Abstract
Despite the widely accepted view that transcription of gid and mioC is required for efficient initiation of cloned oriC, we show that these transcriptions have very little effect on initiation of chromosome replication at wild-type chromosomal oriC. Furthermore, neither gid nor mioC transcription is required in cells deficient in the histone-like proteins Fis or IHF. However, oriC that is sufficiently impaired for initiation by deletion of DnaA box R4 requires transcription of at least one of these genes. We conclude that transcription of mioC and especially gid is needed to activate oriC only under suboptimal conditions. We suggest that either the rifampicin-sensitive step of initiation is some other transcription occurring from promoter(s) within oriC, or the original inference of transcriptional activation derived from the rifampicin experiments is incorrect.
Keywords: DNA replication, oriC, transcriptional activation
Initiation of replication of the Escherichia coli chromosome occurs at a unique site, oriC. Initiation depends primarily on DnaA protein, which binds to five 9-bp repeats within oriC and facilitates duplex melting at three A+T-rich 13-mers (see Fig. 1). DnaB helicase subsequently enters the opened duplex and leads to priming and chain elongation reactions resulting in bidirectional replication. oriC is a complex regulatory region containing recognition sites for many positive and negative regulatory proteins, including Fis, IHF, SeqA, and IciA, some or all of which are thought to help precisely regulate initiations occurring at oriC (see ref. 1 for review).
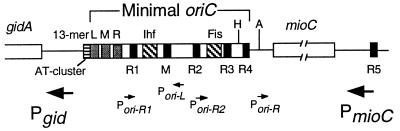
Figure 1.
The minimal oriC and surrounding transcription. The position of the six DnaA boxes R1–R5 and M; 13-mer repeats L, M, and R; A+T-rich cluster; and binding sites for IHF and Fis proteins are indicated. Large arrows represent location and direction of major promoters near oriC. Small arrows represent weaker promoters within oriC. H, HindIII (+244); A, AccI (+285) (ref. 1 and references therein).
Early physiology experiments by Lark (2) and Messer (3) first suggested that RNA polymerase (RNAP) is somehow involved in initiation at oriC. This was inferred from findings that rifampicin (an inhibitor of RNAP) inhibits a new round of DNA synthesis at a time when protein synthesis is no longer required. Genetic evidence as well suggests a role of RNAP in initiation. Mutations in rpoB and rpoC, which encode the β and β′ subunits of RNAP, respectively, have been shown to increase copy numbers of both the chromosome and chimeric plasmids (oriC plasmids) carrying both an oriC site and a ColE1-type replication origin (4, 5). Further, specific rpoB mutations have been shown to suppress the temperature sensitivity phenotype of certain dnaA(Ts) mutations (6). Despite the evidence suggesting an involvement of RNAP, a specific transcription event required for initiation has not been identified.
Of promoters possibly involved in transcriptional activation of initiation, one likely candidate is the promoter of gidAB. The gid promoter (Pgid) is situated just counterclockwise of oriC (Fig. 1) and transcription proceeds leftward away from oriC. oriC plasmids in which the gid promoter has been inactivated exhibit decreased transformation efficiency and stability as well as decreased replication in vitro (7, 8). The twin-domain model of Liu and Wang (9) predicts that an actively transcribing RNAP generates local domains of increased negative supercoiling behind it and positive supercoiling in front of it. It has been postulated that transcription from Pgid facilitates duplex opening in the 13-mer region by increasing the local negative supercoiling. This idea is supported by the findings that gid or kan transcription stimulates initiation of an oriC plasmid only when transcription is oriented away from oriC (7) and that Plac transcription entering oriC is inhibitory (10). In an alternative model, helix destabilization may be propagated from an R-loop formed in the vicinity of oriC, to the 13-mer region, thus activating initiation (11, 12). Also implicated in initiation control is transcription originating from the mioC promoter (PmioC). mioC is located clockwise of oriC (Fig. 1), and transcription proceeds leftward either reading through or occasionally terminating within oriC (13). Mutation of the mioC promoter decreases copy number and stability of minichromosomes (14, 15).
A remarkable feature of PmioC and Pgid is their periodicity within the cell cycle (16, 17). It was found that just prior to initiation gid transcription peaks and mioC transcription is shut off, whereas just after initiation mioC inhibition is relieved and gid transcription declines. These findings support the idea that gid transcription is activating and mioC transcription is inhibitory. On the other hand, it is not clear whether these promoters are regulated to control initiation or as a consequence of initiation.
