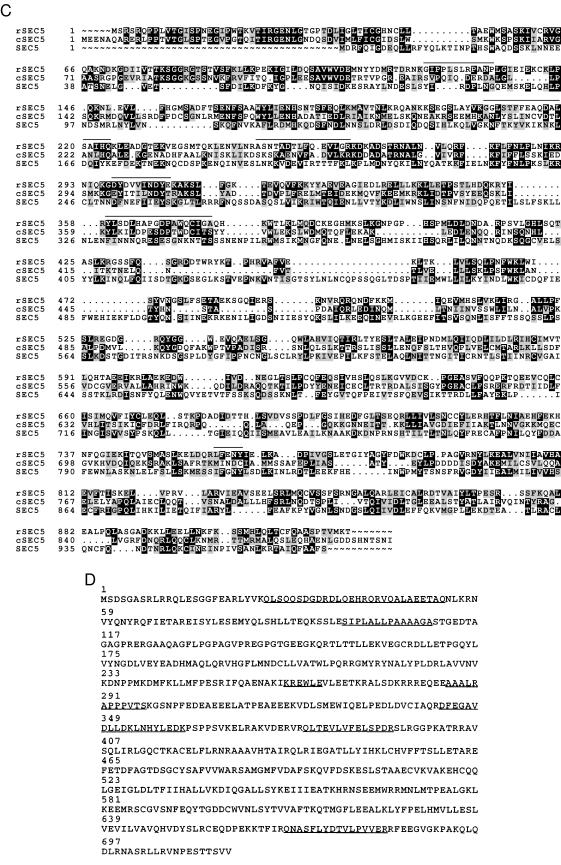
Figure 2.
Comparisons of the predicted amino acid sequences of mammalian exocyst subunits with respective Caenorhabditis elegans and yeast homologs. Predicted amino acid sequences of rexo70, rsec15, and rsec5 derived from rat cDNA analyses are compared with the homologous sequences in the database with the pileup program of GCG. Identical residues are in a black box with white letters and similar residues are shaded. Dotted regions represent gaps. Lines above the amino acid sequences indicate the peptides determined by amino acid sequencing of the subunits of the purified mammalian complex. (A) rexo70: Accession numbers of C. elegans and yeast homologs are U80437 and P19658. The smallest sum probability for the homology between the C. elegans and rat exo70 is 2.6 × 10−24. pileup analysis indicates that rat and C. elegans share 25% identity and 42% similarity, rat and yeast share 19% identity and 34% similarity, and C. elegans and yeast share 17% identity and 30% similarity. (B) rsec15: Accession numbers of C. elegans and yeast homologs are U41026 and Z72755, respectively. The smallest sum probability for the homology between the C. elegans and rat exo70 is 2.2 × 10−153. pileup analysis indicates that rat and C. elegans share 36% identity and 50% similarity, rat and yeast share 18% identity and 34% similarity, and C. elegans and yeast share 16% identity and 30% similarity. An asterisk (∗) at the end of the amino acid sequences of C. elegans homolog indicates the first possible stop codon in the genomic sequence. (C) rsec5: Accession numbers of C. elegans and yeast homologs are Z68319 and Z50046. The smallest sum probability for the homology between the C. elegans and rat exo70 is 3.2 × 10−61. pileup analysis indicates that rat and C. elegans share 28% identity and 46% similarity, rat and yeast share 18% identity and 31% similarity, and C. elegans and yeast share 13% identity and 27% similarity. (D) exo84: The 716 amino acid protein contains the sequences of seven peptides determined from amino acid sequencing of the 84-kDa band. The peptide sequences are underlined.
The rat exocyst 79-kDa subunit is homologous to yeast exo70 (Fig. 2A). rexo70 is encoded by an ORF that predicts a protein of 653 amino acids with a molecular mass of 75 kDa. The smallest sum probability for the homology of the rat and yeast exo70 sequences is 2.2 × 10−7 corresponding to 23% identity and 33% similarity with 17 gaps introduced into the sequence when the gap weight is set at 12 and length weight is set to 1 with the bestfit program of GCG (Wisconsin Package version 9.0). This homology is 26.5 SDs greater than the mean quality score attained by aligning 100 randomized rexo70 sequences. By using Coils 2.1 program, a region of 30 amino acids (residues 5–34) at the amino terminus of the protein is predicted to have a high probability (between 0.88 and 1.0) of forming a coiled-coil domain, a structure that may be involved in protein–protein interactions (16).
