Abstract
Golgi membranes in Drosophila embryos and tissue culture cells are found as discrete units dispersed in the cytoplasm. We provide evidence that Golgi membranes do not undergo any dramatic change in their organization during the rapid mitotic divisions of the nuclei in the syncitial embryo or during cell division postcellularization. By contrast, in Drosophila tissue culture cells, the Golgi membranes undergo complete fragmentation during mitosis. Our studies show that the mechanism of Golgi partitioning during cell division is cell type-specific.
A major question in biology that remains poorly understood is the mechanism by which organelles such as the Golgi are partitioned into daughter cells during cell division. In mammalian tissue culture cells stacks of Golgi cisternae are located in the pericentriolar region through association with the microtubule organizing center (MTOC). At the onset of mitosis, protein transport along the secretory pathway is blocked and Golgi stacks break down into small vesicular structures (1, 2). The vesiculated Golgi membranes (VGMs) are found dispersed throughout the cytoplasm. VGMs are somehow partitioned into the daughter cells, where after cytokinesis, they assemble into stacks of cisternae followed by the reactivation of the secretory pathway (3, 4). Similarly, the nuclear envelope also breaks down into small vesicles at the onset of mitosis, which then assemble into a nucleus around the chromatin in the cytoplasm of each daughter cell after cytokinesis (5).
The mechanism of organelle partitioning described above is not universal. For example, in the budding yeast Saccharomyces cerevisiae, Golgi units (a unit is composed of tubulovesicular membranes) are found randomly dispersed in the cytoplasm. At the time of budding the nucleus undergoes a closed type of division while the Golgi in the new bud is acquired by migration of intact individual units (6). In plant cells, Golgi stacks also are found dispersed in the cytoplasm (i.e., are not pericentriolarly located). During cell division, although the nuclear envelope vesiculates, the Golgi stacks remain intact analogous to the budding yeast (7). Why Golgi membranes vesiculate in the cytoplasm of mammalian tissue culture cells with each mitotic cycle but not in S. cerevisiae or in plant cells is not known. In mammalian cells there are about 30–40 stacks interconnected to make a single Golgi complex in the pericentriolar region. Then why are stacks not partitioned into daughter cells as their counterparts in the budding yeast or plant cells? What is it that determines the fragmentation of Golgi stacks into VGMs during mitosis?
MATERIALS AND METHODS
The Drosophila melanogaster (Canton-S) flies were maintained in population cages, and the eggs were harvested by a standard procedure (8).
Monoclonal Antibodies.
Golgi membranes were isolated from 0- to 12-hr Drosophila embryos by using sucrose density gradient centrifugation as described before (9) and then used to immunize mice. Hybridomas were prepared from the mouse splenocytes by using SP 2/0 myelomas essentially as described previously (10). The resulting monoclonal antibodies were characterized as being anti-Golgi based on the fact that they colocalized with rabbit anti Drosophila β-COP antibodies by fluorescence microscopy in Drosophila S2 cells.
Immunofluorescence Microscopy on Drosophila Tissue Culture Cells and Embryos.
Syncitial Drosophila embryos were harvested for 0–2 hr. Postcellularized embryos were harvested for 0–2 hr and then aged for 3 hr. The embryos were dechorionated, fixed for 20 min with 4% formaldehyde, and devitillinized (11). The embryos were stained for fluorescence microscopy and visualized by confocal microscopy as described previously (12).
RESULTS AND DISCUSSION
We addressed the mechanism of Golgi partitioning in Drosophila melanogaster, which offers a range of distinct modes of mitotic divisions. After fertilization, the zygotic nucleus undergoes 14 very rapid mitotic divisions without cytokinesis. The syncytial embryo containing about 6,000 nuclei then undergoes a synchronous cellularization process to form a cellularized blastoderm (13). The transition from syncytium to cellularized embryo is accompanied by a tremendous growth in plasma membrane, exemplified by the highly villus appearance of plasma membrane, which, before cellularization, appeared smooth (14, 15). We have shown previously that numerous Golgi units are found in the developing oogonium and remain cortically located throughout the early embryonic events (12). Based on our observations we proposed previously that the cortical location of the Golgi membranes throughout early embryogenesis may be indicative of an active participation in the sorting and transport of newly synthesized proteins to provide for the rapid growth of plasma membrane. To comply with the high demand for protein sorting and transport, these Golgi membranes, unlike their mammalian counterparts, may not break down and assemble with each mitotic cycle.