Until recently, all knowledge of the cis requirements for oriC initiation has been based almost exclusively on information obtained from cloned oriC, using either minichromosomes or oriC plasmids. However, recent findings have made it obvious that conditions on the chromosome are not matched by plasmids, and therefore, cloned oriC sites do not represent reliable models of chromosomal initiation. For instance, while inactivation of the mioC promoter on a minichromosome has been shown to result in a dramatic decrease in replication activity, this same mutation, when placed on the E. coli chromosome, had little or no effect on initiation (18). We recently found that the deletion of DnaA box R4 (Fig. 1), which completely blocks initiation of an oriC plasmid, is tolerated on the chromosome (19). To examine the effects of oriC modifications on the chromosome, we have developed a genetic system by which oriC modifications are systematically transferred from an oriC plasmid onto the E. coli chromosome (19). Using this system, we have investigated the contributions of gid and mioC transcription to initiation in their native location. We also examined the effects of gid and mioC transcription in cells that have been compromised for initiation by mutation of the genes encoding Fis and IHF proteins, or by deletion of the DnaA box R4. We show that disruption of transcription from Pgid and PmioC has a very modest effect on initiation, even in the absence of Fis or IHF protein. In the absence of DnaA box R4, however, the presence of at least one of these transcriptions becomes essential for growth.
MATERIALS AND METHODS
Media and Growth Conditions.
Cells were grown at 37°C, with aeration by shaking, in LB medium (20) supplemented with 0.1% glucose except for rnhA mutants, which were grown in CAA medium (21). Antibiotics were added at the following concentrations: ampicillin, 40 μg/ml; chloramphenicol, 50 μg/ml; tetracycline, 20 μg/ml; and kanamycin, 55 μg/ml.
E. coli strains and plasmids.
Strains are listed in Table 1. All oriC modified strains were constructed by using an in vivo replacement system previously described (19). Briefly, mutated oriC sites were transferred from an oriC plasmid onto the chromosome of a specialized λ transducing phage (λ10.2) by a double-crossover event, then from the phage onto the AQ7664 chromosome by a second double-crossover event. The exchange of wild-type oriC with the oriC mutations was confirmed by Southern blot hybridization (21) for all clones. For flow cytometric analysis, oriC modifications (and asnA101::cat) were transferred from AQ7664 into AQ9555 by phage P1 transduction, by selecting for chloramphenicol resistance and screening for kanamycin sensitivity. Modified oriC configurations on the chromosome were reconfirmed by Southern blot hybridization for all transductants. Promoter mutant derivatives of fis and himA mutant strains were created by moving fis::kan and himA::tet alleles into promoter mutant strains by P1 transduction.
Table 1.
E. coli strains
| Strain | Genotype | Ref. and/or source |
|---|---|---|
| AQ634 | F−ilv metB his-29 pro trp9605 thyA deoB (or -C) | 26 |
| AQ2178 | AQ634 polA1 | 26 |
| AQ3529 | supF (λcI857 Sam7) | Lab collection |
| AQ7626 | supF (λcI857 Sam7/λ10.2) | Lab collection |
| AQ7664 | AQ634 gidA95::kan | 19 |
| AQ7996 | fis::kan | As WM2016 (27) |
| AQ7998 | himA::tet | As WM2017 |
| AQ9552 | Wild type | As MG1655 (28) |
| AQ9555 | AQ9552 gidA95::kan | 19 |
| AQ9648 | AQ9552 asnA101::cat | This work |
| AQ9652 | AQ9552 Pgid-103 asnA101::cat | This work |
| AQ10033 | AQ634 rnhA224 gidA95::kan | 19 |
| AQ10293 | AQ9552 PmioC112 asnA101::cat | This work |
| AQ10614 | AQ9552 Pgid-103 PmioC112 asnA101::cat | This work |
| AQ11296 | AQ10033 Pgid-103 PmioC112 asnA101::cat | This work |
| AQ11297 | AQ10033 oriC207::bla Pgid-103 PmioC112 asnA101::cat | This work |
All oriC plasmids used are derivatives of pDB101 (Fig. 2A) (19). pDB103 carries a mutation (Pgid-103) in the −10 region of the gid promoter (Fig. 2B) that was constructed by digestion with HinfI, brief digestion with mung bean nuclease, and religation (7). pDB109 was constructed by deletion of the BglII(+38)–AccI(+285) oriC fragment of pDB101, and insertion of a bla gene into the deleted region (19). pDB112, which carries PmioC112, an EcoRV(+722)–MluI(+777) deletion in the mioC promoter (Fig. 2B), was constructed by replacing the HindIII(+244)–BglII(+2006) fragment of pDB101 with the same from pALO47 (15). pDB113 carries both gid and mioC promoter mutations and was constructed by replacing the SmaI(−47)–XhoI(+417) fragment of pDB112 with the SmaI(−47)–XhoI(+417) fragment from pDB103. pDB123 contains deletions in the open reading frames of gidA and mioC resulting in inactive gene products, without affecting promoter activity. To create the 305-bp mioC deletion (ΔmioC121), pDB101 was digested with XhoI and EcoRV, filled in with Klenow enzyme, and religated to form pDB121. pDB121 was then digested with SnaBI and SacII, blunt-ended, and religated to create the 253-bp gidA deletion (ΔgidA122), resulting in pDB123. The DnaA box R4 deletion (oriC226) was constructed by deleting the oriC domain between the HindIII (+244) and AccI (+285) sites, followed by Klenow enzyme treatment and religation, just like the construction of the previously reported R4 deletion (oriC207::bla) (19) except for omission of the insertion of a bla gene in the deleted region. pRNH-Km is a derivative of pHK (22) carrying a lacZ′–′rnhA fusion which encodes a fusion protein that has near normal RNase HI activity (23). Construction of pLacIq, which contains a lacIq gene fragment, was previously described (23).
