Abstract
Sik, the mouse homologue of the breast tumor kinase Brk, is expressed in differentiating cells of the gastrointestinal tract and skin. We examined expression and activity of Sik in primary mouse keratinocytes and a mouse embryonic keratinocyte cell line (EMK). Calcium-induced differentiation of these cells has been shown to be accompanied by the activation of tyrosine kinases and rapid phosphorylation of a 65-kDa GTPase-activating protein (GAP)-associated protein (GAP-A.p65). We demonstrate that Sik is activated within 2 min after calcium addition in primary keratinocytes and EMK cells. In EMK cells, Sik binds GAP-A.p65, and this interaction is mediated by the Sik Src homology 2 domain. Although Sik directly complexes with GAP-A.p65, overexpression of wild-type or kinase defective Sik in EMK cells does not lead to detectable changes in GAP-A.p65 phosphorylation. These data suggest that Sik is not responsible for phosphorylation of GAP-A.p65. GAP-A.p65 may act as an adapter protein, bringing Sik into proximity of an unidentified substrate. Overexpression of Sik in EMK cells results in increased expression of filaggrin during differentiation, supporting a role for Sik in differentiation.
Sik is a nonmyristoylated Src-related intracellular tyrosine kinase with Src homology 2 (SH2) and SH3 domains and a very short unique amino terminus (1, 2). Its expression is epithelial-specific and developmentally regulated and was first detected at mouse embryonic day 15.5 in the differentiating granular layer of the skin (2). In adult skin, Sik is restricted to the differentiating suprabasal layers. Sik is the mouse homologue of the breast tumor kinase Brk, which is expressed in differentiating cells of normal human colon and skin (X. Llor and A.L.T., unpublished results). Increased levels of Brk expression have been found in breast tumors (3, 4) and some metastatic melanoma cell lines (5).
To begin to determine the function of Sik, we examined its role during differentiation of cultured mouse keratinocytes. In low Ca2+ medium, these cells remain undifferentiated. Addition of Ca2+ to levels found in standard medium induces tyrosine kinase activity (6), desmosome formation, cell stratification, inhibition of cell proliferation (7, 8), and expression of differentiation markers (9, 10). Cornified envelopes form and cells are shed into the medium (8). Ca2+-induced in vitro differentiation mimics in vivo differentiation, where increased levels of intracellular Ca2+ have been detected in the differentiating layers of skin (11). Inhibitors of tyrosine kinases interfere with Ca2+-induced keratinocyte differentiation, underscoring an important role for these proteins (6).
Within 5 min after addition of Ca2+ to undifferentiated keratinocytes, a 65-kDa protein that binds to the Ras-GTPase activating protein (GAP) is tyrosine-phosphorylated (6). GAP binds to a variety of phosphorylated proteins including p190 (12) and the GAP-associated protein p62 (13), which was recently cloned and named Dok (14, 15). Dok is a substrate of several kinases including v-Abl (14), and it is constitutively phosphorylated in Bcr-Abl-expressing hematopoietic cell progenitors of chronic myelogenous leukemia patients (15). Dok contains a putative pleckstrin homology domain that may mediate protein–protein interactions and binding to lipids and target the protein to the membrane (14, 15).
Although previously identified as the protein now known as Dok (16), it has been suggested that the 65-kDa protein (GAP-A.p65) that is rapidly phosphorylated in response to Ca2+ in keratinocyte cultures is distinct (17). GAP-A.p65 is not recognized by the monoclonal antibody 2C4 that specifically reacts with Dok (17). It does not bind poly(U)-Sepharose and is distinct from SAM68, an RNA binding protein that is a substrate for c-Src during mitosis (17). It was found that Ca2+ addition induces rapid phosphorylation of GAP-A.p65 but not Dok or SAM68 in the mouse C5N keratinocyte cell line (17).
Two distinct tyrosine kinase activities, the first appearing within minutes and the second hours after Ca2+ addition, are associated with keratinocyte differentiation. The latter activity belongs at least in part to the Src-family kinase Fyn, which is activated hours after Ca2+ addition and has been shown to play a role in the normal differentiation response (18). Kinases responsible for the early tyrosine kinase activity and the rapid phosphorylation of GAP-A.p65 have not been identified. In this study, we examined Sik activity after Ca2+ addition to mouse keratinocytes, the association of Sik with GAP-A.p65, and the role of Sik during keratinocyte differentiation.
MATERIALS AND METHODS
Cells and Antibodies.
The EMK embryonic mouse keratinocyte cell line was a gift of K. Turksen (Loeb Medical Research Institute, Ontario, Canada). Primary keratinocytes were isolated from newborn Sencar mice (19) and used within 1 week of plating. The retrovirus packaging cell line BOSC23 (20) was grown in gpt selection medium (21).
