Abstract
The genetic properties of the non-Mendelian element, [URE3], suggest that it is a prion (infectious protein) form of Ure2p, a mediator of nitrogen regulation in Saccharomyces cerevisiae. Into a ure2Δ strain (necessarily lacking [URE3]), we introduced a plasmid overproducing Ure2p. This induced the frequent “spontaneous generation” of [URE3], with properties identical to the original [URE3]. Altering the translational frame only in the prion-inducing domain of URE2 shows that it is Ure2 protein (and not URE2 RNA) that induces appearance of [URE3]. The proteinase K-resistance of Ure2p is unique to [URE3] strains and is not seen in nitrogen regulation of normal strains. The prion-inducing domain of Ure2p (residues 1–65) can propagate [URE3] in the absence of the C-terminal part of the molecule. In contrast, the C-terminal part of Ure2p cannot be converted to the prion (inactive) form without the prion-inducing domain covalently attached. These experiments support the prion model for [URE3] and extend our understanding of its propagation.
A prion is an infectious protein without an essential nucleic acid component. Extensive evidence shows that prion protein (PrP) is necessary for transmissible spongiform encephalitis, a uniformly fatal disease of many mammalian species, including scrapie (sheep), Creutzfeldt–Jakob disease (humans), and mad cow disease (reviewed in refs. 1–4). However, doubts remain about the protein-only model of these diseases (5). PrP produced in Escherichia coli, yeast, or other microorganisms never has been shown to be infectious (6). The scrapie form of PrP (PrPSc) is highly aggregated and apparently impossible to obtain in really pure form from animals. The best preparations require 105 PrP molecules to infect one mouse. Transmissible agent free of detectable PrPSc has been reported (7). It is still possible that PrP is the receptor for a virus, as all of the evidence for the critical role of PrP in scrapie show that it is necessary, but none show that it is sufficient.
We proposed that two non-Mendelian genetic elements of yeast, [URE3] and [PSI], are actually prions (infectious protein forms) of the Ure2 and Sup35 proteins, respectively (8). This proposal has been supported by biochemical evidence that Ure2p is altered in [URE3] strains (9) and that Sup35p is aggregated in [PSI] strains (10). That overexpression of the chaperone Hsp-104 cures [PSI] also suggests it is a prion (11).
The non-Mendelian genetic element [URE3] was discovered by Lacroute in a variant able to take up ureidosuccinate (USA) on medium containing ammonium (12). The phenotype of the dominant [URE3] was essentially the same as that of recessive mutants in the chromosomal URE2 gene (13, 14). URE2 is part of a system in yeast that blocks synthesis of proteins needed for utilization of poor nitrogen sources when a good nitrogen source is available (15, 16). The accidental structural similarity of USA, an intermediate in uracil biosynthesis, and allantoate, a poor nitrogen source for yeast, results in control of USA uptake (DAL5) by this nitrogen regulation system, providing a simple selection for inactivity of Ure2p (17, 18).
The three genetic properties of [URE3] that made us propose it was a prion are as follows (8): (i) Reversible curability. [URE3] is efficiently cured by growth on media containing 5 mM guanidine HCl, but from these cured, purified [ure-o] strains again arise [URE3] derivatives (8). (ii) Overexpression of URE2 increases the frequency with which [URE3] arises in a normal strain by 100-fold (8). (iii) Recessive ure2 mutations and the presence of [URE3] (dominant) produce the same phenotype, and yet propagation of [URE3] requires a normal URE2 gene (8, 12, 19). Each of these properties is expected of a prion, but not of a nucleic acid replicon (8), as reviewed in refs. 4 and 20. The same genetic properties were known for [PSI], and we suggested that, by analogy, [PSI] is a prion form of Sup35p (8).
Although these genetic properties and the fact that Ure2p and Sup35 are altered in [URE3] and [PSI] strains, respectively, are predicted by the hypothesis that [URE3] and [PSI] are prion forms of Ure2p and Sup35p, they do not constitute proof of this conclusion. Here we demonstrate “spontaneous generation” of [URE3] in a ure2Δ strain, showing that [URE3] is not a dominant mutant of a nucleic acid replicon that depends on Ure2p for its replication. We show it is the Ure2 protein, not URE2 RNA, that induces formation of [URE3]. Because alternate heritable states can occur because of regulatory circuits (ref. 21; reviewed in ref. 22), and Ure2p is a transcription regulator, it is critical to examine the relationship of nitrogen metabolism and [URE3]. We show that nitrogen regulation and [URE3] can be dissociated, that the alteration of Ure2p in [URE3] strains is not a consequence of the altered nitrogen regulation, and that the nitrogen state of the cells does not affect the generation or stability of [URE3]. Finally, we examine the roles of the domains of Ure2p in the propagation of [URE3].
METHODS
Cytoduction.
