Abstract
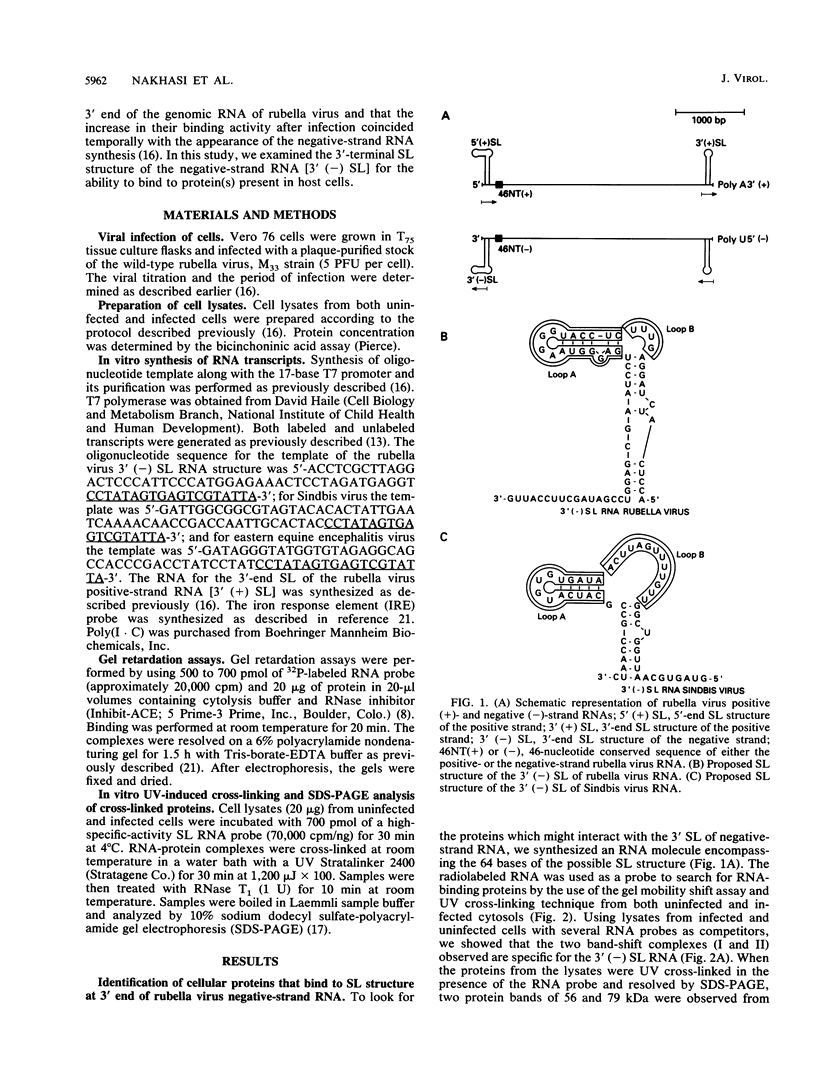
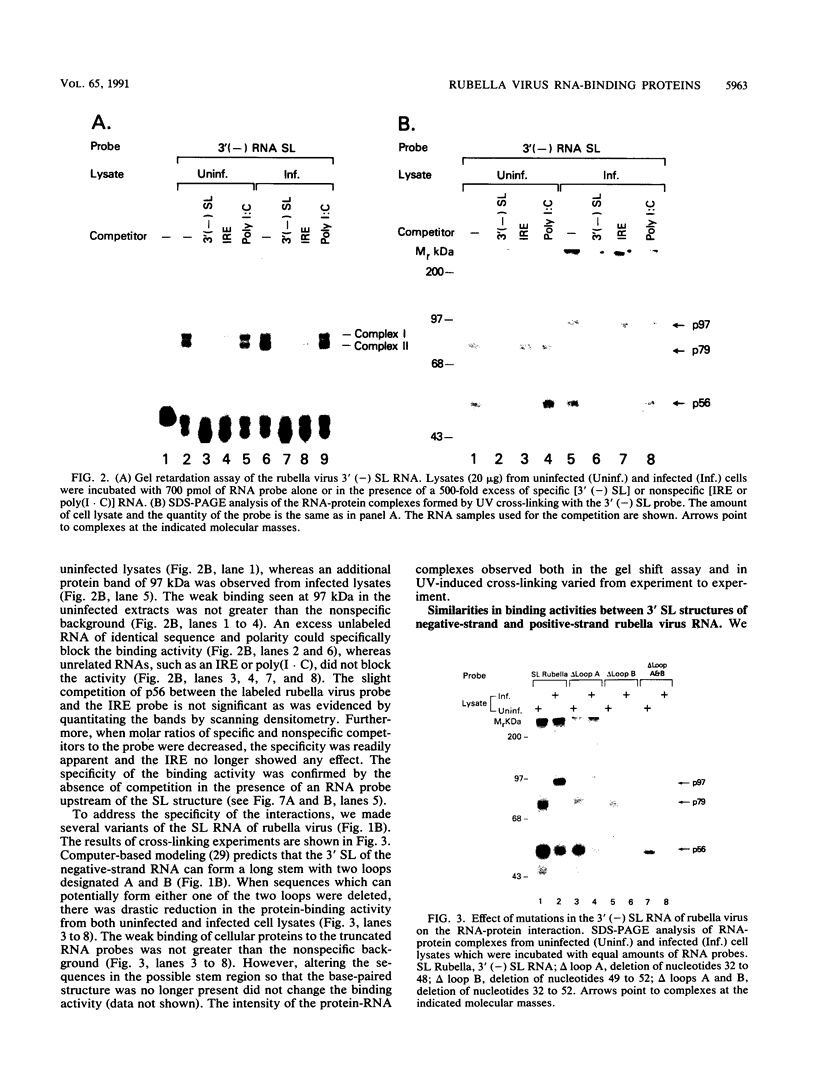
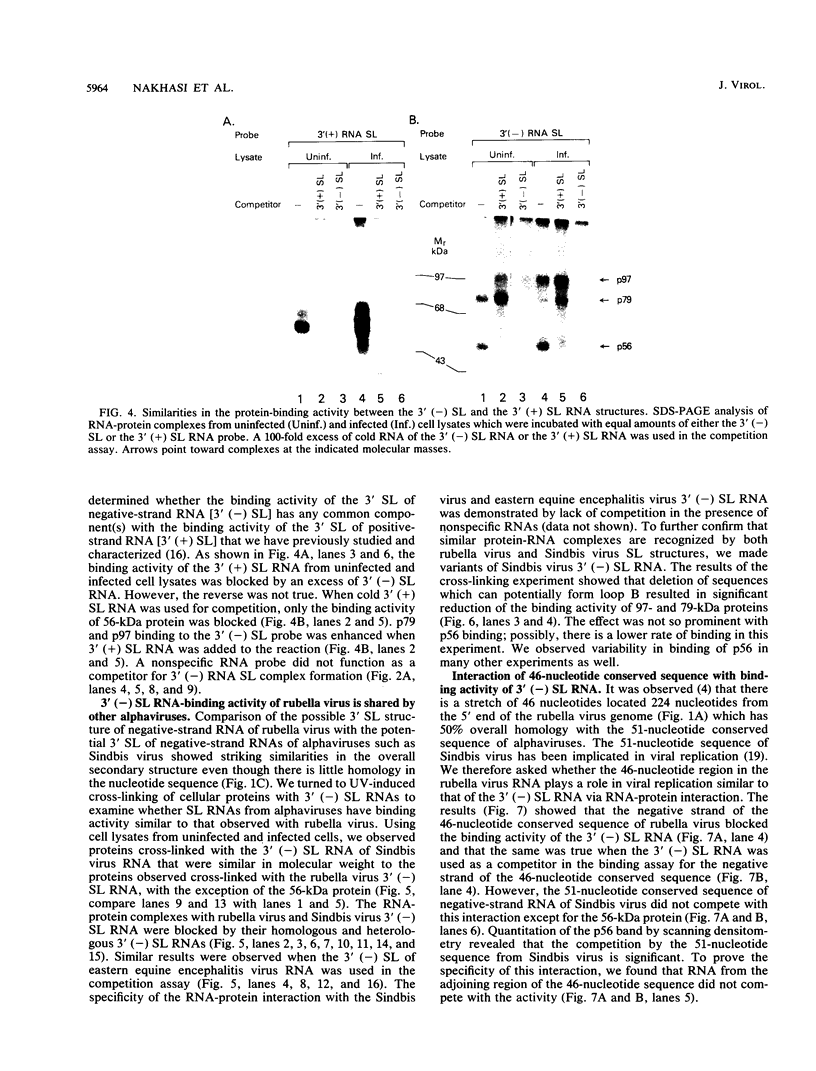
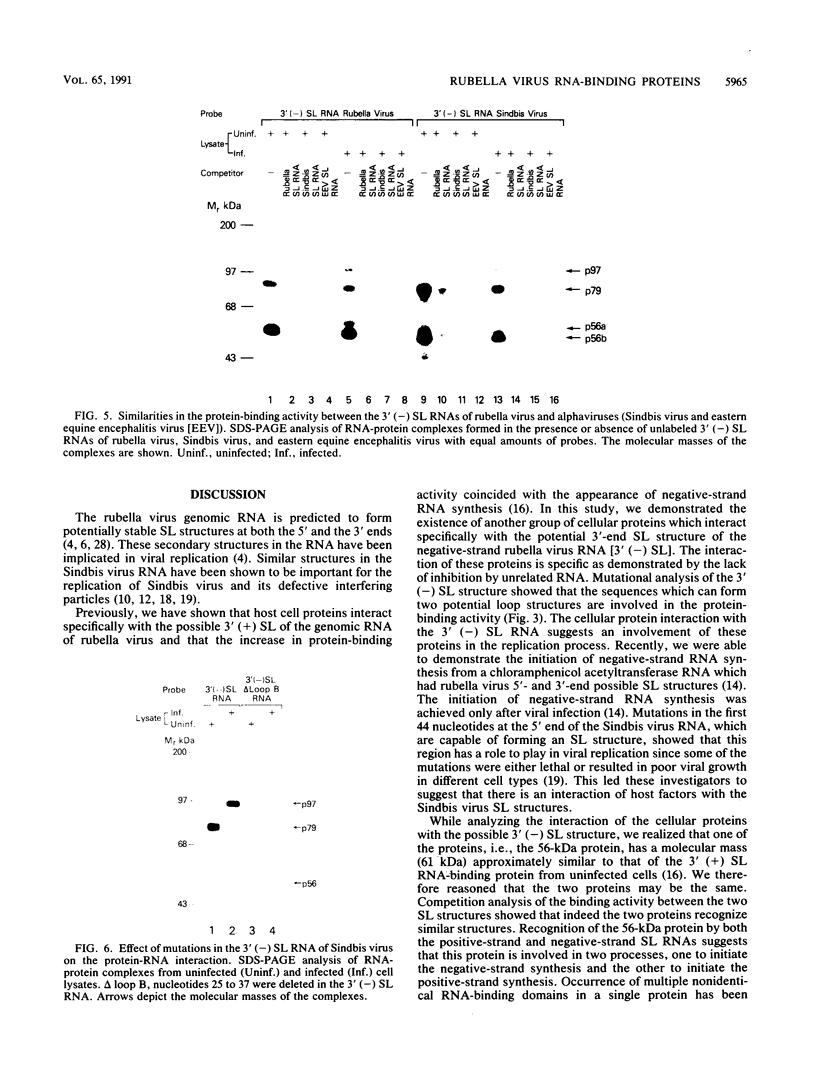
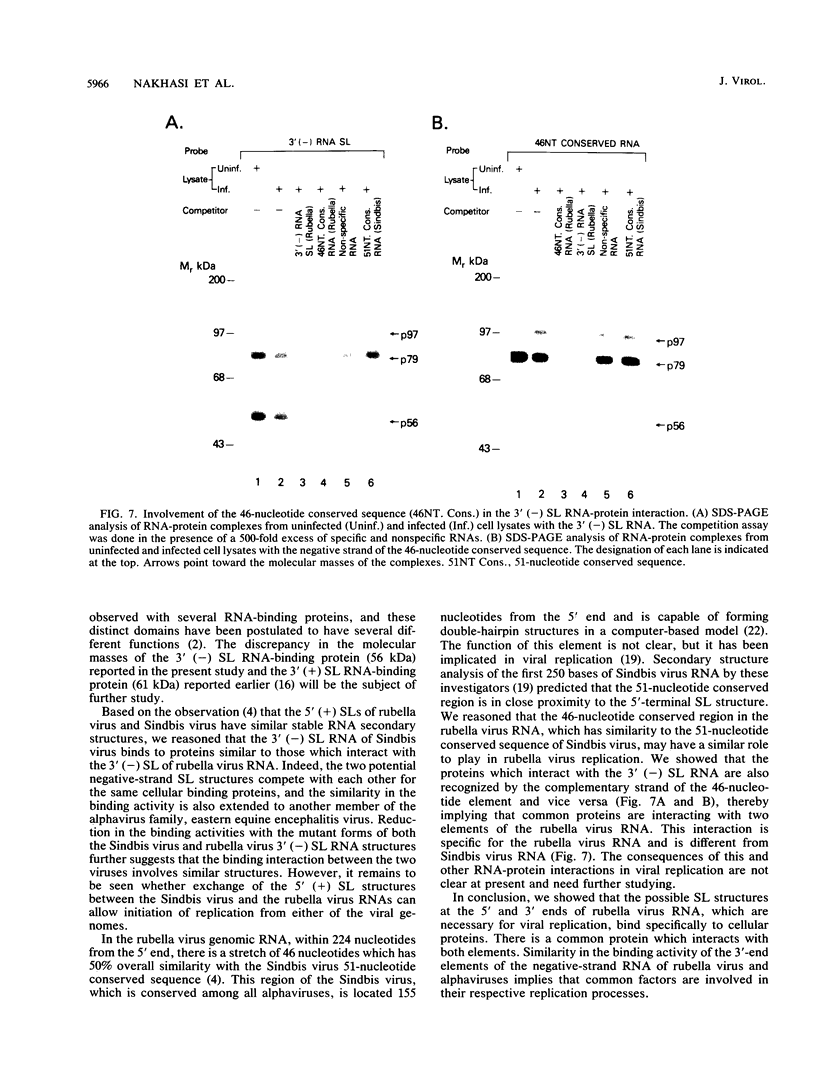
At the 5' end of the rubella virus genomic RNA, there are sequences that can form a potentially stable stem-loop (SL) structure. The complementary negative-strand equivalent of the 5'-end SL structure of positive-strand rubella virus RNA [5' (+) SL structure] is thought to serve as a promoter for the initiation of positive-strand synthesis. We screened the negative-strand equivalent of the 5' (+) SL structure (64 nucleotides) and the adjacent region of the negative-strand RNA for their ability to bind to host cell proteins. Specific binding to the 64-nucleotide-long potential SL structure of three cytosolic proteins with relative molecular masses of 97, 79, and 56 kDa was observed by UV-induced covalent cross-linking. There was a significant increase in the binding of the 97-kDa protein from cells upon infection with rubella virus. Altering the SL structure by deleting sequences in either one of the two potential loops abolished the binding interaction. The 56-kDa protein also appeared to bind specifically to an SL derived from the 3' end of positive-strand RNA. The 3'-terminal structure of rubella virus negative-strand