Abstract
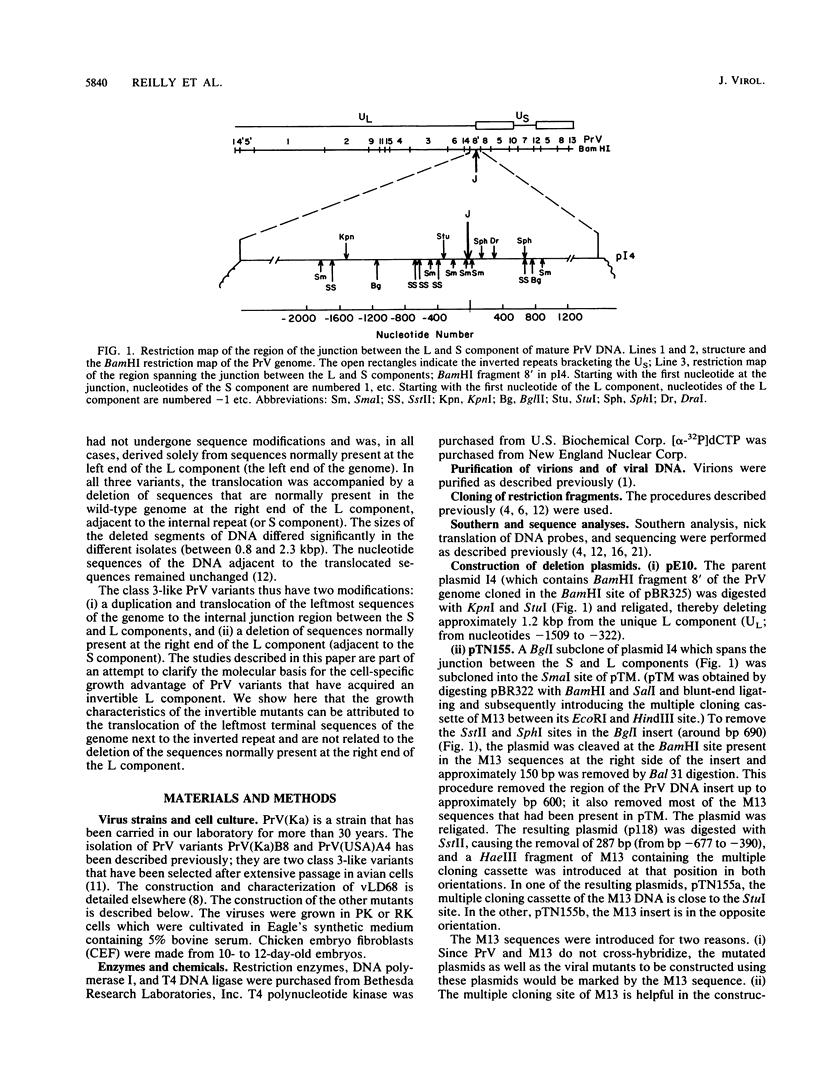
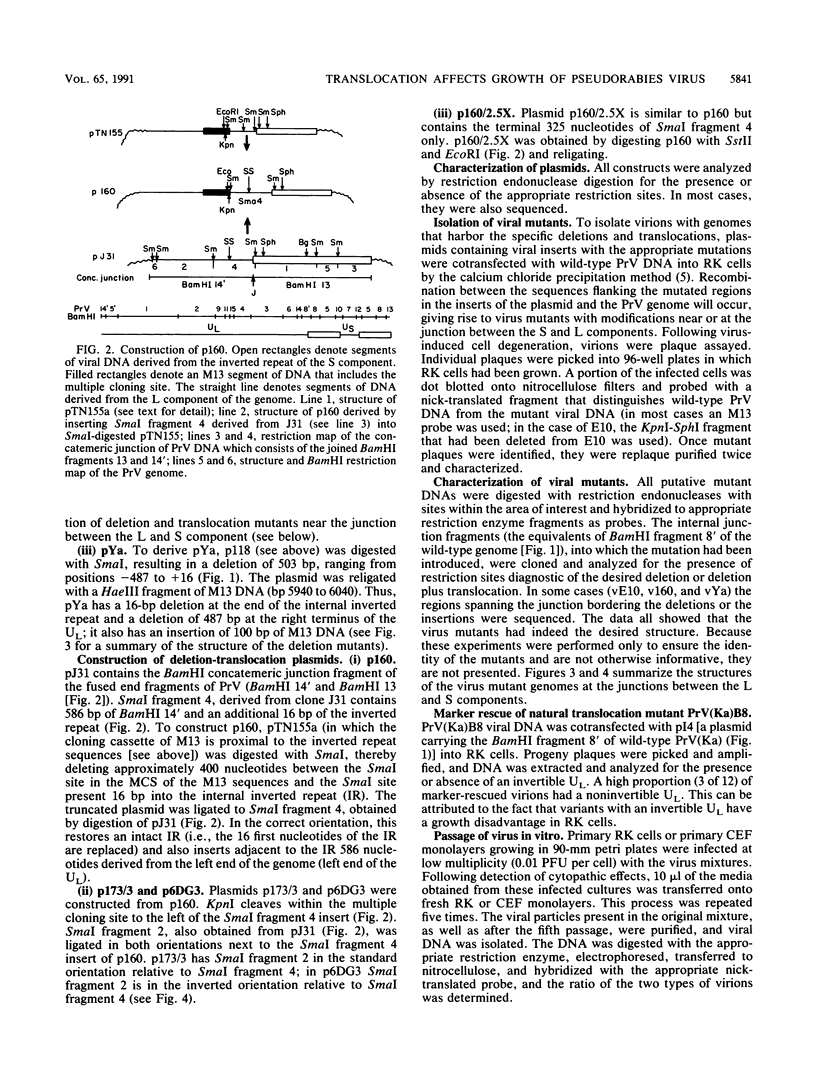
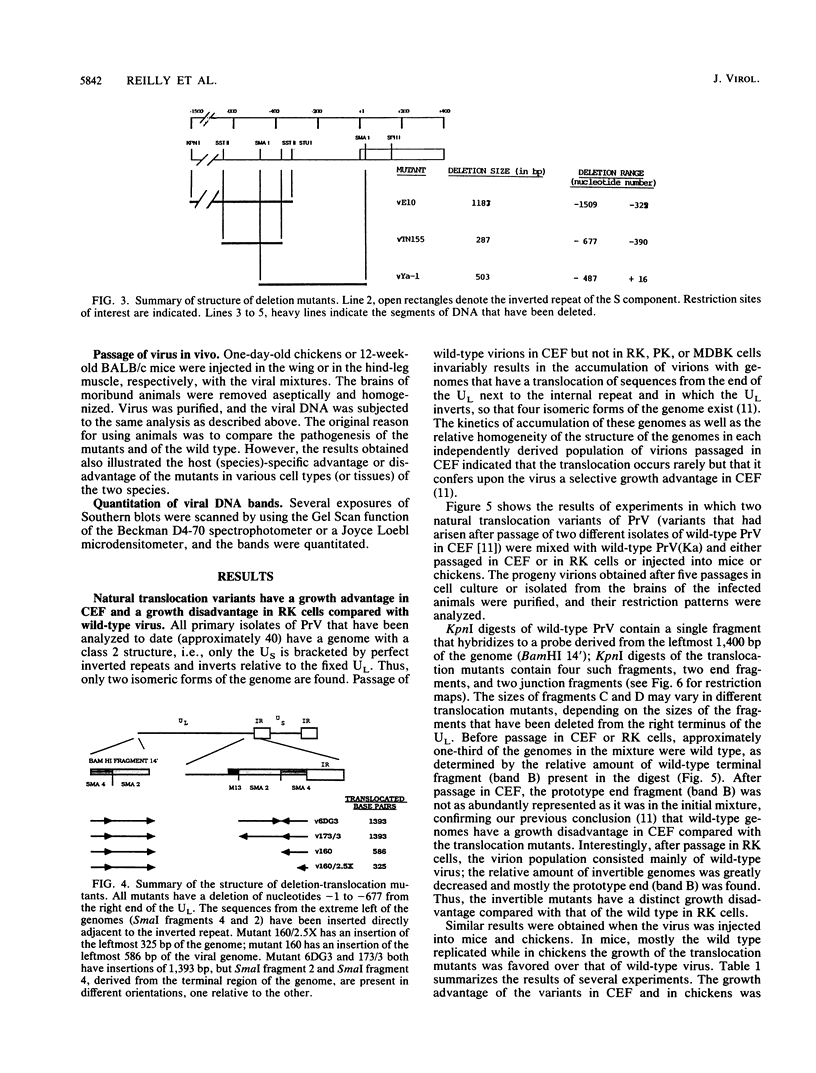
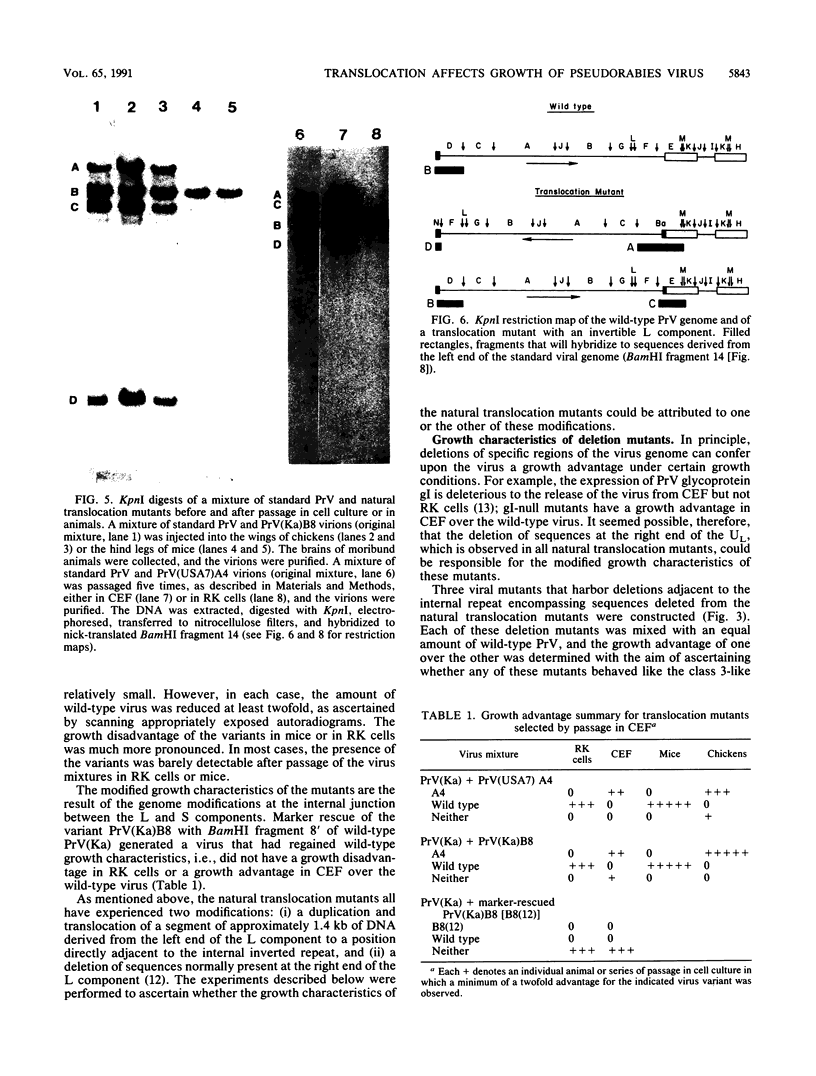
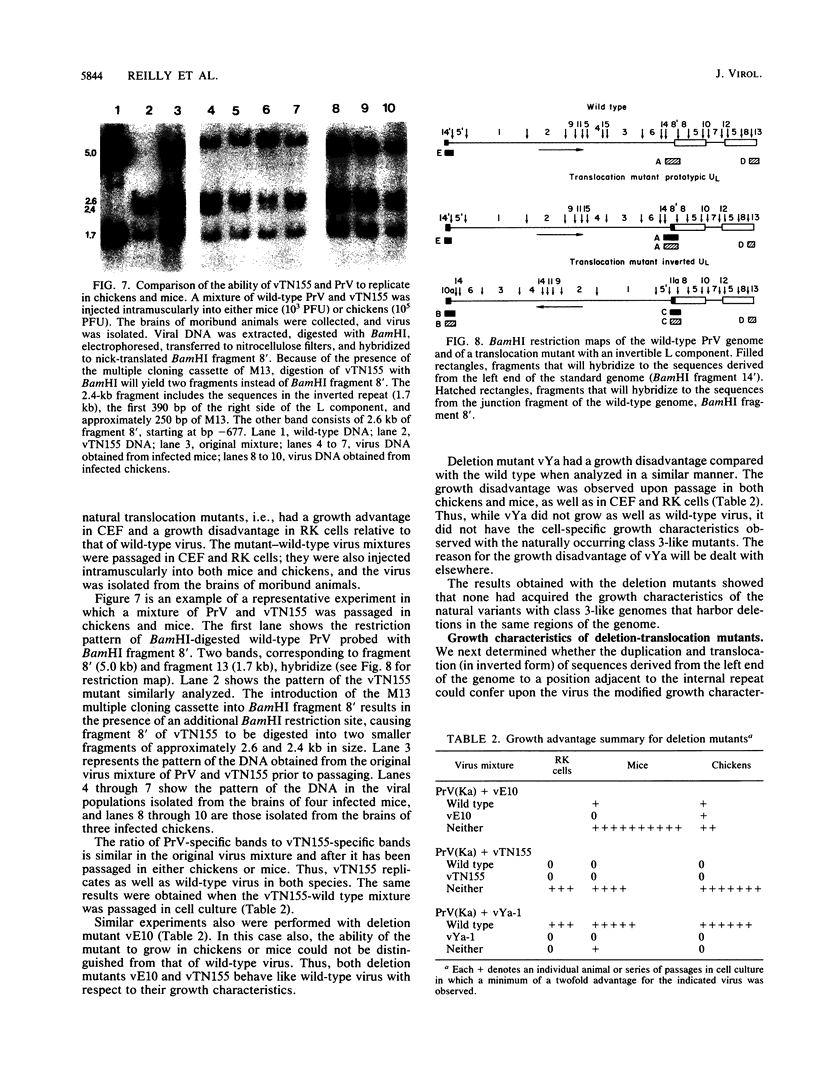
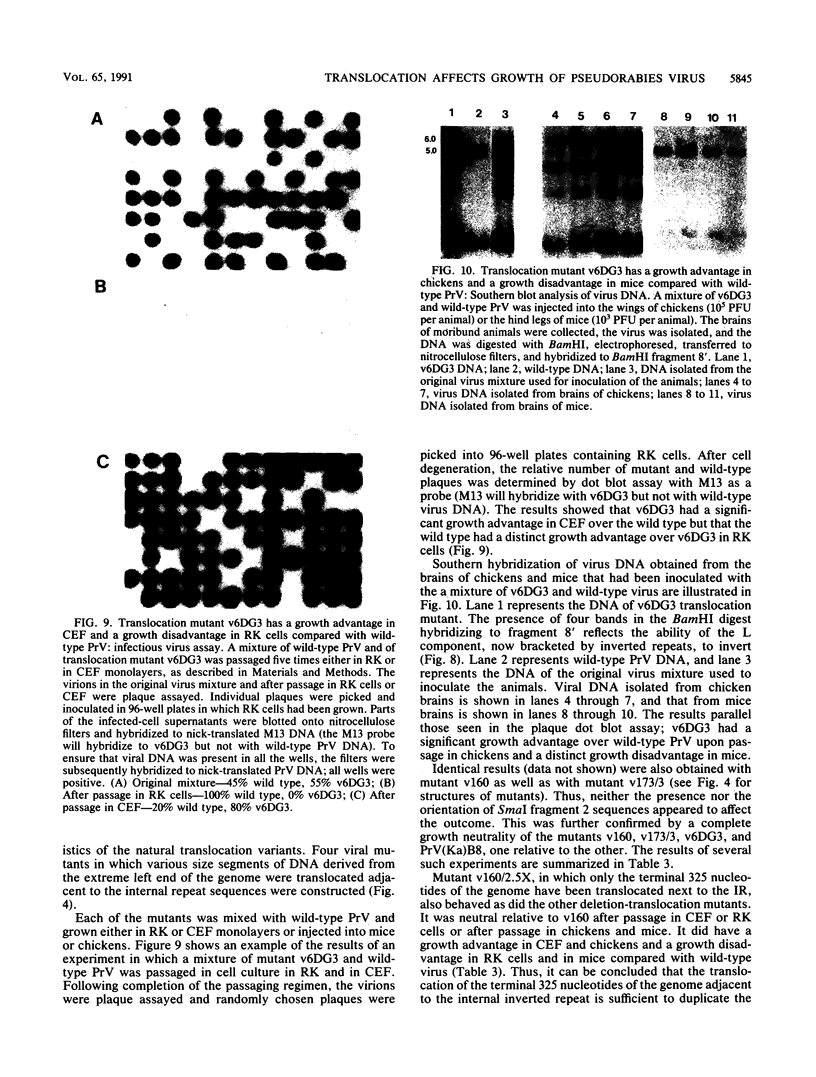
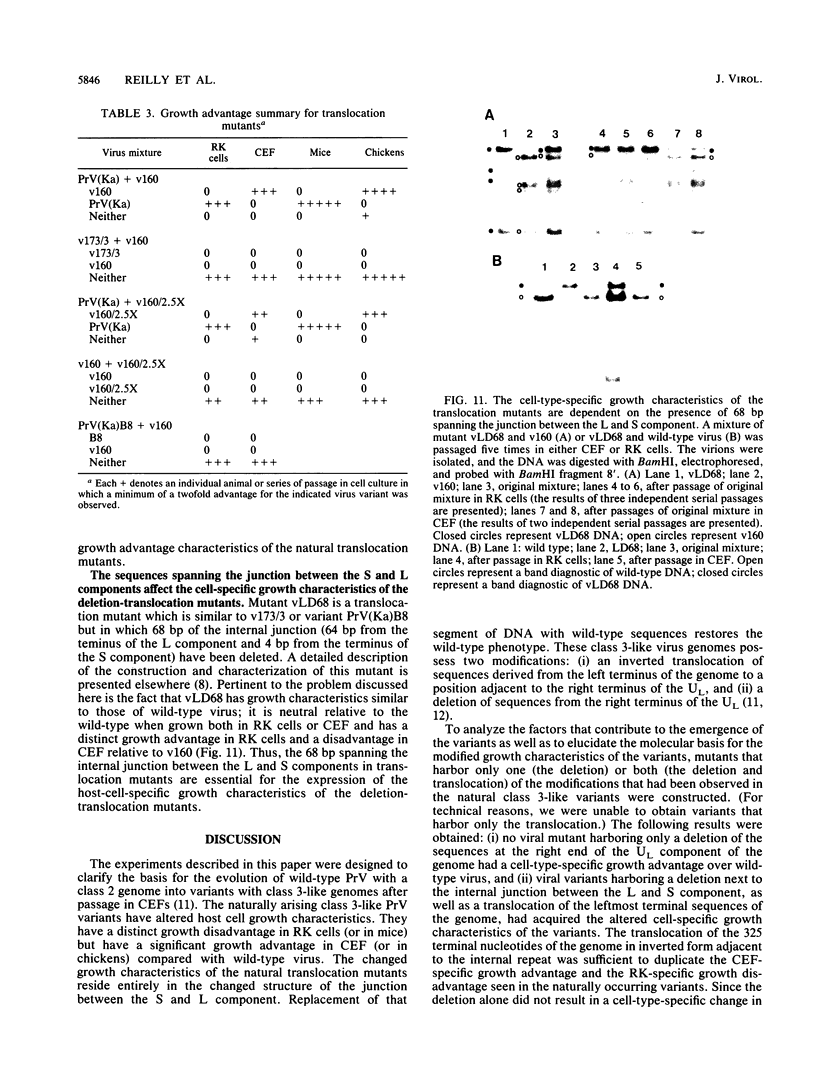
Pseudorabies virus is a herpesvirus which has a class 2 genome. However, under certain growth conditions it acquires a genome with class 3-like characteristics. In these variants, the leftmost sequences of the long (L) component of the viral genome have been duplicated and translocated to the right of the L component next to the short (S) component, resulting in an L component that is bracketed by inverted repeats. Consequently, the L component can invert and is found in two orientations relative to the S component. The translocation is accompanied invariably by a deletion of sequences that are normally present in the wild-type genome at the right end of the L component. The virion variants with an invertible L component have