Cytoplasm can be transferred from one strain (the donor) to another (the recipient) by using the kar1 mutant that fails to undergo karyogamy (23). Donor (ρ+) and recipient (ρo) cells are mixed in water, mated on rich medium for 7 hr, and streaked for single colonies on medium selecting against the donor. Cytoductants are identified as those with the recipient nuclear genotype, but able to grow on glycerol (ρ+) (24).
Spontaneous Generation.
Strain 4444–3A (= MATα his3 ura2 leu2 ure2::URA3) was transformed with p645, a LEU2-2 μm DNA plasmid in which URE2 is under control of the GAL1 promoter (8). Leu+ transformants shown to be USA− (i.e., Ure2+) were grown on galactose (Gal) + Ura + His and then plated on Gal + USA + His to select [URE3] derivatives. Six USA+ clones were purified on Gal + USA + His and then tested for [URE3] maintenance on dextrose (Dex) + Ura + His, Gal + Ura + His, and Gal + Ura + His guanidine. For four of the six clones (U1, U3, U15, and U19), only cells streaked on Gal + Ura + His maintained the USA+ phenotype (as tested on Gal + USA + His). U15 and U19 were used further as donors in cytoduction experiments. Cells of each donor were taken from Gal + USA + His, mixed with a similar number of cells of strain 3748 (MATa kar1 ura2 ade2 ρo), and incubated on yeast extract/peptone/adenine/dextrose or yeast extract/peptone/adenine/galactose for 8 hr. The mating mixture then was streaked on Dex + Ura + Ade. Cytoductants, those able to grow on yeast extract/peptone/glycerol but not on Dex + Ura, were purified on yeast extract/peptone/dextrose and tested by replicaplating on Dex + Ura + Ade and Dex + USA + Ade.
Plasmid Constructions.
Plasmid p530 (YEp351URE2) and p644 (YEp351G-URE2) are LEU2-2 μm DNA shuttle plasmids with URE2 under its own or the GAL1 promoter (8). To construct plasmids with a base removed from codon 44 of URE2 or a base added to codon 80 or both, pGAL1::URE2 (p644) was used as template in the Kunkel method (25) with the mutagenic oligonucleotides −1@44 = CTATTGTTATTATTATTATTA TTACACCTGTTG and +1@80 = CGATGTTGTTCTAAGG T TATTCTTGATATTATTCTCG. Four mutant plasmids were obtained with oligo −1@44, called −FS1D, E2+1, E2+7, and E2+15. One mutant was obtained from p644 with oligo +1@80 and called E2+3. Two mutants obtained by using E2+7 as template with oligo +1@80 had both frameshifts and were named E2+7−1 and E2+7−3. All constructs were checked by sequencing. All constructs were tested for induction of [URE3] in strain 3469 in two or more experiments, and typical values are shown in Table 1.
Table 1.
Nitrogen source does not affect frequency of [URE3]
| Strain | Nitrogen source | Carbon source | [URE3]/106 cells |
|---|---|---|---|
| 3385 | Proline | Galactose | 1,000 |
| Ammonia | Galactose | 1,200 | |
| Proline | Dextrose | 13 | |
| Ammonia | Dextrose | 9 | |
| 3469 | Proline | Galactose | 48 |
| Ammonia | Galactose | 176 | |
| Proline | Dextrose | 2 | |
| Ammonia | Dextrose | 2 |
Strain 3385 (MATa kar1 ura2 leu2 his- [ure-o]) and strain 3469 (MATa/MATα kar1/kar1 ura2/ura2 leu2/leu2 trp1/+ +/his-) were transformed with p644 (YEp351G-URE2) carrying URE2 under control of the GAL1 promoter (8). Transformants were grown on the indicated media for 2 days. Yeast nitrogen base without amino acids (YNB) (ammonia nitrogen source) or YNB without ammonia +2 mg/ml l-proline contained either 2% dextrose or 2% galactose + 2% raffinose. After growth, cells were suspended and plated on dextrose-ammonia with USA in place of uracil to measure [URE3] clones.
pDM12 is a HIS3–CEN plasmid carrying the URE2 gene under its own promoter with residues 2–65 deleted and was described (9). p576 is the plasmid obtained from p532 by mutagenesis with oligo UB and contains the entire URE2 gene (8). pDM14 is p576 cut with EagI and religated, removing amino acid residues 66 to the C terminus. pDM16 is pDM12 with the SalI–XbaI fragment replaced by the 2.65-kb SalI–XbaI fragment of p576.
Northern Blots.
Transformants of strain 3718 (ure2Δ) were grown overnight in yeast extract/peptone/dextrose and for 4 hr in SGal + His + Ura. Nucleic acids were extracted by agitation with glass beads in the presence of 1% SDS and phenol-chloroform. The template for making probes was pH2 (kindly provided by Herman K. Edskes, National Institutes of Health), made by inserting URE2 on a BamHI–EcoRI fragment between the same sites of KS+. Probes were made with T3 RNA polymerase by using BamHI-cut pH2 and the Ambion Megascript kit.
RESULTS
Spontaneous Generation of [URE3].