Polyclonal anti-Sik antibodies sc-915 and sc-916 were obtained from Santa Cruz Biotechnology. Immunoblotting was performed with a combination of sc-915 and sc-916 for increased sensitivity. The sc-916 antibody was used for all Sik immunoprecipitation experiments.
Antibodies obtained commercially include polyclonal anti-GAP, Fyn, and Src (Santa Cruz Biotechnology), the anti-phosphotyrosine antibody RC-20H (Transduction Laboratories, Lexington, KY), monoclonal anti-β-actin (Sigma), mouse involucrin, and keratin 1 (Babco, Richmond, CA). Antibodies received as gifts, include RQ19 against Rak (R. Craven and E. T. Liu, Univ. of North Carolina), anti- loricrin (E. Fuchs, Univ. of Chicago), anti-p85 regulatory subunit of human PI-3 kinase (L.C. Cantley, Harvard Medical School), and anti-mouse filaggrin (G.P. Dotto, Harvard Medical School).
Immunoprecipitations, in Vitro Kinase Assays, Immunoblotting, and Northern Blots.
Immunoprecipitations, immune complex protein kinase assays, and immunoblotting were done as described (22). One milligram of total cellular protein was used for immunoprecipitations with the anti-Sik and Rak antibodies, and 200 μg was used for immunoprecipitations with all other antibodies. Quantitative analysis of 32P incorporation was done by scanning dried gels with a Betascope Blot Analyzer (Betagen, Waltham, MA). For immunoblotting, cells were lysed in 1× SDS sample buffer and 30 μg of total protein per lane was subjected to SDS/PAGE. RNA was isolated using TRIzol (GIBCO/BRL), and Northern blot hybridizations were performed as described (2). A rat keratinocyte transglutaminase cDNA probe was a gift of R. H. Rice (Univ. of California, Davis).
Sik–Glutathione S-Transferase (GST) Fusion Proteins, in Vitro Binding, and Overlay Assays.
An NcoI–XhoI fragment containing the full-length sik cDNA (2) was inserted into the pGEX-KG expression vector (23). The GST–Sik SH3 domain was amplified by using oligonucleotides 5′-CAAGCAGGCCACAGCTGACT-3′ and 5′-GCGCTCGAGTCACACAGTTTCCTTCTCAGCCA-3′ and the sik cDNA as template. This was digested by NcoI and XhoI and subcloned into pGEX-KG. For cloning of the GST–Sik SH3/2 domain, the NcoI–Eco47III fragment of sik cDNA was subcloned into pet25b(+) (Novagen) by using NcoI and a filled-in NotI site. The NcoI–XhoI fragment from this plasmid was then subcloned into pGEX-KG. The GST–Sik SH2 domain construct was made by using PCR and the oligonucleotides 5′-GCGCCATGGAACCGTGGTTCTTTGGTT-3′ and 5′-GCTAGTTATTGCTCAGCGG-3′ and the pet25b(+) plasmid containing the Sik SH3/2 domains as a template. The amplified fragment was digested with NcoI and XhoI and ligated into pGEX-KG. Integrity of constructs was verified by sequencing. Fusion proteins were expressed as described (23). GST–GRB2 fusion protein was purchased from Santa Cruz Biotechnology.
In vitro protein binding assays and the overlay assay were performed as described (22). GST fusion proteins were labeled with biotin by using the protein biotinylation module RPN 2202 (Amersham) and the recommended protocol, but 2 mM DTT was added to all solutions.
Phosphopeptide Mapping.
Confluent EMK cells were labeled for 3 h with 32Pi (Amersham) at 1.5 mCi/ml (1 Ci = 37 GBq) in phosphate-free chelex-treated DMEM with 4% dialyzed fetal calf serum. Ca2+ was added 10 min prior to cell lysis. 32P-labeled Sik-A.p65 and GAP-A.p65 were separated by PAGE, transferred to nitrocellulose, and visualized by autoradiography. The 65-kDa protein bands were excised and the phosphotryptic peptide mapping procedure was performed as described (22).
Transfections and Infections.
The sik cDNA was subcloned into pAlter (Promega) by using BamHI and KpnI sites. The 3′ nontranslated region was deleted by partial digestion with SstI and self-ligation. Next, the sik coding sequence was cloned into the EcoRI site of the pLXSN retroviral expression vector (24). The oligonucleotide-mediated altered sites in vitro mutagenesis system (Promega) was used for preparation of the mutant kinase-deficient sik cDNA. The oligonucleotide used for mutagenesis 5′-GTGGCTGTGATGGTGATCTCT-3′ has a A → T substitution that changes the lysine at position 219 of wild-type Sik to methionine. Transfections and retroviral infection of EMK cells were done as described (20).