a growth advantage over wild-type virus in chicken embryo fibroblasts and chickens; they also have a growth disadvantage in mice or rabbit kidney cells. The changed growth characteristics of the variants reside entirely in the changed structure of the junction between the S and L components. Replacement of that region of the DNA with wild-type sequences restores the wild-type phenotype. To determine whether the modified growth characteristics of the variants are related to the translocation or to the deletion, mutants that have a deletion or that have a deletion as well as a translocation similar to those observed in the variants were constructed, and the growth characteristics of these mutants were determined. We show that the modified growth characteristics of the mutants with an invertible L component can be attributed to the translocation of the leftmost terminal sequences of the genome next to the inverted repeat; they are not related to the deletion of the sequences normally present at the right end of the L component. The translocation of the leftmost 325 bp of the genome is sufficient to confer upon the virus the modified cell-type-specific growth characteristics. Furthermore, the modified growth characteristics are contingent upon the presence of 68 bp spanning the internal junction between the L and S components.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ben-Porat T., Demarchi J. M., Kaplan A. S. Characterization of defective interfering viral particles present in a population of pseudorabies virions. Virology. 1974 Sep;61(1):29–37. doi: 10.1016/0042-6822(74)90239-6. [DOI] [PubMed] [Google Scholar]
- Ben-Porat T., Rixon F. J., Blankenship M. L. Analysis of the structure of the genome of pseudorabies virus. Virology. 1979 Jun;95(2):285–294. doi: 10.1016/0042-6822(79)90484-7. [DOI] [PubMed] [Google Scholar]
- Ben-Porat T., Veach R. A., Ladin B. F. Replication of herpesvirus DNA. VI. Virions containing either isomer of pseudorabies virus DNA are infectious. Virology. 1980 Apr 30;102(2):370–380. doi: 10.1016/0042-6822(80)90104-x. [DOI] [PubMed] [Google Scholar]