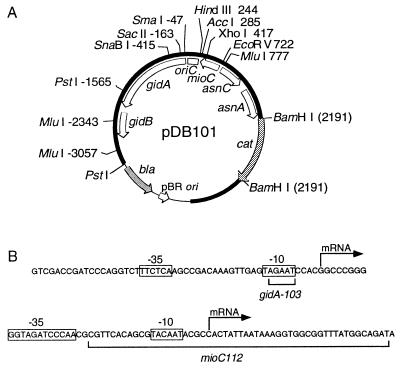
Figure 2.
Structure of oriC plasmid and promoter sequences. (A) The plasmid (pDB101) from which other oriC plasmids are derived is shown with relative restriction sites and genetic map (open arcs). Thick (solid) and thin arcs denote the oriC region and vector sequences, respectively. Shaded arcs denote antibiotic markers. All oriC coordinates are according to refs. 29 and 30. (B) DNA sequences of the gid (7) and mioC (14) promoter regions with promoter mutations are shown. The −10 and −35 consensus homologies and transcriptional start sites are indicated.
oriC Plasmid Copy Number Determination.
AQ2178 cells harboring each oriC plasmid were grown to approximately 2 × 108 cells per ml in LB + glucose in the presence of chloramphenicol (50 μg/ml). A 5-ml sample of the culture was collected and total DNA was extracted as previously described (21). The DNA (≈9 μg) was digested with PstI, electrophoresed in a 0.8% agarose gel, blotted onto a nylon filter, and hybridized with a [32P]dCTP-labeled probe consisting of a 714-bp oriC region fragment from MluI(−3057) to MluI(−2343). The blot was then exposed to a PhosphorImager screen (Molecular Dynamics) and the relative band intensities were quantified.
Flow Cytometry.
Cell preparation and flow cytometry with an Argus flow cytometer (Skatron, Lier, Norway) were performed essentially as previously described (24, 25).
RESULTS
Effects of Promoter Mutations on Cloned oriC.
Promoter deletion mutations (Fig. 2B) were introduced into pDB101 (Fig. 2A), a derivative of pBR322 carrying a 6.6-kb oriC fragment. gid and mioC promoter mutations have been reported to reduce the frequency of replication initiated from cloned oriC (7, 8, 14, 15). It is known, however, that the origin activity of cloned oriC is affected by various factors such as the type of cloning vectors, the cloning sites, the orientation of oriC with respect to the vector, and the size of cloned fragments (refs. 1, 18, and 19 and references therein). We therefore reexamined the effects of promoter mutations on our plasmids that contain relatively long chromosomal sequences at both sides of oriC. The examination took advantage of the fact that the replication origin of pBR322, but not oriC, requires the polA gene product (DNA polymerase I) for initiation. Thus, derivatives of pDB101 can transform and be maintained in polA mutant cells when they contain active oriC.