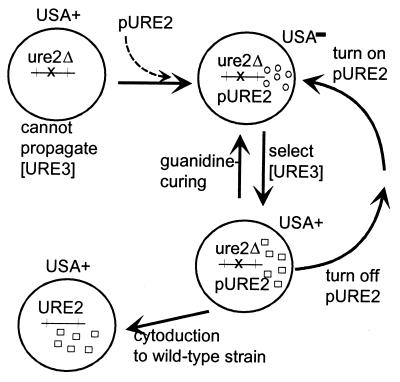
Could [URE3] be a dominant mutant form of a nucleic acid replicon that depends on Ure2p for its replication? Suppressive petites, for example, are dominant deletion mutants of mtDNA, which depend, like mtDNA itself, on chromosomal genes (e.g., PET18), for their replication. Although the dominant mutant derivatives of mtDNA can be isolated in a normal host, they cannot be isolated from a pet18 host that then was made PET18+, because the parent replicon is gone. We began with a ure2Δ strain (4444–3A) that must lack [URE3] and its putative parent genome because it cannot propagate [URE3] even if it is introduced into the strain (8, 19). We introduced p645 carrying the URE2 gene under control of the galactose-inducible GAL1 promoter (Fig. 1). Transformants grew on USA on glucose medium, but not on galactose medium, indicating that functional Ure2p is being expressed only on galactose. Rare USA+ colonies of these transformants were selected on galactose plates as potential cases of spontaneous generation of [URE3]. Two colonies (U15 and U19) were purified and analyzed in more detail. On galactose medium, these strains could be cured of [URE3] by growing in the presence of 5 mM guanidine HCl. Additionally, growing these USA+ cells for several generations on dextrose medium resulted in reversion to a USA− phenotype when returned to galactose, indicating that maintenance of [URE3] requires continuous expression of Ure2p. Finally, for both strains the USA+ trait was found to be transmissible by cytoplasmic transfer to the recipient strain 3748.
Figure 1.
Spontaneous generation of [URE3]. Strain 4444–3A (MATα his3 ura2 leu2 ure2::URA3) was transformed with p645 (YEp351G-URE2; ref. 8) in which URE2 is under control of the GAL1 promoter on a LEU2 plasmid. The expression of Ure2p in this strain on galactose made it unable to use USA in place of uracil. Colonies that grew on USA were candidates to have had spontaneous generation of [URE3]. The USA+ trait of colonies U15 and U19 was cytoducible to the URE2 recipient strain 3748, showing that U15 and U19 carried [URE3]. Colonies U15 and U19 were streaked for single colonies on GalUraHis plates containing 5 mM guanidine HCl. Each of nine Leu+ single colonies of both U15 and of U19 were unable to grow on USA. Colonies of U15 and U19 grown on Dex + Ura + His (to shut off Ure2p synthesis) also had become unable to grow on Gal + USA + His.
Ure2 Protein Overproduction Induces [URE3] Formation.
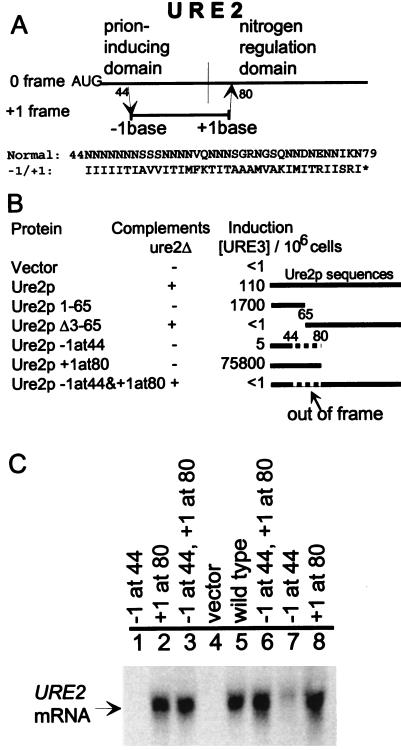
Overexpression of URE2 induces formation of [URE3], but the gene itself in high copy is not sufficient for this effect (8, 9). We distinguish protein effects from those of RNA by introducing frameshift mutations in the URE2 ORF, which minimally change RNA structure or steady-state mRNA levels, but dramatically alter the protein sequence (Fig. 2). Removing a single nucleotide from codon 44, inside the prion domain, shifts the reading frame to produce a peptide that has the correct first 43 amino acids attached to the amino acid sequence shown in Fig. 2A, followed at codon 81 by a stop codon. This construct does not detectably induce [URE3] formation in a strain with a normal chromosomal copy of URE2, but the low levels of mRNA (Fig. 2C, lanes 1 and 7) make it impossible to distinguish an effect on mRNA levels from an effect on protein sequence. Adding a single nucleotide in codon 80 to the normal gene results in a peptide with the correct first 80 residues followed immediately by a stop codon. This peptide has dramatic [URE3]-inducing ability, even better than that of residues 1–65 alone (Fig. 2B). Clearly the added nucleotide at residue 80 does not adversely affect prion-inducing ability.
Figure 2.