RESULTS
Rapid Activation of Sik During Ca2+-Induced Differentiation.
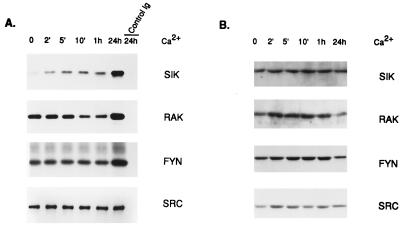
Activities of Sik, Rak, Fyn, and Src were measured after Ca2+ addition to primary keratinocytes by using immunoprecipitation coupled with in vitro kinase assays and enolase as an exogenous substrate for tyrosine phosphorylation (Fig. 1A). Significant activation of Sik occurred rapidly, with Sik activity increasing approximately 2-fold within 2 min after Ca2+ addition. Sik activity continued to increase as differentiation proceeded, and by 24 h its activity increased 5- to 7-fold. Protein levels were determined by immunoblotting and the amount of each protein did not change significantly (Fig. 1B).
Figure 1.
Sik is rapidly activated during differentiation of primary mouse keratinocytes. (A) Immunoprecipitations at different times after Ca2+ addition with Sik-, Rak-, Fyn-, and Src-specific polyclonal antibodies or control rabbit Ig followed by in vitro kinase assays using enolase as a substrate. Reaction products were separated by SDS/PAGE and visualized by autoradiography. (B) Immunoblot of total cellular protein isolated at different times after Ca2+ addition probed with Sik-, Rak-, Fyn-, and Src-specific antibodies. Primary antibodies were detected with horseradish peroxidase-conjugated anti-rabbit antibodies and ECL.
Rak (or Frk) (25, 26) is a nonmyristoylated intracellular kinase that shares significant homology with Sik (2). It is expressed in epithelial cell lines and tissues (25). We did not detect Rak activation at early stages of keratinocyte differentiation. By 24 h Rak activity increased approximately 2-fold. The apparent decrease in Rak activity at 10 min and 1 h was not consistently observed.
We detected a 5- to 6-fold increase in Fyn tyrosine kinase activity by 24 h after Ca2+ addition as reported (18). We did not detect significant activation of Fyn after 1 h. In agreement with Calautti et al. (18), no increase in mouse Src activity was detected after Ca2+ addition to primary keratinocytes.
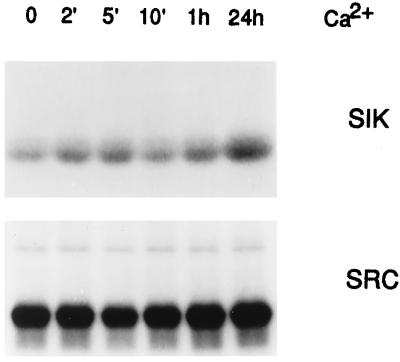
We also examined Sik activity in a cell line derived from embryonic mouse keratinocytes (EMK cells). Sik is rapidly activated in EMK cells after Ca2+ addition, and Src activity remains constant (Fig. 2). This spontaneously immortalized cell line remains undifferentiated in low Ca2+ medium. EMK cells are capable of differentiating after addition of Ca2+, and this is accompanied by stratification and induction of differentiation markers (see Fig. 8).
Figure 2.
Sik is activated during Ca2+-induced differentiation of EMK cells. Protein extracts from EMK cells at different times after Ca2+ addition were immunoprecipitated with Sik- or Src-specific antibodies, and in vitro kinase assays were performed by using enolase as a substrate. Reaction products were analyzed by SDS/PAGE followed by autoradiography.
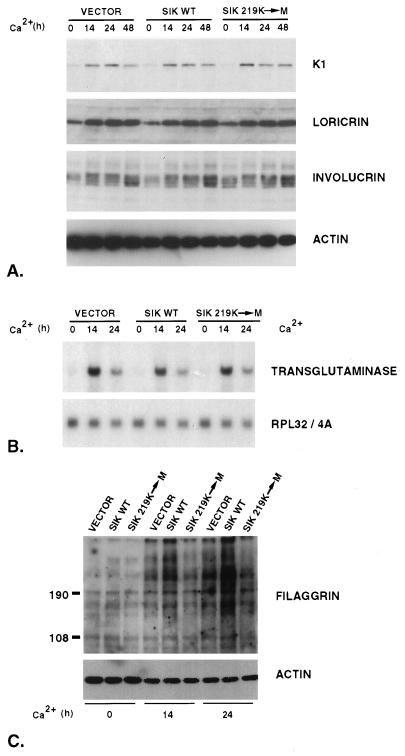
Figure 8.