We created deletions in the promoter regions of gid and mioC (Fig. 2B), and the inactivation of promoter activities was confirmed by a greater than 500-fold drop in β-galactosidase activity expressed from gidA–lacZ and mioC–lacZ fusion plasmids in which the promoters were mutated. As shown in Table 2, pDB112 carrying the mioC promoter mutation (PmioC112) alone exhibited no detectable decrease in the efficiency of transformation of polA mutant cells compared with pDB101. The copy number of pDB112 in the mutant cells, however, was reduced from that of pDB101 (Fig. 3). pDB103 carrying the gid promoter mutation (Pgid-103) alone, on the other hand, showed a decreased transformation efficiency, and the copy number of this plasmid was further reduced from that of pDB112. Simultaneous inactivation of both promoters (pDB113) had an even greater effect than Pgid-103 alone; only a fraction of cloned oriC initiated plasmid replication in polA mutant cells. These results are in a general agreement with a current view of the importance of these transcriptions on cloned oriC: (i) Cloned oriC requires gid transcription for efficient initiation; and (ii) mioC transcription, although less significant in the presence of gid transcription, does compensate for the absence of it. The residual oriC activity observed with pDB113 may depend on other transcriptions detected within and/or near oriC (Fig. 1).
Table 2.
Transformation frequencies of polA1 cells with oriC plasmids
| Plasmid | mioC | gid | Ratio of transformants (polA−/polA+) relative to pDB101 |
|---|---|---|---|
| pDB101 | + | + | 1.00 |
| pDB112 | 112 | + | 1.09 ± 0.12 |
| pDB103 | + | 103 | 0.32 ± 0.02 |
| pDB113 | 112 | 103 | 0.08 ± 0.01 |
| pDB123 | ▵ | ▵ | 0.83 ± 0.09 |
+, Wild type; ▵, gene deletion mutant.
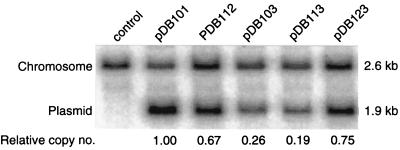
Figure 3.
Copy numbers of oriC plasmids carrying promoter mutations. polA1 cells (AQ2178) were transformed with the indicated modified oriC plasmids and grown in the presence of chloramphenicol (50 μg/ml). Total DNA was extracted from exponentially growing cells and digested with PstI and electrophoresed in a 0.8% agarose gel. DNA was blotted, and probed with a 714-bp MluI(−2343) to MluI(−3057) 32P-labeled oriC DNA fragment. Band intensities were quantified by using a PhosphorImager, and the plasmid-to-chromosome ratio relative to pDB101 is indicated under each lane. For relevant genotypes of plasmids, see Table 2. Control lane is untransformed AQ2178.
To examine the unlikely possibility that the initiation defects of the promoter mutant plasmids were caused by reduced levels of gid and mioC gene product (neither of which have any known function), we constructed a gene deletion mutant plasmid (pDB123) carrying large deletions in the open reading frames of both gidA and mioC, leaving the promoter regions intact. This plasmid had a transformation efficiency (Table 2) and copy number (Fig. 3) severalfold higher than pDB113, suggesting that cloned oriC requires gid and mioC transcription per se, for efficient replication, rather than their gene products. The slightly decreased replication of pDB123 compared with the wild-type plasmid (pDB101) may be due to the decreased distance between oriC and the mioC promoter in the gene deletion mutant plasmid.
Promoter Mutations Have Little Effect on Initiation at Chromosomal oriC.
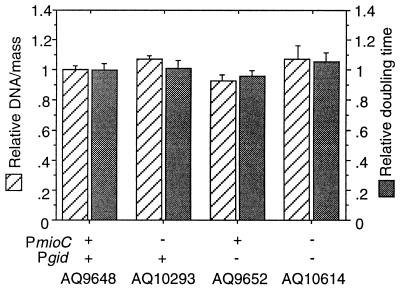
Using a genetic system previously described (19), we replaced the wild-type promoters on the chromosome with modified ones from the oriC plasmids described above. Growth characteristics of the resulting promoter mutants were then examined by flow cytometry. Analysis of exponentially growing cells showed that the DNA/mass ratio and doubling times of the promoter mutant cells were comparable to those of wild type (Fig. 4), suggesting that initiation from chromosomal oriC is not affected by either promoter mutation. Notably, the absence of gid transcription appears to cause a slight decrease in DNA/mass, whereas inhibition of mioC transcription may slightly increase DNA content. However, while these divergences are generally repeatable, they are within the range of experimental error and may be insignificant. As mentioned, the cell cycle-specific regulation of gid and mioC transcription suggests that these transcriptions affect the precise timing of initiation. Flow cytometry revealed that inhibition of gid and/or mioC transcription resulted in no detectable loss of the high degree of initiation synchrony exhibited by wild-type cells (data not shown). We conclude that these transcription defects have no effect on initiation, at least under otherwise optimal conditions for initiation.