RNA shared the same protein-binding activity with similar structures in alphaviruses, such as Sindbis virus and eastern equine encephalitis virus. A possible role for the host proteins in the replication of rubella virus and alphaviruses is discussed.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andino R., Rieckhof G. E., Baltimore D. A functional ribonucleoprotein complex forms around the 5' end of poliovirus RNA. Cell. 1990 Oct 19;63(2):369–380. doi: 10.1016/0092-8674(90)90170-j. [DOI] [PubMed] [Google Scholar]
- Bandziulis R. J., Swanson M. S., Dreyfuss G. RNA-binding proteins as developmental regulators. Genes Dev. 1989 Apr;3(4):431–437. doi: 10.1101/gad.3.4.431. [DOI] [PubMed] [Google Scholar]
- Clarke D. M., Loo T. W., Hui I., Chong P., Gillam S. Nucleotide sequence and in vitro expression of rubella virus 24S subgenomic messenger RNA encoding the structural proteins E1, E2 and C. Nucleic Acids Res. 1987 Apr 10;15(7):3041–3057. doi: 10.1093/nar/15.7.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez G., Wang C. Y., Frey T. K. Sequence of the genome RNA of rubella virus: evidence for genetic rearrangement during togavirus evolution. Virology. 1990 Jul;177(1):225–238. doi: 10.1016/0042-6822(90)90476-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey T. K., Marr L. D., Hemphill M. L., Dominguez G. Molecular cloning and sequencing of the region of the rubella virus genome coding for glycoprotein E1. Virology. 1986 Oct 15;154(1):228–232. doi: 10.1016/0042-6822(86)90446-0. [DOI] [PubMed] [Google Scholar]