Differentiation marker expression following Ca2+ treatment of stably transfected EMK cells expressing wild-type (SIK WT) or kinase-deficient Sik, where the conserved lysine in the catalytic domain has been changed to methionine (SIK 219K-M). Control cells contain the pLXSN retroviral expression vector alone (VECTOR). (A) Total protein was extracted from cells grown in low Ca2+ medium (0 h) or incubated in high Ca2+ medium for 14, 24, or 48 h. Immunoblotting was performed with keratin 1, loricrin, involucrin, or β-actin antibody. (B) Induction of transglutaminase mRNA expression during EMK cell differentiation. Total RNA extracted from cells grown in low Ca2+ medium (0) or in high Ca2+ medium for 14 or 24 h was hybridized with a probe for rat keratinocyte transglutaminase. As a control, the membrane was stripped and rehybridized with a probe for the ribosomal gene rpL32/4A (RPL32/4A) (31). (C) Increased filaggrin levels in differentiating cells overexpressing wild-type Sik. Equal amounts of total proteins from cells grown in low Ca2+ medium (0 h) or in high Ca2+ medium for 14 or 24 h were separated by SDS/PAGE, transferred to Immobilone-P membranes, and probed with anti-filaggrin antibody. As a control, the membrane was stripped and probed with β-actin antibody. Size markers (kDa) are at the left.
Sik Associates with a Rapidly Phosphorylated 65-kDa GAP-Associated Protein Through Its SH2 Domain.
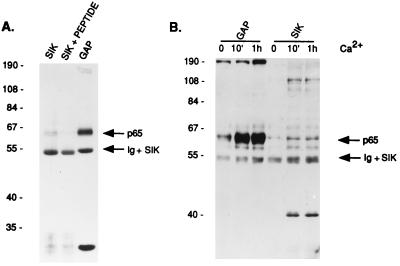
To identify tyrosine-phosphorylated proteins that associate with Sik, we examined proteins in Sik immunoprecipitates by immunoblotting with anti-phosphotyrosine antibody. In differentiated EMK cells, the only prominent tyrosine phosphorylated protein that coimmunoprecipitates with Sik is approximately 65 kDa (Fig. 3A). This protein comigrates with a tyrosine-phosphorylated protein that coimmunoprecipitates with GAP, p65 (GAP-A.p65). In controls where the anti-Sik antibody was incubated with an excess of Sik peptide, no 65-kDa protein was coprecipitated. It was previously shown that GAP-A.p65 becomes rapidly tyrosine-phosphorylated during Ca2+-induced differentiation of the mouse C5N keratinocyte cell line (17). Our data demonstrate that the tyrosine phosphorylation of GAP-A.p65 and the protein that associates with Sik is up-regulated during Ca2+-induced differentiation of EMK cells (Fig. 3B).
Figure 3.
Sik-associated and GAP-associated 65-kDa proteins comigrate and are tyrosine-phosphorylated during differentiation. (A) Proteins from EMK cells incubated for 5 days in high Ca2+ medium were immunoprecipitated with anti-Sik antibodies with or without peptide block or with anti-GAP antibodies. Tyrosine-phosphorylated proteins were detected by using anti-phosphotyrosine antibodies. Sik protein comigrates with the Ig background band. Molecular size markers (kDa) are indicated on the left. (B) Tyrosine-phosphorylated proteins precipitated with anti-Sik or anti-GAP antibodies at different times after Ca2+ addition were detected with anti-phosphotyrosine antibodies.
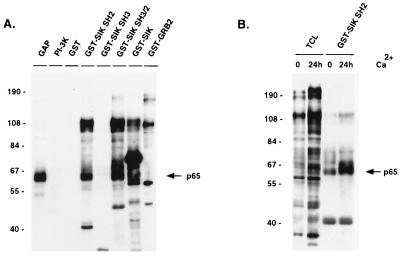
Fusion proteins containing the Sik SH2 domain, the SH3 domain, both the SH2 and SH3 domains and full-length Sik were incubated with lysates from Ca2+-treated EMK cells and then with glutathione-Sepharose. Phosphotyrosine-containing proteins associated with the GST–Sik fusions were detected by anti-phosphotyrosine antibody immunoblotting (Fig. 4A). The smallest bands visible in the GST–SIK SH2, GST–SIK SH3, and GST–SIK SH3/2 lanes are the GST fusion proteins themselves. A 65-kDa protein was one of the major tyrosine-phosphorylated proteins that specifically associated with the Sik SH2 domain. Binding was not detected with GST alone or with the GST–Sik SH3 and GST–GRB2 fusion proteins. Other proteins interacting with the Sik SH2 domain are approximately 100 and 120 kDa in size. The Sik SH3 domain did not bind significantly to phosphotyrosine-containing proteins. In addition to binding all proteins that bound the Sik SH2 domain alone, the Sik SH3/2 domain also bound an additional protein of approximately 68 kDa. The major tyrosine-phosphorylated protein seen after binding of proteins with full-length GST–Sik is the GST–Sik fusion protein itself, although binding of the 65-kDa protein is also detected. After Ca2+ addition, increased tyrosine phosphorylation of many proteins in total cell lysates was observed (Fig. 4B). There is a significant increase in tyrosine phosphorylation of the 65-kDa protein associated with the Sik SH2 domain by 24 h after Ca2+ addition (Fig. 4B).