Figure 4.
DNA synthesis and growth rates of promoter mutants. Cells were grown in LB plus glucose at 37°C with aeration. DNA/mass values were obtained from flow cytometric analysis of exponentially growing cells. Doubling times were determined from cell counts with a particle counter. Values are averages of three independent analyses plotted relative to AQ9648 (18.8-min doubling time). Error bars indicate the standard deviation.
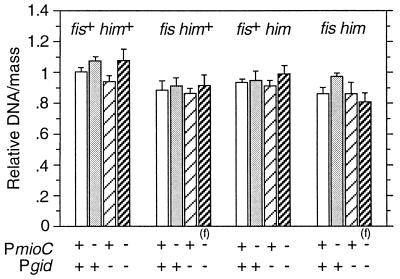
fis or himA Mutations Do Not Enhance the Effect of the Promoter Mutations on Chromosomal Initiation.
For replication of cloned oriC, gid and mioC transcriptions become essential under nonoptimal conditions such as lack of histone-like proteins (11, 31). We introduced gid and mioC promoter mutations onto the chromosome of fis::kan and himA::tet mutant cells, which are deficient in Fis and IHF, respectively, and the resulting mutants were examined for growth characteristics by flow cytometry. Surprisingly, fis::kan and himA::tet mutations had little effect on requirements of chromosomal oriC for gid or mioC transcription. DNA/mass ratios (Fig. 5) were not significantly affected by either gid or mioC promoter mutations. Note that the slight effects on DNA synthesis caused by inhibition of mioC and gid transcription (Fig. 4) are generally reflected in fis and himA mutants. Exacerbation of the severe asynchrony phenotype of fis and himA mutants (32) was not detected when the promoter mutations were introduced (data not shown). Interestingly, cell size was significantly increased in fis::kan cells carrying either Pgid-103 or PmioC112 mutation, and cells with all three mutations exhibited extreme filamentation (data not shown).
Figure 5.
DNA synthesis of fis::km himA::tet cells carrying promoter mutations. Cells were grown and DNA/mass values were obtained as in Fig. 4. An (f) indicates that these cells exhibit extreme filamentation during exponential growth. Values shown are averages of three independent analyses. Error bars indicate the standard deviation.
Transcription from Either gid or mioC Is Essential for Initiation with a Suboptimal oriC Sequence.
Previously we showed that cells carrying a deletion of DnaA box R4 on the chromosome are viable (19). These cells replicate inefficiently as indicated by decreased DNA/mass and presence of the asynchrony phenotype. We considered that local transcription might be required to activate this truncated oriC. To test this hypothesis we created oriC plasmids carrying a deletion of DnaA box R4 (oriC207::bla) combined with either or both Pgid-103 and PmioC112. Transfer of the R4 mutated oriC combined with a single promoter mutation onto the chromosome was successful. Cells carrying a Pgid promoter mutation exhibited severely retarded growth (48.6-min doubling time) as indicated by a more than 2-fold increase in doubling time compared with oriC207::bla alone (23.2 min). The absence of mioC transcription had a much smaller effect on the ΔR4 mutant (32.4-min doubling time).
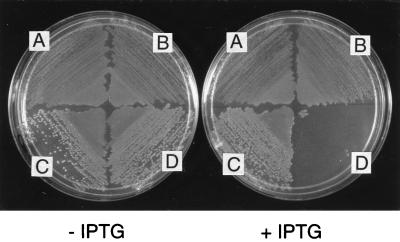
Several attempts to transfer the triple mutation oriC207::bla Pgid-103 PmioC112 from oriC plasmid onto the chromosome were fruitless, suggesting that E. coli could not accommodate such an extensive oriC modification. In rnhA224 mutant cells (AQ10033), an alternative replication system, constitutive stable DNA replication (cSDR) is activated and thus oriC defects are suppressed (33). Using this strain, we successfully transferred the triple mutant oriC onto the chromosome. We could transfer the mutant oriC into an rnhA224 mutant by P1 transduction but not into wild-type cells, further confirming the lethality of the triple mutant. To verify the dependence of the growth of the resulting rnhA224 oriC207::bla Pgid-103 PmioC112 mutant (AQ11297) on cSDR, the mutant strain was transformed with pRNH-Km, which produces an active RNase HI protein from a lac promoter. The transformants did not grow in the presence of 5 mM isopropyl β-d-thiogalactoside (IPTG), whereas the vector plasmid (pHK) had no effect on cell growth (Fig. 6). Furthermore, a DnaA box R4+ counterpart (AQ11296) was not sensitive to IPTG. We conclude that the oriC triple mutant is inviable in the presence of RNase HI. In addition, the OriC− phenotype of this strain could not be rescued by introduction of a plasmid (pDB109) carrying wild-type mioC and gid genes (data not shown). This suggests that the R4 deletion mutant requires an activating transcription event per se, rather than gid or mioC gene product.