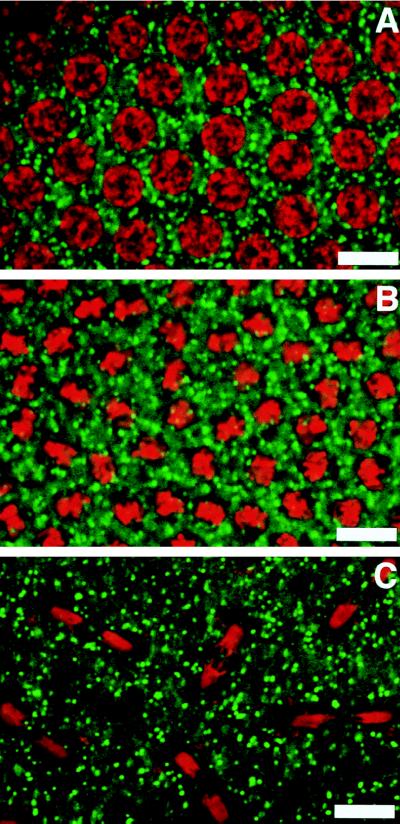
To test this hypothesis, we now have screened a large population of early embryos in the precellularization stages of embryogenesis. Fig. 1A shows an embryo in the interphase stage as evidenced by the uncondensed DNA (visualized by staining with propidium iodide). Fig. 1B shows an embryo in which the DNA is condensed and appears in metaphase. In these embryos the Golgi membranes, visualized by using a monoclonal antibody against an integral membrane protein of the Drosophila Golgi, appear as discrete punctate structures around the condensed DNA. Fig. 1C shows an embryo in anaphase. In these embryos the Golgi membranes still appear as discrete punctate structures. This same result was obtained by using monoclonal antibodies to three other Drosophila Golgi integral membrane proteins (data not shown). Thus, at the light microscopic level the Golgi membranes appear as discrete punctate structures around the nuclear material regardless of the cell-cycle stage. In some older embryos the Golgi appeared to be slightly larger than in younger embryos (compare Fig. 1 A with C). Whether or not this apparent size difference represents a real biological phenomenon (e.g., Golgi cisternae forming into more complicated structures such as Golgi stacks) will require future studies by immunoelectron microscopy. The lack of vesiculation of the Golgi membranes parallels the lack of vesiculation of the nuclear membrane in these embryos, which has been demonstrated previously (16, 17).
Figure 1.
Organization of Golgi membranes in precellularized embryos. (A) Nonmitotic nuclei. (B) Metaphase nuclei. (C) Late anaphase nuclei. The Golgi membranes were stained with a Golgi-specific antibody that recognizes a 120-kDa integral Golgi membrane protein. DNA was stained with propidium iodide. The Golgi membranes appear randomly dispersed around the nuclear material, and the organization remains unchanged during the mitotic cycle. (Bars = 10 μ.)
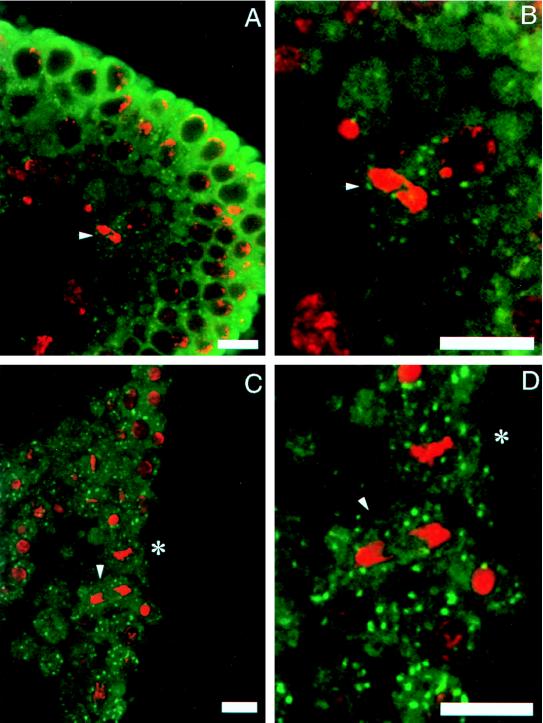
We next visualized Golgi membranes in embryos after cellularization and gastrulation. In these embryos the nuclei have lost the mitotic synchrony characteristic of younger embryos, such that cells in various stages of cell cycle can be captured at one time. Cells in interphase contain Golgi membranes distributed around the nuclear periphery as in the precellularized embryos (Fig. 2). Fig. 2 also shows cells in the metaphase stage of the cell cycle, as is evident by the condensed DNA (arrowheads). The Golgi membranes in these cells remain as discrete structures around the condensed DNA. Therefore, there appears to be no difference in the distribution of Golgi membranes during the cell cycle in the embryos before or after cellularization.
Figure 2.