Figure 4.
Sik-associated p65 binds to the SH2 domain of Sik. (A) Tyrosine-phosphorylated proteins immunoprecipitated with GAP or phosphatidylinositol 3-kinase (PI-3K) antibodies or with GST fusion proteins 10 min after Ca2+ addition were detected by using anti-phosphotyrosine antibodies coupled to horseradish peroxidase and ECL. Tyrosine-phosphorylated proteins binding the PI-3K regulatory subunit (PI-3K) or GST–GRB2 were examined as controls for specificity of 65-kDa protein binding to Sik. In addition to cellular proteins, the fusion proteins themselves are visible (smallest bands in GST–SIK SH2, SH3, and SH3/2 lanes, and the major band in the GST–Sik lane). (B) Increased tyrosine phosphorylation of proteins in total cell lysates (TCL) and proteins that bind the GST–Sik SH2 fusion protein after Ca2+ addition to EMK cells. Phosphotyrosine-containing proteins were detected as described in A. There is a significant increase in the tyrosine phosphorylation of the 65-kDa protein associated with Sik (arrow).
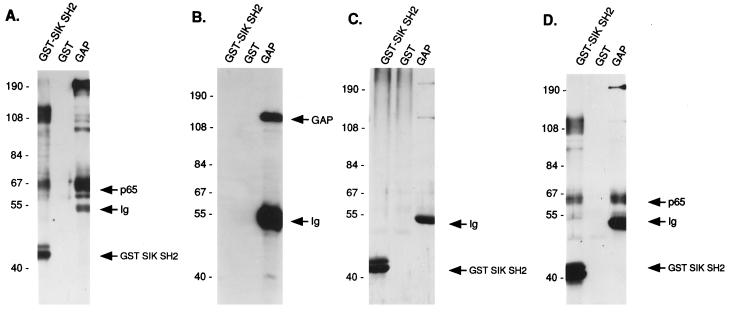
Direct Sik binding to GAP-A.p65 was tested by using an overlay assay. The GST–Sik SH2 domain and anti-GAP antibody were used to precipitate the 65-kDa tyrosine-phosphorylated proteins from extracts of EMK cells incubated in high Ca2+ medium for 10 min. Precipitated proteins were probed with biotinylated GST or GST–Sik SH2 fusion protein (Fig. 5 C and D). As a control, parallel blots were probed with anti-phosphotyrosine (Fig. 5A) or anti-GAP antibody (Fig. 5B). The tyrosine-phosphorylated GAP-associated and Sik-associated 65-kDa proteins comigrate. The Sik SH2 domain bound with a similar affinity to the 65-kDa protein precipitated with the GST–Sik SH2 fusion protein, and the 65-kDa protein immunoprecipitated with anti-GAP antibody (Fig. 5D). GST alone did not bind to the 65-kDa protein (Fig. 5C). Thus, Sik binds directly to GAP-A.p65 through it SH2 domain. GAP was not precipitated by the GST–Sik SH2 fusion protein, although GAP precipitated with anti-GAP antibody was readily detectable (Fig. 5B). We were also unable to detect GAP in anti-Sik immunoprecipitates (data not shown).
Figure 5.
Sik SH2 domain binds directly to GAP-associated p65 in an overlay assay. Precipitations were done with keratinocyte lysates at 10 min after Ca2+ addition by using anti-GAP antibody or GST or the GST–Sik SH2 domain. Precipitates were washed and split into four samples that were subjected to SDS/PAGE and transferred to Immobilone-P membranes. Filters were probed with anti-phosphotyrosine antibody (A), anti-GAP antibody (B), biotin-labeled GST (C), or biotin labeled GST–Sik SH2 domain fusion protein (D). Primary antibodies were detected as described. Bound GST and GST fusion proteins were detected by preformed avidin and biotinylated horseradish peroxidase complex and ECL.