The rat exocyst 96-kDa subunit is a homolog of yeast sec15 (Fig. 2B). The cDNA encoding rsec15 predicts an 822-aa protein with a predicted molecular mass of 95 kDa. The smallest sum probability for the homology between the rat and yeast sec15 sequences is 1.3 × 10−7, and the bestfit program reveals 24% identity and 36% similarity with 26 gaps introduced into the sequence when the gap weight is set at 11 and length weight is set to 1. The quality score for this homology is 51 SDs greater than the mean quality score obtained after comparison to 100 randomized rsec15 sequences. A region of 22 amino acids (residues 85–106) at the amino terminus of the protein has a probability of >0.59 to form a coiled-coil domain.
Characterization of the rat 102-kDa subunit reveals its homology to sec5 (Fig. 2C). rsec5 is encoded by an ORF of 924 amino acids long with a predicted molecular mass of 104 kDa and an isoelectric point of 7.07. The smallest sum probability for the homology between the yeast and rat sec5 is 5.5 × 10−6 that is caused by the 21% identity and 32% similarity obtained after the introduction of 24 gaps in the sequences. The quality score for this homology is 16 SDs beyond the mean randomized quality score when the gap weight is set at 11 and length weight is set to 1. A region of 21 amino acids (residues 240–260) at the amino terminus of the protein has a high probability (>0.94) to form a coiled-coil domain. The three previously identified members of the mammalian exocyst complex, rsec6, rsec8, and rsec10, also contain regions predicted to form coiled-coil domains suggesting that many of the subunits in this complex may interact with each other via these domains (11, 13). Predicted coiled-coil domains are prominent in other membrane trafficking proteins including the SNAREs suggesting these interactions may be used for intercomplex interactions as well.
A set of cDNA clones was characterized that encodes seven peptide sequences derived from the 84-kDa subunit (Fig. 2D). The predicted protein has a molecular mass of 81 kDa and we tentatively name this protein exo84 because it corresponds to the 84-kDa protein in the complex. Like other proteins of the complex there are regions predicted to form coiled-coils, however, unlike the other proteins this sequence is not significantly similar to any of the previously characterized components of the yeast exocyst complex. A hypothetical 85.5-kDa protein in the yeast genome data base (GenBank accession no. Z35971) shows homology to the mammalian sequence and may be the functional equivalent. It will be interesting to see whether mutations in this yeast gene result in a defect in secretion similar to other subunits of the complex. Therefore, we hypothesize that exo84 is a subunit of the exocyst that had either (i) dissociated from the yeast complex during purification, (ii) escaped characterization in the yeast work, or (iii) is a subunit specific to the mammalian complex. Additional work is necessary to test this hypothesis and definitively prove that this protein is an authentic member of the complex.
Tissue Distribution of Mammalian rexo70, rsec15, and rsec5 Transcripts.
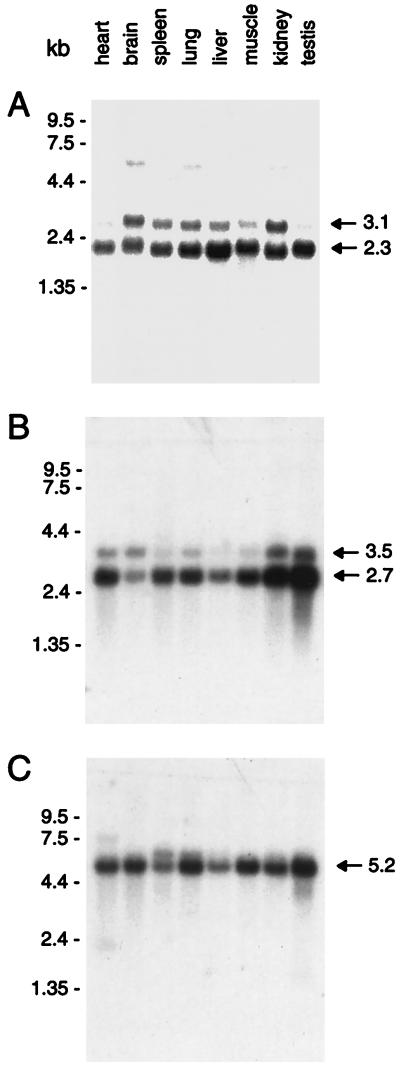
The distribution of transcripts encoding the mammalian rexo70, rsec15, and rsec5 was investigated by using mRNA blotting (Fig. 3). The larger transcript (3.1 kb) of rexo70 is expressed in a pattern similar to rsec8 being most abundant in kidney and brain. The smaller transcript of 2.3 kb is more evenly expressed with the most abundant level of mRNA found in liver. Although the relationship of the two transcripts to each other is not known, a broad tissue distribution is expected based on the wide functional role expected for this complex of proteins in exocytosis. The minor 5-kb RNA band observed in brain, lung, and kidney may represent an unspliced RNA. rsec15 also appears to be represented by two transcripts of 3.5 kb and 2.7 kb and, like rsec6, the transcript is more abundantly expressed in testis. rsec5 is somewhat more evenly expressed in tissues although testis again shows higher levels of expression. A single predominant mRNA of 5.2 kb encodes rsec5.