- Frey T. K., Marr L. D. Sequence of the region coding for virion proteins C and E2 and the carboxy terminus of the nonstructural proteins of rubella virus: comparison with alphaviruses. Gene. 1988;62(1):85–99. doi: 10.1016/0378-1119(88)90582-3. [DOI] [PubMed] [Google Scholar]
- Gatignol A., Buckler-White A., Berkhout B., Jeang K. T. Characterization of a human TAR RNA-binding protein that activates the HIV-1 LTR. Science. 1991 Mar 29;251(5001):1597–1600. doi: 10.1126/science.2011739. [DOI] [PubMed] [Google Scholar]
- Hentze M. W., Rouault T. A., Harford J. B., Klausner R. D. Oxidation-reduction and the molecular mechanism of a regulatory RNA-protein interaction. Science. 1989 Apr 21;244(4902):357–359. doi: 10.1126/science.2711187. [DOI] [PubMed] [Google Scholar]
- Ho-Terry L., Cohen A. Rubella virion polypeptides: characterization by polyacrylamide gel electrophoresis, isoelectric focusing and peptide mapping. Arch Virol. 1982;72(1-2):47–54. doi: 10.1007/BF01314449. [DOI] [PubMed] [Google Scholar]
- Kuhn R. J., Hong Z., Strauss J. H. Mutagenesis of the 3' nontranslated region of Sindbis virus RNA. J Virol. 1990 Apr;64(4):1465–1476. doi: 10.1128/jvi.64.4.1465-1476.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis R., Huang H., Schlesinger S. Engineered defective interfering RNAs of Sindbis virus express bacterial chloramphenicol acetyltransferase in avian cells. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4811–4815. doi: 10.1073/pnas.84.14.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis R., Weiss B. G., Tsiang M., Huang H., Schlesinger S. Deletion mapping of Sindbis virus DI RNAs derived from cDNAs defines the sequences essential for replication and packaging. Cell. 1986 Jan 17;44(1):137–145. doi: 10.1016/0092-8674(86)90492-7. [DOI] [PubMed] [Google Scholar]
- Milligan J. F., Groebe D. R., Witherell G. W., Uhlenbeck O. C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987 Nov 11;15(21):8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakhasi H. L., Meyer B. C., Liu T. Y. Rubella virus cDNA. Sequence and expression of E1 envelope protein. J Biol Chem. 1986 Dec 15;261(35):16616–16621. [PubMed] [Google Scholar]
- Nakhasi H. L., Rouault T. A., Haile D. J., Liu T. Y., Klausner R. D. Specific high-affinity binding of host cell proteins to the 3' region of rubella virus RNA. New Biol. 1990 Mar;2(3):255–264. [PubMed] [Google Scholar]