Figure 6.
Lethal effect of expression of an rnhA+ gene on rnhA224 PmioC112 oriC207::bla Pgid-103 mutant. Cells were transformed with pRNH-Km (carries lacZ::rnhA gene fusion) or pHK (vector) and selected for kanamycin resistance. Transformants were then transformed with pLacIq, selecting for tetracycline resistance. Equal amounts of cells were spread on minimal plates with or without 5 mM IPTG as indicated. (A) rnhA224 PmioC112 Pgid-103/pHK, pLacIq. (B) rnhA224 PmioC112 Pgid-103/pRNH-Km, pLacIq. (C) rnhA224 oriC207::bla PmioC112 Pgid-103/pHK, pLacIq. (D) rnhA224 oriC207::bla PmioC112 Pgid-103/pRNH-Km, pLacIq.
To rule out the possibility that any transcription reading through the bla terminators (transcription is oriented leftward into oriC) is responsible for the oriC lethality of the oriC207::bla Pgid-103 PmioC112 mutant, we constructed an identical R4 deletion mutant except without bla gene insertion. When combined with both gid and mioC promoter mutations, this construct strictly required the presence of an rnhA224 mutation for growth (data not shown).
DISCUSSION
Much evidence exists suggesting a role of gid and mioC transcription in the initiation of replication. oriC plasmids and minichromosomes have been shown to be very sensitive to mutations in these promoters. In concordance with previous findings, our data indicate that an oriC plasmid requires transcription from both promoters for efficient replication. Transcription occurring from Pgid was more important, although a plasmid carrying mutations in both promoters (pDB113) was still able to replicate; initiating at about 8–19% efficiency compared with a wild-type plasmid (Table 2, Fig. 3). In spite of the fact that promoter inactivation had a significant effect on replication of an oriC plasmid, we found that initiation at chromosomal oriC did not require transcription from either promoter. Blocking these transcriptions had almost no detectable effect on the rate of growth or DNA synthesis (Fig. 4). This is in agreement with the recent finding that inactivation of the mioC promoter has no effect on initiation of chromosome replication (18). Findings that oriC plasmids and minichromosomes are much more sensitive to promoter mutations is likely a reflection of the small size of plasmids relative to the chromosome and hence a decreased ability to balance superhelical changes over a much shorter domain.
Transcription from mioC was suggested to regulate the timing of initiations by inhibiting additional oriC firings after negative factors such as sequestration have become exhausted (16, 17). Inconsistent with this suggestion, the present and previous (18) data show no defects in initiation synchrony of PmioC mutants. Our data are in agreement with recent findings that replication initiation in synchronized dnaC2(Ts) mutants is unaffected by the absence of mioC transcription (34). In addition, we found that inhibition of gid transcription had no effect on initiation synchrony. As suggested, the periodicity of these transcriptions within the cell cycle may be a consequence of initiation, rather than a regulator of it (34).
We also found that inhibition of mioC and gid transcription does not affect growth in the absence of the two histone-like proteins Fis and IHF. Whereas DNA synthesis was slightly lower in fis and himA mutants, inactivation of mioC and gid transcription had little or no additional effect (Fig. 5). This is surprising, given that a minichromosome carrying an inactive mioC promoter cannot replicate in the absence of IHF protein (31). In contrast, transcription from either Pgid or PmioC became obligatory when the right-most DnaA box, R4, was deleted (Fig. 6). This is, to our knowledge, the first observed case of transcriptional activation of chromosomal oriC. The DnaA box ΔR4 mutant requires transcription from at least one of the promoters, possibly due to a reduced ability to melt the 13-mer region when fewer DnaA-binding sites are available. This hypothesis is consistent with findings that overexpression of DnaA protein results in significant replication in the presence of rifampicin (35, 36). We propose that gidA and mioC transcription becomes essential only when oriC is under suboptimal conditions. Consistent with this proposal, replication of an oriC template in vitro also requires RNAP, but only under conditions that make unwinding of the origin difficult (e.g., extreme concentrations of HU protein, reduced negative superhelicity, or reduced temperature) (11, 12). Furthermore, phage λ DNA replication in vitro has been found to require transcription when HU protein is present (37). More recent studies indicate that transcription initiated either upstream or downstream of oriλ can activate initiation of λ DNA replication, most likely by enhancing localized negative supercoiling in the A+T-rich region of the origin (S.-H. Chung and R. McMacken, personal communication).