The organization of Golgi membranes in cellularized embryos. The mitotic cells are shown by arrowheads in A–D. B and D are higher magnifications of A and C, respectively. The distribution of Golgi membranes in mitotic and nonmitotic cells appears similar by immunofluorescence microscopy by using Golgi monoclonal antibody. The DNA is stained with propidium iodide.
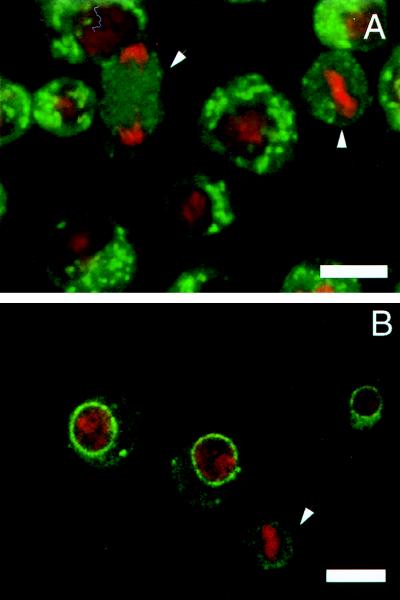
We thus far have demonstrated that the Golgi membranes do not break down with each mitotic cycle regardless of whether the mitotic cycle accompanies cytokinesis. In two previous examples (yeast and plants) in which the Golgi membranes did not fragment during mitosis, the Golgi in interphase cells were not pericentriolarly located. We then reasoned that perhaps Golgi membranes only break down in cells in which they are pericentriolarly located through association with the microtubule organizing center. Therefore, a priori, in the Drosophila tissue culture cells (S2 cells), where the Golgi membranes are randomly distributed in the cytoplasm, the Golgi stacks should not undergo any fragmentation during mitosis. Fig. 3A shows that the Golgi membranes in these tissue culture cells appear randomly distributed in the cytoplasm in interphase cells. In mitotic cells (marked by arrowheads), however, the Golgi membranes are no longer visible as discrete punctate structures but are found as diffusely scattered in the cytoplasm. This same result was also obtained by using another Drosophila tissue culture cell line, Kc (data not shown). We also followed the fate of the nuclear membrane during mitosis by the monoclonal antibody that previously had been shown to react with the nuclear membrane in Drosophila embryos (17). In interphase tissue culture cells, the nuclear envelope is intact; in mitotic cells, the staining appears as diffusely dispersed throughout the cell cytoplasm (Fig. 3B; the mitotic cell is marked by an arrowhead). In Drosophila tissue culture cells, the Golgi membranes thus undergo a very dramatic change in morphology during mitosis much like their mammalian counterparts, despite the fact that Golgi membranes are not pericentriolarly located. It should be pointed out that this lack of pericentriolar localization has also been observed in cells from several adult Drosophila tissues (data not shown).
Figure 3.
The organization of Golgi membranes in Drosophila tissue culture cells. (A) In interphase cells the Golgi membranes appear as discrete units dispersed in the cytoplasm. In mitotic cells (marked with arrowheads) the Golgi membranes appear completely fragmented and diffusely dispersed in the cytoplasm. (B) The nuclear envelop stained with a specific monoclonal antibody appears fragmented in mitotic cells (arrowhead). The DNA was stained with propidium iodide. Thus, both the Golgi membranes and the nuclear envelope break down during mitosis in the Drosophila tissue culture cells.
Why is it then that Golgi membranes undergo a dramatic change in the organization in some cells and not in others during mitosis? The change in the organization obviously is not linked to their intracellular location: both pericentriolar-localized mammalian Golgi and randomly localized Drosophila tissue culture Golgi fragment, whereas randomly localized plant and yeast Golgi do not. Nor is the change in organization linked with events regulating the organization of nuclear envelop (see Table 1). Therefore, these studies show that there is not a unifying mechanism between species for partitioning of Golgi membranes during cell division. Indeed, our data show that two distinctly different systems can exist within a single species. Could it be that the change in the organization of the Golgi during mitosis is linked to an imbalance in membrane trafficking along the secretory pathway? There certainly is evidence in support of this proposal: in mammalian tissue culture cells but not in plant cells or in S. cerevisiae, protein transport from the ER is blocked during mitosis. In fact, blocking transport from the ER by reagents such as RabGDP, brefeldin A, or ilimaquinone is known to perturb the organization of Golgi membranes even in nondividing mammalian tissue culture cells (18–20).
Table 1.