The Ure2 protein induces [URE3] generation. (A) One base was deleted in codon 44 of URE2, or inserted in codon 80, or both. This changed the reading frame for most of the prion domain. The insertion at codon 80 makes it a UAA codon. Deleting a base from codon 44 produces a fusion peptide that terminates … IISRIPter after residue 80, with reduced mRNA levels (compare lanes 1 and 7 with 5 in C). (B) Induction of [URE3] was tested by introducing each plasmid into strain 3469 (= MATa/MATα ura2/ura2 kar1/kar1 leu2/leu2 trp1/+ +/his− [ure-o]). Mixed transformant clones were spotted on SGal + Ura plates. After 3–5 days cells were suspended in water, cell number was measured, and dilutions were plated on SD + USA. After 5 days at 30°C clones were counted. Ten or more clones from each sample were tested for curing by growth on SD + Ura + 5 mM guanidine. Complementation of ure2Δ was carried out by introducing each plasmid into strain 3718 (= MATα his3 leu2 ura2 ure2::URA3), growing transformants on SGal + His + Ura and then replicaplating to SGal + His + USA. Values shown are USA+ clones per 106 cells plated. To the right are shown diagramatically the protein sequences expressed in the various constructs. The solid line represents Ure2p sequences and the dashed line the normally out-of-frame sequences that are expressed because of the frameshift mutations. Vector = p554, Ure2p = p646, Ure2p 1–65 = p680, Ure2p Δ3–65 = p682, Ure2p −1at44 = average of three plasmids containing the −1 frameshift at codon 44: E2+1, E2+7, E2+15; Ure2p+1at80 = +1 frameshift at codon 80: E2+3; Ure2p−1at44&+1at80 = average of two plasmids with both frameshifts: E+7–3, E+7–1. (C) Northern blot hypbridization of URE2 mRNA. Lane 1, −FS1D (−1 frameshift at residue 44). Lane 2, E2+3 (+1 at 80). Lane 3, E2+7–3 (both frameshifts). Lane 4, p554 (vector). Lane 5, p644 (wild type). Lane 6, E2+7–1 (both frameshifts). Lane 7, E2+1 (−1 frameshift at residue 44). Lane 8, E2+3 (+1 frameshift at residue 80).
When a mutant URE2 is prepared with both the single nucleotide deletion in codon 44 and the addition in codon 80 restoring the reading frame beyond this point, a protein is produced that has an intact nitrogen regulation ability, but also has a substitution of most of the prion domain (Fig. 2A). Ability to complement ure2Δ is normal, but this construct does not significantly induce conversion to [URE3] (Fig. 2B). The mRNA from this construct is present at the same steady-state level as for the wild-type URE2 (Fig. 2C) and is altered only in two nucleotides, at least one of which does not adversely affect prion induction. This shows that it is the protein product of URE2, and not the RNA transcript, that induces [URE3] formation.
Nitrogen Source and [URE3].
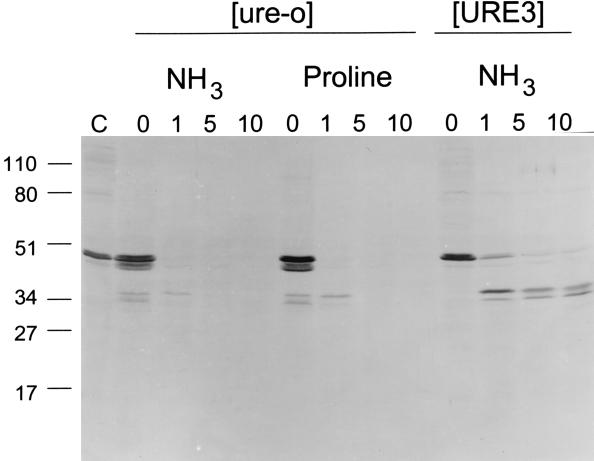
Positive or negative feedback loops regulating transcription are capable of giving rise to alternate heritable states (ref. 21; reviewed in ref. 22). One alternative explanation for [URE3] is that Ure2p can be shut off as part of such a regulatory circuit, and that the protease resistance of Ure2p is a characteristic of its “off” state. Because Ure2p is shut off when cells grow on a poor nitrogen source, we tested whether Ure2p protease sensitivity is altered by the nitrogen source. Cells grown on ammonia (a good, repressing nitrogen source) or on proline (a poor, derepressing nitrogen source) showed no difference in Ure2p protease resistance (Fig. 3). This shows that the protease-resistance of Ure2p in [URE3] strains (9) is an abnormality in [URE3] strains that leads to derepression of nitrogen metabolism, not a concomitant of normal derepression.
Figure 3.