To determine whether the 65-kDa proteins that associate with the Sik SH2 domain and GAP are identical, phosphotryptic peptide mapping was performed. Confluent EMK cells were labeled for 3 h with 32Pi at 1.5 mCi/ml and then treated with Ca2+ for 10 min. Labeled 65-kDa proteins that bound to the GST–Sik SH2 domain or coimmunoprecipitated with GAP were separated by SDS/PAGE, transferred to nitrocellulose membrane, visualized by autoradiography, and then excised from the membrane (Fig. 6A). The phosphorylated proteins were digested with trypsin and separated by electrophoresis and chromatography. The phosphotryptic peptides generated by digestion of Sik-A.p65 and GAP-A.p65 appear identical (Fig. 6B). Separation of a mixture of the Sik-associated and GAP-associated proteins revealed a completely overlapping pattern of peptides, suggesting that the two proteins are identical.
Figure 6.
Phosphotryptic peptide maps of Sik-associated p65 and GAP-A.p65 appear identical. (A) Confluent EMK cells were labeled for 3 h with 32Pi at 1.5 mCi/ml, followed by addition of Ca2+ for 10 min and lysis. Proteins precipitated from the cell lysates with GAP antibodies or the GST–Sik SH2 domain fusion protein were separated by SDS/PAGE, transferred to Immobilone-P membrane, and visualized by autoradiography. (B) The 65-kDa proteins from A were excised and phosphotryptic peptide maps were generated. Maps of Sik-associated p65 (Sik-A.p65), GAP-associated p65 (GAP-A.p65), and a mixture of these two proteins are shown (MIX). Sample origins are indicated by arrows, and the anodes and cathodes for electrophoresis are indicated by the + and − signs.
Overexpression of Sik Does Not Alter GAP-A.p65 Phosphorylation but Results in Increased Filaggrin Expression.
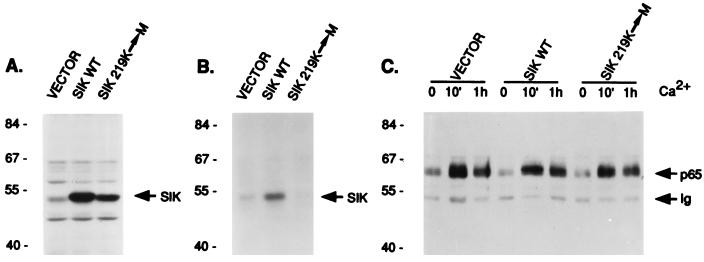
To determine whether Sik is responsible for phosphorylation of GAP-A.p65, we examined tyrosine phosphorylation of GAP-A.p65 in EMK cell lines overexpressing wild-type Sik and kinase defective Sik. A conserved lysine at position 219 in the putative ATP binding region of Sik was changed to methionine, because mutation generally produces a kinase-defective enzyme (27). Stable pools of EMK cells containing the pLXSN retroviral vector alone or with wild-type or kinase-deficient Sik were selected by using G418. Immunoblot analysis revealed a significant increase in the amount of Sik protein in the cell lines transfected with wild-type and kinase-deficient sik cDNAs (Fig. 7A). Transfected cells were incubated in high Ca2+ medium for 1 h and Sik activity was determined by using the immune complex-in vitro phosphorylation assay. Sik activity increased in cells transfected with wild-type Sik and decreased in cells transfected with kinase-negative Sik (Fig. 7B). The decrease in total Sik activity in cells expressing mutant Sik may be due to a dominant negative role of kinase-deficient enzyme. Antibodies used for Sik immunoprecipitations precipitated both endogenous and recombinant Sik proteins.
Figure 7.
Rapid phosphorylation of GAP-A.p65 is not affected by overexpression of wild-type Sik or kinase-deficient Sik. Stable EMK cell lines containing the pLXSN vector alone (VECTOR) or expressing wild-type (SikWT) or kinase-deficient Sik (Sik 219K-M) were selected. (A) Expression of transfected Sik was detected by immunoblotting of total cellular proteins from the stable cell lines with anti-Sik antibody. (B) Sik kinase activity was measured by immunoprecipitation followed by in vitro kinase assays using total protein from EMK cell lines incubated for 1 h in high Ca2+ medium. Autophosphorylated Sik was detected by autoradiography. (C) Phosphorylation of GAP-A.p65 at 10 min and 1 h after Ca2+ addition to EMK cell lines was measured by immunoprecipitation with anti-GAP antibody followed by immunoblotting with anti-phosphotyrosine.
No change in phosphorylation of GAP-A.p65 was detected in EMK cells overexpressing wild-type and mutant Sik (Fig. 7C). We also detected no change in phosphorylation of the 65-kDa Sik-associated protein from these cells that bound the GST–Sik SH2 domain fusion protein (data not shown). These data suggest that Sik is unlikely to be the primary tyrosine kinase responsible for phosphorylating GAP-A.p65 in vivo.