- DeMarchi J. M., Lu Z. Q., Rall G., Kupershmidt S., Ben-Porat T. Structural organization of the termini of the L and S components of the genome of pseudorabies virus. J Virol. 1990 Oct;64(10):4968–4977. doi: 10.1128/jvi.64.10.4968-4977.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Harper L., Demarchi J., Ben-Porat T. Sequence of the genome ends and of the junction between the ends in concatemeric DNA of pseudorabies virus. J Virol. 1986 Dec;60(3):1183–1185. doi: 10.1128/jvi.60.3.1183-1185.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honess R. W., Watson D. H. Unity and diversity in the herpesviruses. J Gen Virol. 1977 Oct;37(1):15–37. doi: 10.1099/0022-1317-37-1-15. [DOI] [PubMed] [Google Scholar]
- Kupershmidt S., DeMarchi J. M., Lu Z. Q., Ben-Porat T. Analysis of an origin of DNA replication located at the L terminus of the genome of pseudorabies virus. J Virol. 1991 Nov;65(11):6283–6291. doi: 10.1128/jvi.65.11.6283-6291.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomniczi B., Blankenship M. L., Ben-Porat T. Deletions in the genomes of pseudorabies virus vaccine strains and existence of four isomers of the genomes. J Virol. 1984 Mar;49(3):970–979. doi: 10.1128/jvi.49.3.970-979.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomniczi B., Gielkens A., Csobai I., Ben-Porat T. Evolution of pseudorabies virions containing genomes with an invertible long component after repeated passage in chicken embryo fibroblasts. J Virol. 1987 Jun;61(6):1772–1780. doi: 10.1128/jvi.61.6.1772-1780.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z. Q., DeMarchi J. M., Harper L., Rall G. F., Ben-Porat T. Nucleotide sequences at recombinational junctions present in pseudorabies virus variants with an invertible L component. J Virol. 1989 Jun;63(6):2690–2698. doi: 10.1128/jvi.63.6.2690-2698.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettenleiter T. C., Schreurs C., Zuckermann F., Ben-Porat T. Role of pseudorabies virus glycoprotein gI in virus release from infected cells. J Virol. 