Nitrogen source does not affect protease-resistance of Ure2p. [ure-o] and [URE3] derivatives of strain 3389/p530 (MATa kar1 ura2 leu2 his− YEp351URE2) from Leu plates were grown to midlog phase in 2% dextrose with uracil and histidine and either ammonia (yeast nitrogen base; Difco) or proline (0.2% in yeast nitrogen base without ammonia; Difco) as nitrogen source. Cell lysates were prepared and treated as described (9). Reactions contain 2.5 mg/ml protein. After removing an aliquot as a PK− control, proteinase K was added to the remainder to a final concentration of 45 μg/ml, and reactions were transferred to 37°. Aliquots were removed at indicated time points and transferred to sample buffer containing 3 mM phenylmethylsulfonyl fluoride and frozen on dry ice. Samples were run on 12% polyacrylamide/SDS gels, and immunoblotted with anti-Ure2p antibody. Molecular mass size markers are indicated. C = 15 μg of protein from the [URE3] strain, no treatment. 0 = PK−; 1, 5, 10 = minutes of proteinase K digestion. [ure-o] means absence of the [URE3] genetic element.
One might expect that changed nitrogen status would affect the frequency with which [URE3] arises via the interaction of Ure2p with glutamine (the putative indicator of cellular nitrogen status) or Gln3p (a positive transcription factor mediating many of Ure2p’s effects). However, we were unable to detect a substantial effect of nitrogen source on [URE3] generation (Table 1). Moreover, growth of the [URE3] strain 3687 on proline or glutamate as a nitrogen source did not result in loss of [URE3] (data not shown).
The N-Terminal Prion Domain Is Sufficient for [URE3] Propagation.
The USA uptake phenotype (USA+ or USA−) is only an indirect measure of whether the [URE3] genetic element is present or not. The C-terminal domain of Ure2p is necessary for nitrogen regulation (9, 26), so in its absence a strain will be USA+ whether [URE3] is present or not. To overcome this problem, we used the method of sequential cytoductions (27). [URE3] was passed by cytoduction from strain 1 to ure2Δ strain 2 in which some part (or all) of Ure2p was expressed from a plasmid. The phenotype of strain 2 was noted, but the test for whether strain 2 could propagate [URE3] was to pass its cytoplasm to a third strain carrying a normal chromosomal URE2 and to examine its phenotype (Table 2).
Table 2.
Prion domain of Ure2p is sufficient for [URE3] propagation
| Strain 1 URE2 | Strain 2 ure2Δ | Strain 3 URE2 | ||
|---|---|---|---|---|
| 1 3560 USA+/[URE3] | → | pURE2 (38 of 43 USA+) | → | D350-3CC (16 all USA+) |
| 2 3560 USA+/[URE3] | → | pURE2C (22 all USA−) | → | 3748 (20 all USA−) |
| 3 3560 USA+/[URE3] | → | pURE2N (29 all USA+) | → | 3748 (42 of 57 USA+) |
| 4 3560 USA+/[URE3] | → | pURE2N + pURE2C (24 all USA−) | → | D350-3CC (15 of 16 USA+) |
Cytoplasm is transferred from [URE3] cells (strain 1) to a ure2Δ strain (strain 2) producing all or part of Ure2p from a plasmid. Strain 2 is USA+ with no plasmid. pURE2 is pDM16 expressing the entire Ure2p, pURE2N is pDM14 expressing residues 1-65 of Ure2p (the prion-inducing domain, and pURE2C is pDM12 expressing residues 66–354, the nitrogen regulation domain (9). The strain 2 cytoductants are tested for their phenotypes and used as cytoduction donors to strain 3, which is URE2. The USA phenotype of strain 3 reveals whether [URE3] is propagating in strain 2. → indicates transfer of cytoplasm by cytoduction. Strain 1 (3560) = MATa ura2 kar1-1 hisx leu2 p530 (YEpURE2) [URE3]. Strain 2 (4444-3A) = MATα ura2 ure2Δ his3 leu2 ρ0 with various plasmids. Strain 3 (3748) = MATa ura2 kar1-1 ade2 ρo [ure-o] or (D350-3CC) = MATa ura2 kar1-1 leu2 trp1 can1 ρo [ure-o].
We expressed Ure2p(1–65) from pDM14 in a ure2Δ strain (4444–3A). Into this strain, expressing only the prion-inducing domain, we introduced [URE3] by cytoduction. The cytoductants generated were grown for 15–20 generations and then used as cytoduction donors to a [ure-o] third strain to determine whether [URE3] persists in the strain expressing only the N-terminal domain. Nearly all of these final recipients became [URE3] (Table 2). This shows that the N-terminal domain is sufficient for propagation of [URE3]. It also shows that the transcription regulation part of Ure2p is not necessary for [URE3] propagation, thus arguing against a regulatory circuit model of [URE3].
The C-Terminal Nitrogen Regulatory Domain Is Insensitive to [URE3].
To determine whether the C-terminal nitrogen regulation domain of Ure2p (residues 66–354) is capable of propagating [URE3] in the absence of the N-terminal domain, we introduced [URE3] into a strain carrying a chromosomal ure2Δ mutation and expressing only the C-terminal domain from a plasmid (Table 2, line 2). These cytoductants, which were all USA−, were used as donors to a normal recipient to determine whether they were carrying [URE3]. None of these final recipients were USA+, indicating that they did not receive [URE3]. Thus, the C-terminal domain is not sufficient to propagate [URE3].