The induction of several differentiation markers was examined after Ca2+ addition to the EMK cell lines that overexpress wild-type and kinase-defective Sik, including keratin K1, loricrin, involucrin, and keratinocyte transglutaminase. No significant differences in expression of these markers was detected in cells over expressing wild-type and kinase-defective Sik (Fig. 8 A and B). In contrast, expression of filaggrin, a protein expressed in the granular layer and a component of terminally differentiated cornified cells, was significantly higher in cells overexpressing wild-type Sik, at 14 and 24 h after Ca2+ addition (Fig. 8C). Filaggrin serves to aggregate keratin filaments and is first synthesized as a very large precursor protein (profilaggrin), which is cleaved into multiple subunits (28, 29). The multiple bands detected by immunoblotting reflect the proteolytic processing of the protein. Cells overexpressing kinase-defective Sik also appear to express slightly less filaggrin than cells containing the vector alone at 24 h.
DISCUSSION
Ca2+-induced differentiation of cultured keratinocytes is accompanied by early and late tyrosine kinase activities (18). Fyn activation corresponds with the later activity and is important for normal differentiation (18). Herein we demonstrate that two additional tyrosine kinases, Rak and Sik, are also activated during Ca2+-induced differentiation of mouse keratinocytes. Like Fyn, Rak is activated at later stages of differentiation. Rak is an epithelial-cell-specific nuclear tyrosine kinase that associates with the retinoblastoma protein and has growth-suppressing properties (30).
Sik represents the first early kinase to be identified during keratinocyte differentiation. It is activated within 2 min after Ca2+ addition. Although Sik directly binds to GAP-A.p65, it does not appear responsible for the rapid phosphorylation of GAP-A.p65 in EMK cells, suggesting that other unidentified tyrosine kinases are also rapidly activated. It is possible that binding of Sik to phosphorylated GAP-A.p65 may result in changes in the conformation of Sik, leading to its activation. The association of Sik with a GAP-associated protein may link Sik to the Ras pathway.
Currently, the relationship between GAP-A.p65 and Dok is not clear, and it is still possible that these two GAP binding proteins are related. Although v-Abl has been shown to phosphorylate Dok, c-Abl does not appear responsible for GAP-A.p65 phosphorylation, because no change in c-Abl activity was noted after Ca2+ addition to keratinocytes (18). A role for Dok as a docking protein for membrane localization has been proposed (14, 15, 22), and GAP-A.p65 may also act as an adapter or docking protein linking Sik to its substrate in the cell.
After Ca2+ addition, levels of filaggrin expression were markedly induced in cells overexpressing wild-type Sik. In fyn-deficient mouse keratinocytes, filaggrin levels were suppressed after Ca2+-induced differentiation, indicating that proper expression of filaggrin is dependent on the activities of tyrosine kinases (18). Filaggrin, which is expressed in the granular cell layer, is the major component of keratohyalin granules. Sik expression is first detected in the mouse embryo at 15.5 days of gestation, in the forming granular layer of the skin (2). The coincidental induction of Sik expression with formation of the granular layer and the ability of Sik to modulate filaggrin levels supports a physiologically relevant role for Sik during skin differentiation.
Acknowledgments
This work was supported by National Institutes of Health Grant DK44525 and a Schweppe Foundation Career Development Award (A.L.T.).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: SH, Src homology; GST, glutathione S-transferase.