Our findings, however, do not readily explain the sensitivity of wild-type cells to rifampicin (2, 3). gid and mioC transcription represents the bulk of RNAP activity around oriC, but the possibility still exists that one or more of the several other promoters detected within or in the close vicinity of the minimal oriC (Fig. 1) may be responsible for transcriptional activation of oriC. Having eliminated the two prime suspects of transcription representing the rifampicin sensitive step in initiation, we should now begin to seriously reexamine the original inference of transcriptional activation. In an in vitro initiation system, either RNAP or DnaG primase (which is insensitive to rifampicin) is capable of activating replication. However, DNA synthesis in both systems is sensitive to rifampicin, suggesting a second unknown capacity of rifampicin (38). It is known that incubation of cells with rifampicin results in a decrease in the sedimentation rate of nucleoids, suggesting a decrease in the overall extent of supercoiling of the nucleoids (39, 40). We suggest the possibility that rifampicin, which shuts down all transcriptions on the chromosome, brings about a drastic global change in the nucleoid structure, severely altering the topological structure of oriC. In other words, the rifampicin effect may be nonspecific, and not due to the inhibition of a particular promoter such as Pgid or PmioC.
Acknowledgments
We are grateful to W. Messer and C. Gross for the gifts of plasmids and E. coli strains. This work was supported by Grant GM22092 from the National Institutes of Health and by Grant BIR-9218818 from the National Science Foundation to T.K. and by a grant from the Norwegian Cancer Society to E.B.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: RNAP, RNA polymerase; IPTG, isopropyl β-d-thiogalactoside.
References
- 1.Messer W, Weigel C. In: Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Neidhardt F C, Curtiss R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1579–1601. [Google Scholar]
- 2.Lark K G. J Mol Biol. 1972;64:47–60. doi: 10.1016/0022-2836(72)90320-8. [DOI] [PubMed] [Google Scholar]
- 3.Messer W. J Bacteriol. 1972;112:7–12. doi: 10.1128/jb.112.1.7-12.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rasmussen K V, Atlung T, Kerszman G, Hansen G E, Hansen F G. J Bacteriol. 1983;154:443–451. doi: 10.1128/jb.154.1.443-451.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanaka M, Ohmori H, Hiraga S. Mol Gen Genet. 1983;192:51–60. doi: 10.1007/BF00327646. [DOI] [PubMed] [Google Scholar]
- 6.Atlung T. Mol Gen Genet. 1984;197:125–128. doi: 10.1007/BF00327932. [DOI] [PubMed] [Google Scholar]
- 7.Asai T, Takanami M, Imai M. EMBO J. 1990;9:4065–4072. doi: 10.1002/j.1460-2075.1990.tb07628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogawa T, Okazaki T. Mol Gen Genet. 1991;230:193–200. doi: 10.1007/BF00290668. [DOI] [PubMed] [Google Scholar]
- 9.Liu L F, Wang J C. Proc Natl Acad Sci USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanaka M, Hiraga S. Mol Gen Genet. 1985;200:21–26. doi: 10.1007/BF00383307. [DOI] [PubMed] [Google Scholar]
- 11.Baker T A, Kornberg A. Cell. 1988;55:113–123. doi: 10.1016/0092-8674(88)90014-1. [DOI] [PubMed] [Google Scholar]
- 12.Skarstad K, Baker T A, Kornberg A. EMBO J. 1990;9:2341–2348. doi: 10.1002/j.1460-2075.1990.tb07406.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rokeach L A, Zyskind J W. Cell. 1986;46:763–771. doi: 10.1016/0092-8674(86)90352-1. [DOI] [PubMed] [Google Scholar]
- 14.Stuitje A R, de Wind N, van der Spek J C, Pors T H, Meijer M. Nucleic Acids Res. 1986;14:2333–2344. doi: 10.1093/nar/14.5.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Løbner-Olesen A, Atlung T, Rasmussen K V. J Bacteriol. 1987;169:2835–2842. doi: 10.1128/jb.169.6.2835-2842.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Theisen P W, Grimwade J E, Leonard A C, Bogan J A, Helmstetter C E. Mol Microbiol. 1993;10:575–584. doi: 10.1111/j.1365-2958.1993.tb00929.x. [DOI] [PubMed] [Google Scholar]