Ure2p C Terminus and N Terminus Act Independently Unless Covalently Linked.
Expressing the Ure2p N-terminal and C-terminal fragments from separate plasmids in the same ure2Δ strain resulted in the USA− phenotype. When [URE3] was introduced into this strain, it remained USA− (Table 2, line 4). When these cytoductants were used as cytoduction donors to a wild-type strain, the recipients became USA+, implying that they acquired [URE3] (Table 2, line 4). This shows that the Ure2p C-terminal domain does not interfere with the propagation of [URE3]. It also shows that [URE3] is not a self-supporting physiological state based on nitrogen repression. Further, this shows that the Ure2p C-terminal domain is inactivated only by the prion form of the N terminus when they are covalently attached.
DISCUSSION
We provide further support for the prion model of [URE3] and present evidence concerning the detailed mechanism by which the signal is transmitted from molecule to molecule.
Spontaneous Generation of [URE3].
We show that [URE3] can be produced de novo in a strain that could not have had the element before. This spontaneous generation of [URE3] goes beyond our original demonstration that overexpression increases the frequency of [URE3] in a normal strain. If [URE3] generation were equivalent to suppressive petite generation, that is, a dominant mutant of a nucleic acid replicon normally present in the cell, whose propagation depends upon Ure2p, then our spontaneous generation experiment would have failed, because the normal replicon would have been lost in the ure2Δ host before pURE2 was introduced. Spontaneous generation of [URE3] is a property expected of prions, because merely expressing the normal protein should allow the prion to develop, without the introduction of any infectious element from outside the cell.
Attempts to produce this phenomenon in mice by introduction of an overexpressing transgene of normal or mutant PrP have produced disease, but so far no material infectious for normal mice (28, 29). Demonstration of spontaneous generation of [PSI] is complicated by the C-terminal domain of the molecule being essential for growth.
Ure2p Is Sufficient to Induce Formation of [URE3].
That it is overexpression of Ure2p and not URE2 mRNA that is responsible for inducing [URE3] previously was suggested based on the lack of [URE3] induction by nonsense mutants located early in URE2. However, nonsense-mediated decay of mRNA could be prematurely degrading URE2 mRNA in these mutants (30). Indeed, a single frameshift mutation at codon 44 results in much lower URE2 mRNA levels. We constructed a full-length URE2, active in nitrogen regulation, but with altered prion domain sequence, by deleting a single nucleotide in codon 44 and inserting a compensating nucleotide in codon 80. The RNA sequence, structure, and amount are minimally changed, but the prion-inducing activity is eliminated. This strongly argues that it is the protein overproduction, and not the RNA, that is inducing formation of [URE3].
This experiment is, in some ways, the genetic equivalent of purifying the protein and showing that protein alone is sufficient to transmit the [URE3] trait. Of course, it will still be important to show that Ure2p purified from a [URE3] strain is more efficient at transmitting [URE3] than is Ure2p from a wild-type strain.
As discussed above, overexpression of PrP has not yet been shown to produce scrapie (with infectious material for normal mice), so the equivalent experiment to that we describe here has not yet been possible in mammals. A similar demonstration has been carried out in the [PSI] system by introduction of a frameshift mutation creating a UAA at codon 20 (31). [PSI] appearance was not induced by this construct even though mRNA levels remained relatively high.
Nitrogen Derepression Does Not Make Ure2p Resistant to Protease.
The finding that Ure2p is altered in [URE3] strains supports the prion explanation of [URE3]. However, because Ure2p is intimately involved in nitrogen regulation and [URE3] strains are derepressed for nitrogen catabolic genes, it could be argued that the protease resistance of Ure2p in [URE3] strains is a change Ure2p undergoes whenever nitrogen genes are derepressed, rather than an abnormality that is the cause of the derepression. Cells grown on proline as the nitrogen source are derepressed (15), and yet they do not show protease resistance of Ure2p. Thus, the protease resistance of Ure2p in [URE3] strains is an indication of an alteration of Ure2p that is the cause of altered N-regulation in these strains.
The lac operon, bacteriophage λ, and Drosophila sex determination all provide examples of regulatory loops that can give rise to alternative stable heritable states, a form of epigenetic mechanism (22). We now have two lines of evidence that [URE3] is not such a phenomenon in spite of the known regulatory function of Ure2p: (i) the prion domain of Ure2p is completely separable from the nitrogen regulation domain, and (ii) the protease resistance of Ure2p is not a concomitant of derepressed nitrogen regulation, but is an alteration specific for the [URE3] state.
The Prion Domain Is Sufficient to Transmit [URE3].