- Nakhasi H. L., Thomas D., Zheng D. X., Liu T. Y. Nucleotide sequence of capsid, E2 and E1 protein genes of Rubella virus vaccine strain RA27/3. Nucleic Acids Res. 1989 Jun 12;17(11):4393–4394. doi: 10.1093/nar/17.11.4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niesters H. G., Strauss J. H. Defined mutations in the 5' nontranslated sequence of Sindbis virus RNA. J Virol. 1990 Sep;64(9):4162–4168. doi: 10.1128/jvi.64.9.4162-4168.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niesters H. G., Strauss J. H. Mutagenesis of the conserved 51-nucleotide region of Sindbis virus. J Virol. 1990 Apr;64(4):1639–1647. doi: 10.1128/jvi.64.4.1639-1647.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oker-Blom C., Ulmanen I., Käriäinen L., Pettersson R. F. Rubella virus 40S genome RNA specifies a 24S subgenomic mRNA that codes for a precursor to structural proteins. J Virol. 1984 Feb;49(2):403–408. doi: 10.1128/jvi.49.2.403-408.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault T. A., Hentze M. W., Caughman S. W., Harford J. B., Klausner R. D. Binding of a cytosolic protein to the iron-responsive element of human ferritin messenger RNA. Science. 1988 Sep 2;241(4870):1207–1210. doi: 10.1126/science.3413484. [DOI] [PubMed] [Google Scholar]
- Strauss J. H., Strauss E. G. Evolution of RNA viruses. Annu Rev Microbiol. 1988;42:657–683. doi: 10.1146/annurev.mi.42.100188.003301. [DOI] [PubMed] [Google Scholar]