The organization of the Golgi membranes and nuclear envelope and the fate of protein transport from the ER in dividing and nondividing cells
| Cell type | Interphase
|
Mitosis
|
||
|---|---|---|---|---|
| Golgi organization | Golgi organization | Nuclear envelope | Protein transport from the ER | |
| Mammalian tissue culture cells | Pericentriolar | Fragmented | Fragmented | Blocked |
| S. cerevisiae | Random | Intact | Intact | Not blocked |
| Plants | Random | Intact | Fragmented | Not blocked |
| Drosophila embryos | ||||
| Precellularization | Random | Intact | Intact | ? |
| Postcellularization | Random | Intact | Intact | ? |
| Drosophila tissue culture cells | Random | Fragmented | Fragmented | ? |
In summary, it is clear that Golgi membranes in a selected number of cell types undergo extensive fragmentation during mitosis. However, the reason for this fragmentation remains unclear.
Acknowledgments
We thank Drs. Yukiko Goda and Ben Glick for extensive discussions and for improving the quality of this manuscript. Members of the Malhotra lab are acknowledged for their useful critique of this work. The work in V.M.’s lab is supported by the National Institutes of Health (GM46224) and a California Sea Grant (NA36RGO537). V.M. is an established Investigator of the American Heart Association. We thank Theo Palmer and Brian Dolby for help with imaging and photography, and Dr. Riparbelli for the gift of anti-vimentin antibody. Drs. Rusty Gage and Larry Goldstein are acknowledged for the generous use of their confocal microscope.
References
- 1.Featherstone C, Griffiths G, Warren G. EMBO J. 1987;6:3239–3246. doi: 10.1002/j.1460-2075.1987.tb02641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lucocq J M, Pryde J, Berger E, Warren G J. Cell Biol. 1987;104:865–874. doi: 10.1083/jcb.104.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lucocq J, Berger E G, Warren G J. Cell Biol. 1989;109:463–474. doi: 10.1083/jcb.109.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warren G. Annu Rev Biochem. 1995;62:323–348. doi: 10.1146/annurev.bi.62.070193.001543. [DOI] [PubMed] [Google Scholar]
- 5.Newport J W, Forbes D. J Annu Rev Biochem. 1987;56:535–565. doi: 10.1146/annurev.bi.56.070187.002535. [DOI] [PubMed] [Google Scholar]
- 6.Preuss D, Mulholland J, Franzusoff A, Segev N, Botstein D. Mol Biol Cell. 1992;3:789–803. doi: 10.1091/mbc.3.7.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirose S, Komamine A. New Phytol. 1989;111:599–605. doi: 10.1111/j.1469-8137.1989.tb02353.x. [DOI] [PubMed] [Google Scholar]
- 8.Elgin S C R, Miller D W. In: The Genetics and Biology of Drosophila. Ashburner M, Wright T R F, editors. 2a. London: N.Y.S.F.; 1978. pp. 112–120. [Google Scholar]
- 9.Malhotra V, Serafini T, Orci L, Shepherd J C, Rothman J E. Cell. 1989;58:329–336. doi: 10.1016/0092-8674(89)90847-7. [DOI] [PubMed] [Google Scholar]
- 10.Stanley H A, Reese R T. Proc Natl Acad Sci USA. 1985;82:6272–6275. doi: 10.1073/pnas.82.18.6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ashburner M. Drosophila: A Laboratory Handbook. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 12.Ripoche J, Link B, Yucel J K, Tokuyasu K, Malhotra V. Proc Natl Acad Sci USA. 1994;91:1878–1882. doi: 10.1073/pnas.91.5.1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foe V E, Alberts B E. J Cell Sci. 1983;61:31–70. doi: 10.1242/jcs.61.1.31. [DOI] [PubMed] [Google Scholar]
- 14.Katoh K, Ishikawa H. Protoplasma. 1989;150:83–95. [Google Scholar]
- 15.Loncar D, Singer S. Proc Natl Acad Sci USA. 1995;92:2199–2203. doi: 10.1073/pnas.92.6.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stafstrom J P, Staehelin L A. Eur J Cell Biol. 1984;34:179–189. [PubMed] [Google Scholar]
- 17.Callaini G, Riparbelli M G. Cell Motil Cytoskeleton. 1991;19:1–8. doi: 10.1002/cm.970190102. [DOI] [PubMed] [Google Scholar]
- 18.Peter F, Nuoffer C, Pind S N, Balch W E. J Cell Biol. 1994;126:1393–1406. doi: 10.1083/jcb.126.6.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lippincott-Schwartz J, Donaldson J G, Schweizer A, Berger E G, Hauri H-P, Yuan L C, Klausner R D. Cell. 1990;60:821–836. doi: 10.1016/0092-8674(90)90096-w. [DOI] [PubMed] [Google Scholar]
- 20.Takizawa P A, Yucel J K, Veit B A, Faulkner D J, Deerinck T, Soto G, Ellisman M, Malhotra V. Cell. 1993;73:1079–1090. doi: 10.1016/0092-8674(93)90638-7. [DOI] [PubMed] [Google Scholar]