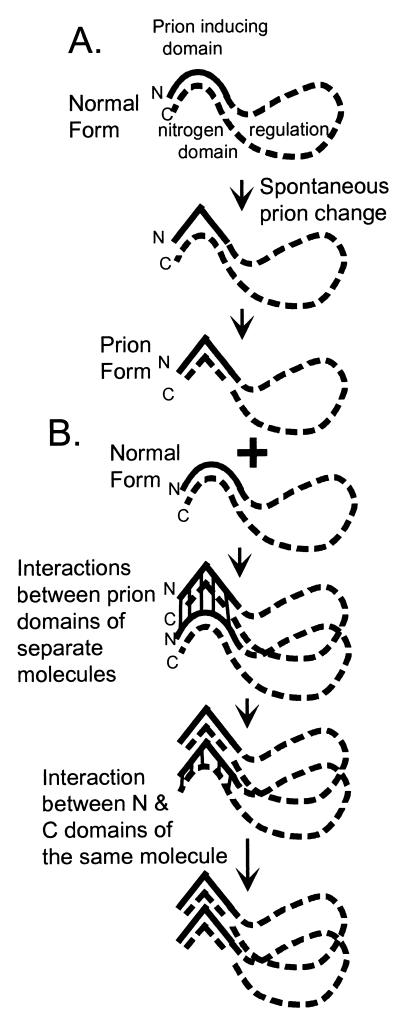
We previously showed that overexpression of Ure2p(1–65) is sufficient to induce the emergence of [URE3] in the presence of the intact Ure2 protein. Here we show that the intact protein in the prion form can convert Ure2p(1–65) into the prion form. More important, Ure2p(1–65) can propagate [URE3] in the absence of the intact protein. This shows that the C-terminal domain is dispensable for prion propagation, and that the [URE3] change can be transmitted from the N terminus of one molecule to the N terminus of another (Fig. 4). The C-terminal domain is sufficient for the nitrogen regulatory effect of Ure2p (9, 26), and [URE3] makes its presence known by its effect on the activity of the C terminus. But [URE3] propagation can occur in a strain with no C terminus. It is detected in these experiments by transfer of cytoplasm into a [ure-o] strain expressing the full-length protein.
Figure 4.
Model of [URE3] prion generation and propagation. (A) Spontaneous change of the N-terminal prion domain produces the prion form of Ure2p with inactivation of the C-terminal domain by an intramolecular interaction with this altered N-terminal domain. (B) Propagation of [URE3] is by N terminus to N terminus intermolecular interactions.
This type of experiment cannot be done in the case of [PSI] because the C-terminal domain of Sup35p is essential for growth. Thus, it remains possible that the C-terminal part of Sup35p is involved in the generation and/or propagation of [PSI]. Some experiments of this type have been carried out in mice, showing that the N-terminal repeats of PrP are not necessary for scrapie propagation (32).
The C-Terminal Domain (Residues 66–354) Is Insensitive to [URE3].
The part of Ure2p lacking the prion domain fails to induce [URE3] when overexpressed (9), and we show here that it is not inactivated by the introduction into the cell of [URE3] from another cell. Thus the [URE3] change cannot be transmitted from N terminus or C terminus of one molecule (the altered Ure2p introduced) to the C terminus of another molecule [the Ure2p(66–354) expressed in the recipient]. The C terminus must, in some sense, interact with the N terminus of the same molecule because C termini with an attached N terminus (intact Ure2p) are inactivated by the [URE3] change. We suggest that the N terminus–C terminus interaction is of sufficiently low affinity that it is only efficient when these two parts of the molecule are attached (Fig. 4). Further, we find that the C-terminal domain does not interfere with the propagation and transmission of [URE3] by N-terminal molecules expressed in the same strain. Thus, the Ure2p N and C termini seem to act independently when they are not covalently attached, but to affect each other profoundly when attached. (i) The N-terminal domain can inactivate the C-terminal domain when they are attached. (ii) Deletions in the C-terminal domain increase the frequency with which the N-terminal domain induces [URE3], an effect that we interpret as the stabilization of the N-terminal domain by the C-terminal part, preventing the prion change (9). (iii) The N-terminal domain improves the nitrogen regulation activity of the C-terminal domain (9).
The putative yeast prions hold the promise of actually proving that such a thing as a prion can exist, and of using yeast molecular genetics to dissect the factors influencing prion generation and propagation. The experiments presented here make critical advances in achieving both of these goals.
Acknowledgments
We thank Don Court for useful discussion.