- Takkinen K., Vidgren G., Ekstrand J., Hellman U., Kalkkinen N., Wernstedt C., Pettersson R. F. Nucleotide sequence of the rubella virus capsid protein gene reveals an unusually high G/C content. J Gen Virol. 1988 Mar;69(Pt 3):603–612. doi: 10.1099/0022-1317-69-3-603. [DOI] [PubMed] [Google Scholar]
- Tsiang M., Weiss B. G., Schlesinger S. Effects of 5'-terminal modifications on the biological activity of defective interfering RNAs of Sindbis virus. J Virol. 1988 Jan;62(1):47–53. doi: 10.1128/jvi.62.1.47-53.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidgren G., Takkinen K., Kalkkinen N., Käriäinen L., Pettersson R. F. Nucleotide sequence of the genes coding for the membrane glycoproteins E1 and E2 of rubella virus. J Gen Virol. 1987 Sep;68(Pt 9):2347–2357. doi: 10.1099/0022-1317-68-9-2347. [DOI] [PubMed] [Google Scholar]
- Waxham M. N., Wolinsky J. S. Immunochemical identification of rubella virus hemagglutinin. Virology. 1983 Apr 15;126(1):194–203. doi: 10.1016/0042-6822(83)90471-3. [DOI] [PubMed] [Google Scholar]
- Zheng D. X., Dickens L., Liu T. Y., Nakhasi H. L. Nucleotide sequence of the 24S subgenomic messenger RNA of a vaccine strain (HPV77) of rubella virus: comparison with a wild-type strain (M33). Gene. 1989 Oct 30;82(2):343–349. doi: 10.1016/0378-1119(89)90061-9. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]