- 17.Ogawa T, Okazaki T. J Bacteriol. 1994;176:1609–1615. doi: 10.1128/jb.176.6.1609-1615.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Løbner-Olesen A, Boye E. Nucleic Acids Res. 1992;20:3029–3036. doi: 10.1093/nar/20.12.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bates D B, Asai T, Cao Y, Chambers M W, Cadwell G W, Boye E, Kogoma T. Nucleic Acids Res. 1995;23:3119–3125. doi: 10.1093/nar/23.16.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab.; 1972. [Google Scholar]
- 21.Magee T R, Asai T, Malka D, Kogoma T. EMBO J. 1992;11:4219–4225. doi: 10.1002/j.1460-2075.1992.tb05516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Asai T, Imai M, Kogoma T. J Mol Biol. 1994;235:1459–1469. doi: 10.1006/jmbi.1994.1101. [DOI] [PubMed] [Google Scholar]
- 23.Hong X, Cadwell G W, Kogoma T. EMBO J. 1995;14:2385–2392. doi: 10.1002/j.1460-2075.1995.tb07233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skarstad K, Steen H B, Boye E. J Bacteriol. 1985;163:661–668. doi: 10.1128/jb.163.2.661-668.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boye E, Løbner-Olesen A. Res Microbiol. 1991;142:131–135. doi: 10.1016/0923-2508(91)90020-b. [DOI] [PubMed] [Google Scholar]
- 26.Ogawa T, Pickett G G, Kogoma T, Kornberg A. Proc Natl Acad Sci USA. 1984;81:1040–1044. doi: 10.1073/pnas.81.4.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koch C, Vanderkerckhove J, Kahmann R. Proc Natl Acad Sci USA. 1988;85:4237–4241. doi: 10.1073/pnas.85.12.4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singer M, Baker T A, Schnitzler G, Deischel S M, Goel M, Dove W, Jaacks K J, Grossman A D, Erickson J W, Gross C A. Microbiol Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meijer M, Beck F, Hansen F G, Bergman H F, Messer W, von Meyenburg K, Schaller H. Proc Natl Acad Sci USA. 1979;76:580–584. doi: 10.1073/pnas.76.2.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sugimoto K, Oka A, Sugisaki H, Takanami M, Nishimura A, Yasuda S, Hirota Y. Proc Natl Acad Sci USA. 1979;76:575–579. doi: 10.1073/pnas.76.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kano Y, Ogawa T, Ogura T, Hiraga S, Okazaki T, Imamoto F. Gene. 1991;103:25–30. doi: 10.1016/0378-1119(91)90386-p. [DOI] [PubMed] [Google Scholar]
- 32.Boye E, Lyngstadaas A, Løbner-Olesen A, Skarstad K, Wold S. In: DNA Replication and the Cell Cycle. Fanning E, Knippers R, Winnedler E L, editors. Berlin: Springer; 1992. pp. 15–26. [Google Scholar]
- 33.Kogoma T, von Meyenburg K. EMBO J. 1983;2:463–468. doi: 10.1002/j.1460-2075.1983.tb01445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bogan J A, Helmstetter C E. J Bacteriol. 1996;178:3201–3206. doi: 10.1128/jb.178.11.3201-3206.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pierucci O, Helmstetter C E, Rickert M, Weinberger M, Leonard A C. J Bacteriol. 1987;169:1871–1877. doi: 10.1128/jb.169.5.1871-1877.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Atlung T, Hansen F G. J Bacteriol. 1993;175:6537–6545. doi: 10.1128/jb.175.20.6537-6545.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mensa-Wilmot K, Carroll K, McMacken R. EMBO J. 1989;8:2393–2402. doi: 10.1002/j.1460-2075.1989.tb08369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ogawa T, Baker T A, van der Ende A, Kornberg A. Proc Natl Acad Sci USA. 1985;82:3562–3566. doi: 10.1073/pnas.82.11.3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pettijohn D E, Hecht R. Cold Spring Harbor Symp Quant Biol. 1973;38:31–41. doi: 10.1101/sqb.1974.038.01.006. [DOI] [PubMed] [Google Scholar]
- 40.Drlica K, Franco R J, Steck T R. J Bacteriol. 1988;170:4983–4985. doi: 10.1128/jb.170.10.4983-4985.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]