1987 Sep;61(9):2764–2769. doi: 10.1128/jvi.61.9.2764-2769.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poffenberger K. L., Tabares E., Roizman B. Characterization of a viable, noninverting herpes simplex virus 1 genome derived by insertion and deletion of sequences at the junction of components L and S. Proc Natl Acad Sci U S A. 1983 May;80(9):2690–2694. doi: 10.1073/pnas.80.9.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Roizman B. The structure and isomerization of herpes simplex virus genomes. Cell. 1979 Mar;16(3):481–494. doi: 10.1016/0092-8674(79)90023-0. [DOI] [PubMed] [Google Scholar]
- Sheldrick P., Berthelot N. Inverted repetitions in the chromosome of herpes simplex virus. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):667–678. doi: 10.1101/sqb.1974.039.01.080. [DOI] [PubMed] [Google Scholar]
- Skare J., Summers W. C. Structure and function of herpesvirus genomes. II. EcoRl, Sbal, and HindIII endonuclease cleavage sites on herpes simplex virus. Virology. 1977 Feb;76(2):581–595. doi: 10.1016/0042-6822(77)90240-9. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stevely W. S. Inverted repetition in the chromosome of pseudorabies virus. J Virol. 1977 Apr;22(1):232–234. doi: 10.1128/jvi.22.1.232-234.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadsworth S., Jacob R. J., Roizman B. Anatomy of herpes simplex virus DNA. II. Size, composition, and arrangement of inverted terminal repetitions. J Virol. 1975 Jun;15(6):1487–1497. doi: 10.1128/jvi.15.6.1487-1497.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie N. M. Physical maps for Herpes simplex virus type 1 DNA for restriction endonucleases Hind III, Hpa-1, and X. bad. J Virol. 1976 Oct;20(1):222–233. doi: 10.1128/jvi.20.1.222-233.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. A., Harper L., Ben-Porat T. cis Functions involved in replication and cleavage-encapsidation of pseudorabies virus. J Virol. 1986 Aug;59(2):318–327. doi: 10.1128/jvi.59.2.318-327.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]