Figure 3.
Multiple tissue Northern blot analysis. Size markers are on the left in kb. (A) rexo70: Two transcripts of 2.3 and 3.1 kb are observed all tissues examined. (B) rsec15: Two transcripts of 2.7 and 3.5 kb are observed in all tissues. (C) rsec5: A major transcript of 5.2 kb is observed in all tissues.
mAbs Specific for rsec6 Reveal Its Tissue Distribution and Subcellular Localization.
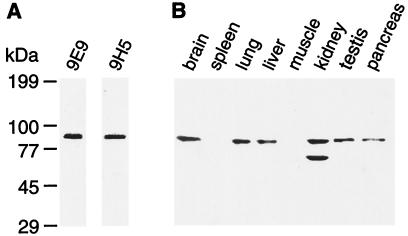
To better understand the distribution and function of the exocyst complex we raised mAbs against the recombinant rsec6 subunit. The specificity of two of these antibodies, 9E9 and 9H5, is demonstrated by using Western blot analysis in whole brain tissue (Fig. 4A). These antibodies recognize a single band in brain tissue and the single rsec6 subunit of the purified complex. mAb 9H5 was then used to investigate the protein distribution in multiple tissues (Fig. 4B). The distribution is similar to that of the rsec6 mRNA previously determined, with the most abundant protein level in kidney followed by brain (11). Although we find a transcript in muscle the protein is not detected. A lower molecular weight immunoreactive band is also observed in kidney and is most likely caused by alternative RNA splicing that generates a smaller protein, or to some protein degradation. The rsec6 protein profile is similar to that observed for rsec8 including the observation of a lower molecular weight rsec8 related protein in kidney.
Figure 4.
Tissue distribution of rsec6 protein. (A) Western blot detection of rsec6 in brain postnuclear supernatant by anti-rsec6 mAbs 9E9 and 9H5. (B) Regional Western blot analysis of rsec6 using anti-rsec6 mAb 9H5. Samples (20 μg) of postnuclear supernatant proteins from the indicated tissues were loaded per lane.
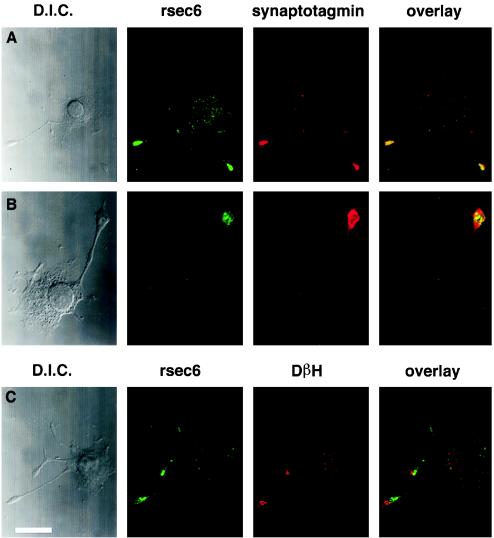
The subcellular distribution of exocyst immunoreactivity was visualized in PC12 cells. In cells that have been differentiated with nerve growth factor for 2 days rsec6 immunoreactivity is enriched in processes that contain synaptotagmin immunoreactivity (Fig. 5 A and B). However, higher magnification micrographs demonstrate a punctate localization distinct from vesicles. In the cell soma, a less intense punctate staining is also observed in many cells (Fig. 5A). Sites of exocytosis can be observed by depolarizing PC12 cells and staining the extracellular membrane for dopamine β-hydroxylase (DβH) (see Materials and Methods), an enzyme stored within the vesicle that is exposed following membrane fusion (Fig. 5C). As shown in Fig. 5C DβH staining does not precisely match the rsec6 immunoreactivity but instead appears to be juxtaposed to the sites of exocytosis. It is likely that the membrane of the vesicles is not enriched in rsec6, therefore fusion of this membrane to the surface may generate an area of staining lacking rsec6 immunoreactivity. Clearly, the vesicle fusion sites are adjacent to the area enriched in the exocyst complex.