References
- 1.Siyanova E Y, Serfas M S, Mazo I A, Tyner A L. Oncogene. 1994;9:2053–2057. [PubMed] [Google Scholar]
- 2.Vasioukhin V, Serfas M S, Siyanova E Y, Polonskaia M, Costigan B L, Liu B, Thomason A, Tyner A L. Oncogene. 1995;10:349–357. [PubMed] [Google Scholar]
- 3.Barker K T, Jackson L E, Crompton M R. Oncogene. 1997;15:799–805. doi: 10.1038/sj.onc.1201241. [DOI] [PubMed] [Google Scholar]
- 4.Mitchell P J, Barker K T, Martindale J E, Kamalati T, Lowe P N, Page M J, Gusterson B A, Crompton M R. Oncogene. 1994;9:2383–2390. [PubMed] [Google Scholar]
- 5.Easty D J, Mitchell P J, Patel K, Florenes A A, Spritz R A, Bennet D C. Int J Cancer. 1997;71:1061–1065. doi: 10.1002/(sici)1097-0215(19970611)71:6<1061::aid-ijc24>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 6.Filvaroff E, Stern D F, Dotto G P. Mol Cell Biol. 1990;10:1164–1173. doi: 10.1128/mcb.10.3.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hennings H, Michael D, Cheng C, Steinert P, Holbrook K, Yuspa S H. Cell. 1980;19:245–254. doi: 10.1016/0092-8674(80)90406-7. [DOI] [PubMed] [Google Scholar]
- 8.Hennings H, Holbrook K A. Exp Cell Res. 1983;143:127–142. doi: 10.1016/0014-4827(83)90115-5. [DOI] [PubMed] [Google Scholar]
- 9.Yuspa S H, Kilkenny A E, Steinert P M, Roop D R. J Cell Biol. 1989;109:1207–1217. doi: 10.1083/jcb.109.3.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hohl D, Lichti U, Breitkreutz D, Steinert P M, Roop D R. J Invest Dermatol. 1991;96:414–418. doi: 10.1111/1523-1747.ep12469779. [DOI] [PubMed] [Google Scholar]
- 11.Menon G K, Grayson S, Elias P M. J Invest Dermatol. 1985;84:508–512. doi: 10.1111/1523-1747.ep12273485. [DOI] [PubMed] [Google Scholar]
- 12.Settleman J, Narasimhan V, Foster L C, Weinberg R A. Cell. 1992;69:539–549. doi: 10.1016/0092-8674(92)90454-k. [DOI] [PubMed] [Google Scholar]
- 13.Ellis C, Measday V, Moran M F. Methods Enzymol. 1995;255:179–192. doi: 10.1016/s0076-6879(95)55022-4. [DOI] [PubMed] [Google Scholar]
- 14.Yamanashi Y, Baltimore D. Cell. 1997;88:205–211. doi: 10.1016/s0092-8674(00)81841-3. [DOI] [PubMed] [Google Scholar]
- 15.Carpino N, Wisniewski D, Strife A, Marshak D, Kobayashi R, Stillman B, Clarkson B. Cell. 1997;88:197–204. doi: 10.1016/s0092-8674(00)81840-1. [DOI] [PubMed] [Google Scholar]
- 16.Filvaroff E, Calautti E, McCormick F, Dotto G P. Mol Cell Biol. 1992;12:5319–5328. doi: 10.1128/mcb.12.12.5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Medema J P, de Vries-Smits A M M, Bos J L. Oncogene. 1995;11:757–762. [PubMed] [Google Scholar]
- 18.Calautti E, Missero C, Stein P L, Ezzell R M, Dotto G P. Genes Dev. 1995;9:2279–2291. doi: 10.1101/gad.9.18.2279. [DOI] [PubMed] [Google Scholar]
- 19.Yuspa S H, Harris C C. Exp Cell Res. 1974;86:95–105. doi: 10.1016/0014-4827(74)90653-3. [DOI] [PubMed] [Google Scholar]
- 20.Pear W S, Nolan G P, Scott M L, Baltimore D. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mulligan R C, Berg P. Proc Natl Acad Sci USA. 1981;78:2072–2076. doi: 10.1073/pnas.78.4.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neet K, Hunter T. Mol Cell Biol. 1995;15:4908–4920. doi: 10.1128/mcb.15.9.4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guan K L, Dixon J E. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 24.Miller D, Garcia V, Lynch C. Methods Enzymol. 1993;217:581–599. doi: 10.1016/0076-6879(93)17090-r. [DOI] [PubMed] [Google Scholar]
- 25.Cance W G, Craven R J, Bergman M, Xu L, Aitalo K, Liu E T. Cell Growth Diff. 1994;5:1347–1355. [PubMed] [Google Scholar]
- 26.Lee J, Wang Z, Luoh S-M, Wood W L, Scadden D T. Gene. 1994;138:247–251. doi: 10.1016/0378-1119(94)90817-6. [DOI] [PubMed] [Google Scholar]
- 27.Thomas J E, Soriano P, Brugge J S. Science. 1991;254:568–571. doi: 10.1126/science.1719633. [DOI] [PubMed] [Google Scholar]
- 28.Resing K A, Walsh K A, Haugen-Scofield J, Dale B A. J Biol Chem. 1989;264:1837–1845. [PubMed] [Google Scholar]
- 29.Manabe M, Sanchez M, Sun T T, Dale B A. Differentiation. 1991;48:43–50. doi: 10.1111/j.1432-0436.1991.tb00241.x. [DOI] [PubMed] [Google Scholar]
- 30.Craven R J, Cance W G, Liu E T. Cancer Res. 1995;55:3969–3972. [PubMed] [Google Scholar]
- 31.Dudov K P, Perry R P. Cell. 1984;37:457–468. doi: 10.1016/0092-8674(84)90376-3. [DOI] [PubMed] [Google Scholar]