ABBREVIATIONS
- PrP
prion protein
- USA
ureidosuccinate
- Dex
dextrose
- Gal
galactose
References
- 1.Prusiner S B. In: Fields Virology. Fields B N, Knipe D M, Howley P M, editors. Vol. 2. New York: Raven; 1996. pp. 2901–2950. [Google Scholar]
- 2.Weissmann C, Fischer M, Raeber A, Bueler H, Sailer A, Shmerling D, Rulicke T, Brandner S, Aguzzi A. Function and Dysfunction in the Nervous System. Vol. 61. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 511–522. [PubMed] [Google Scholar]
- 3.Caughey B, Chesebro B. Trends Cell Biol. 1997;7:56–62. doi: 10.1016/S0962-8924(96)10054-4. [DOI] [PubMed] [Google Scholar]
- 4.Wickner R B. Prion Diseases of Mammals and Yeast: Molecular Mechanisms and Genetic Features. Austin, TX: Landes; 1997. [Google Scholar]
- 5.Chesebro B, Fields B N. In: Fields Virology. Fields B N, Knipe D M, Howley P M, editors. Vol. 2. New York: Raven; 1996. pp. 2845–2849. [Google Scholar]
- 6.Weiss S, Famulok M, Edenhofer F, Wang Y-H, Jones I M, Groschup M, Winnacker E-L. J Virol. 1995;69:4776–4783. doi: 10.1128/jvi.69.8.4776-4783.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lasmezas C I, Deslys J-P, Robain O, Jaegly A, Beringue V, Peyrin J-M, Fournier J-G, Hauw J J, Rossier J, Dormont D. Science. 1997;275:402–405. doi: 10.1126/science.275.5298.402. [DOI] [PubMed] [Google Scholar]
- 8.Wickner R B. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 9.Masison D C, Wickner R B. Science. 1995;270:93–95. doi: 10.1126/science.270.5233.93. [DOI] [PubMed] [Google Scholar]
- 10.Paushkin S V, Kushnirov V V, Smirnov V N, Ter-Avanesyan M D. EMBO J. 1996;15:3127–3134. [PMC free article] [PubMed] [Google Scholar]
- 11.Chernoff Y O, Lindquist S L, Ono B-I, Inge-Vechtomov S G, Liebman S W. Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 12.Lacroute F. J Bacteriol. 1971;106:519–522. doi: 10.1128/jb.106.2.519-522.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drillien R, Lacroute F. J Bacteriol. 1972;109:203–208. doi: 10.1128/jb.109.1.203-208.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drillien R, Aigle M, Lacroute F. Biochem Biophys Res Comm. 1973;53:367–372. doi: 10.1016/0006-291x(73)90671-2. [DOI] [PubMed] [Google Scholar]
- 15.Cooper T G. In: The Molecular Biology of the Yeast Saccharomyces: Metabolism and Gene Expression. Strathern J N, Jones E W, Broach J R, editors. Vol. 2. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. pp. 39–99. [Google Scholar]
- 16.Magasanik B. In: The Molecular and Cellular Biology of the Yeast Saccharomyces. Jones E W, Pringle J R, Broach J R, editors. Vol. 2. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. pp. 283–317. [Google Scholar]
- 17.Turoscy V, Cooper T G. J Bacteriol. 1987;169:2598–2600. doi: 10.1128/jb.169.6.2598-2600.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rai R, Genbauffe F, Lea H Z, Cooper T G. J Bacteriol. 1987;169:3521–3524. doi: 10.1128/jb.169.8.3521-3524.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aigle M, Lacroute F. Mol Gen Genet. 1975;136:327–335. doi: 10.1007/BF00341717. [DOI] [PubMed] [Google Scholar]
- 20.Wickner R B. Annu Rev Genet. 1996;30:109–135. doi: 10.1146/annurev.genet.30.1.109. [DOI] [PubMed] [Google Scholar]
- 21.Novick A, Weiner M. Proc Natl Acad Sci USA. 1957;43:553–566. doi: 10.1073/pnas.43.7.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riggs A D, Porter T N. In: Epigenetic Mechanisms of Gene Regulation. Russo V E A, Martienssen R A, Riggs A D, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 29–45. [Google Scholar]
- 23.Conde J, Fink G R. Proc Natl Acad Sci USA. 1976;73:3651–3655. doi: 10.1073/pnas.73.10.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ridley S P, Sommer S S, Wickner R B. Mol Cell Biol. 1984;4:761–770. doi: 10.1128/mcb.4.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kunkel T A. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coschigano P W, Magasanik B. Mol Cell Biol. 1991;11:822–832. doi: 10.1128/mcb.11.2.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Esteban R, Wickner R B. Genetics. 1987;117:399–408. doi: 10.1093/genetics/117.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsiao K K, Scott M, Foster D, Groth D F, DeArmond S J, Prusiner S B. Science. 1990;250:1587–1590. doi: 10.1126/science.1980379. [DOI] [PubMed] [Google Scholar]
- 29.Westaway D, DeArmond S J, Cayetano-Canlas J, Groth D, Foster D, Yang S-L, Torchia M, Carlson G A, Prusiner S B. Cell. 1994;76:117–129. doi: 10.1016/0092-8674(94)90177-5. [DOI] [PubMed] [Google Scholar]
- 30.Jacobson A, Peltz S W. Annu Rev Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- 31.Derkatch I L, Chernoff Y O, Kushnirov V V, Inge-Vechtomov S G, Liebman S W. Genetics. 1996;144:1375–1386. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fischer M, Rulicke T, Raeber A, Sailer A, Moser M, Oesch B, Brandner S, Aguzzi A, Weissmann C. EMBO J. 1996;15:1255–1264. [PMC free article] [PubMed] [Google Scholar]