Figure 5.
rsec6 colocalizes with vesicles and sites of exocytosis in nerve growth factor-differentiated PC12 cells. (A and B) Comparison of the distribution of rsec6 and the synaptic vesicle marker, synaptotagmin, in PC12 cells that have been differentiated with nerve growth factor for 2 days demonstrated by double immunofluorescence. The antibodies used are listed above their respective fluorescence micrographs. Differential interference contrast micrographs of each cell are shown on the left. A shows that rsec6 immunofluorescence is more intense in the vesicle-rich terminals of growing neurites with lighter labeling in the cell body. (B) A higher magnification view reveals the punctate distribution of rsec6 in the neurite terminal. (C) rsec6 is also found at sites of Ca2+-regulated secretion as demonstrated by the presence of DβH on the cell surface. Antibodies directed against DβH were applied in the absence of detergent to reveal cell-surface sites of recent exocytosis in response to K+/Ca2+-induced depolarization. Subsequent application of antibodies to rsec6 in the presence of detergent reveal the distribution of internal rsec6. The overlay micrograph demonstrates the contrast between intracellular rsec6 localization and cell-surface DβH distribution. Bar (in C) = 7.8 μm (A), 3.3 μm (B), 6.8 μm (C).
DISCUSSION
In this report we present the predicted amino acid sequences of the 79-, 96-, and 102-kDa subunits of the mammalian exocyst complex that correspond to the yeast exo70, sec15, and sec5 genes, respectively. Each of the predicted sequences contains multiple peptide sequences determined from amino acid sequencing of subunits of the purified complex (see Fig. 2). Because the mammalian peptide sequences match amino acid sequences predicted from the cDNAs, we are certain that these cDNA clones encode authentic components of the complex. The C. elegans genome project has progressed to the point where homologs of the three genes are now present in the database. Although the C. elegans proteins are also likely to be present in a similar complex to the exocyst complex we cannot rule out the possibility that the predicted C. elegans sequences represent related genes with other functions. The relatively low homologies of these genes between species suggests that the complex is likely to have a structural function rather than an enzymatic activity. We have also characterized a fourth set of cDNAs defining a new gene, exo84, and additional studies are in progress to test the hypothesis that this protein is a member of the exocyst complex. This gene is also broadly expressed, consistent with the expression patterns of the other members of the complex.
The mRNA expression patterns for the subunits are similar but not identical. For example rsec8 mRNA appears most abundant in kidney, whereas rsec6 mRNA appears most abundant in brain and testis (11). rsec15 and rsec5 are also abundantly expressed in testis but are more evenly expressed in other tissues than rsec8. We find this surprising for a set of genes that are present at a 1:1 stoichiometery in a complex (12). Perhaps multiple genes encode related subunits and the overall expression levels will be similar when summed over the members of the gene family. It is important to keep in mind that the relative protein levels of the different subunits of the mammalian exocyst complex in different tissues remains to be determined. Thus it is not yet clear that mRNA levels are precisely reflective of the relative protein levels. Furthermore, the expression of different size mRNAs may be a reflection of alternative RNA splicing that could add to the complexity of components of the exocyst. If the exocyst subunits are present in a variety of isoforms the seemingly simple complex may actually be very heterogeneous and the functional significance of this structural heterogeneity will need to be further studied. It is also possible that each cell type has a specific subunit assembly process requiring unique levels of component proteins.
The higher levels of rsec5, rsec6, and rsec15 expression in testis is interesting in light of the expression of a rsec6 related gene in testis and sperm. rsec6 is 24% identical to a mammalian protein termed B94, a gene that is highly expressed during periods of embryonic development of numerous tissues but tapers off after birth (17). Cells of such developing tissues require high levels of secretion both for growth and division as well as membrane addition permitting differentiation. Thus, an alternative form of rsec6 may play a role in the constitutive secretion underlying development. Intriguingly, however, a functional role for B94 is suggested by experiments focusing on sperm, a mature cell with high levels of expression. Fluorescence microscopy reveals that B94 is localized to the acrosome, a Golgi-derived compartment analogous to an exocytic granule. B94 immunoreactivity is observed on or near the acrosome membrane suggesting a role in the fusion of the acrosomal and plasma membrane lipid bilayers. sec6 may have diverged early in evolution to negotiate such diverse membrane fusion events. It remains to be determined whether such mammalian homologs of the other exocyst components exist or if the sec6 component, perhaps being substitutable, imparts a diversity of functions on the exocyst.
The antibodies raised against bacterial recombinant rsec6 are monospecific as demonstrated by Western blot analysis. The immunoreactivity is highly localized toward the terminals of PC12 processes in a similar location to synaptotagmin. Moreover, rsec6 immunoreactivity is juxtaposed to exocytotic sites as judged by the surface staining of DβH, an antigen contained in the lumen of the synaptic vesicle. However, while rsec6 immunoreactivity is punctate, it is not exactly colocalized with vesicles as demonstrated by the higher magnification micrograph (Fig. 5B). This result was anticipated by previous biochemical studies demonstrating that the exocyst is not associated with vesicles (12). Because there is no complex similar to the exocyst known to be required for other membrane trafficking steps in the secretory pathway it is possible that the complex is not a critical element of the basic membrane fusion apparatus, but instead plays a role in the specialized steps leading up to exocytosis. Future experiments will be directed at determining the interactions of the exocyst puncta with the cytoskeleton, the vesicle, and/or the plasma membrane. Although the enrichment of rsec6 at sites of vesicle localization and exocytosis suggests a role for the mammalian exocyst complex in one or more forms of secretion, definitive functional studies in higher organisms are necessary.
Acknowledgments
We thank Cindy Adams and Dr. Stephen J Smith for help obtaining confocal images and Jason Bock for help with computers.
ABBREVIATIONS
- SNARE
SNAP receptor
- DβH
dopamine β-hydroxylase
Footnotes
References
- 1.Söllner T, Whiteheart S W, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman J E. Nature (London) 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- 2.Scheller R H. Neuron. 1995;14:893–7. doi: 10.1016/0896-6273(95)90328-3. [DOI] [PubMed] [Google Scholar]
- 3.Südhof T C. Nature (London) 1995;375:645–53. doi: 10.1038/375645a0. [DOI] [PubMed] [Google Scholar]
- 4.Pevsner J, Hsu S-C, Braun J E A, Calakos N, Ting A E, Bennett M K, Scheller R H. Neuron. 1994;13:353–361. doi: 10.1016/0896-6273(94)90352-2. [DOI] [PubMed] [Google Scholar]
- 5.Söllner T, Bennett M K, Whiteheart S W, Scheller R H, Rothman J E. Cell. 1993b;75:409–418. doi: 10.1016/0092-8674(93)90376-2. [DOI] [PubMed] [Google Scholar]
- 6.Katz B. The Release of Neurotransmitter Substances. Liverpool: Liverpool Univ. Press; 1969. [Google Scholar]
- 7.TerBush D R, Novick P. J Cell Biol. 1995;130:299–312. doi: 10.1083/jcb.130.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Novick P, Ferro S, Scheckman R. Cell. 1981;25:461–469. doi: 10.1016/0092-8674(81)90064-7. [DOI] [PubMed] [Google Scholar]
- 9.TerBush D R, Maurice T, Roth D, Novick P. EMBO J. 1996;15:6483–6494. [PMC free article] [PubMed] [Google Scholar]
- 10.Finger F P, Novick P. Mol Biol Cell. 1997;8:647–662. doi: 10.1091/mbc.8.4.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ting, A. E., Hazuka, C. D., Hsu, S.-C., Kirk, M. D., Bean, A. J. & Scheller, R. H. (1995) Proc. Natl. Acad. Sci. USA 92. [DOI] [PMC free article] [PubMed]
- 12.Hsu S C, Ting A E, Hazuka C D, Davanger S, Kenny J W, Kee Y, Scheller R H. Neuron. 1996;17:1209–1219. doi: 10.1016/s0896-6273(00)80251-2. [DOI] [PubMed] [Google Scholar]
- 13.Hazuka C D, Hsu S C, Scheller R H. Gene. 1997;187:67–73. doi: 10.1016/s0378-1119(96)00720-2. [DOI] [PubMed] [Google Scholar]
- 14.Adams M D, Kelley J M, Gocayne J D, Dubnick M, Polymeropoulos M H, Xiao H, Merril C R, Wu A, Olde B, Moreno R F, Kerlavage A R, McCombie W R, Venter J C. Science. 1991;252:1651–1656. doi: 10.1126/science.2047873. [DOI] [PubMed] [Google Scholar]
- 15.Lane R D, Crissman R S, Grinn S. Methods Enzymol. 1986;121:183–192. doi: 10.1016/0076-6879(86)21017-4. [DOI] [PubMed] [Google Scholar]
- 16.Lupas A. Methods Enzymol. 1996;266:513–524. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- 17.Wolf F W, Sarma V, Seldin M, Drake S, Suchard S J, Shao H, O’Shea K S, Dixit V M. J Biol Chem. 1994;269:3633–3640. [PubMed] [Google Scholar]