Abstract
FKBP52 (HSP56, p59, HBI) is the 59-kDa immunosuppressant FK506-binding protein and has peptidyl prolyl isomerase as well as a chaperone-like activity in vitro. FKBP52 associates with the heat shock protein HSP90 and is included in the steroid hormone receptor complexes in vivo. FKBP52 possesses a well conserved phosphorylation site for casein kinase II (CK2) that was previously shown to be associated with HSP90. Here we examined whether FKBP52 is phosphorylated by CK2 both in vivo and in vitro. Recombinant rabbit FKBP52 was phosphorylated by purified CK2. We expressed and purified deletion mutants of FKBP52 to determine the site(s) phosphorylated by CK2. Thr-143 in the hinge I region was identified as the major phosphorylation site for CK2. A synthetic peptide corresponding to this region was phosphorylated by CK2, and the peptide competitively inhibited the phosphorylation of other substrates by CK2. The [32P]phosphate labeling of FKBP52-expressing cells revealed that the same site is also phosphorylated in vivo. FK506 binding to FKBP52 did not affect the phosphorylation by CK2 and, conversely, the FK506-binding activity of FKBP52 was not affected by the phosphorylation. Most importantly, CK2-phosphorylated FKBP52 did not bind to HSP90. These results indicate that CK2 phosphorylates FKBP52 both in vitro and in vivo and thus may regulate the protein composition of chaperone-containing complexes such as those of steroid receptors and certain protein kinases.
The 90-kDa heat shock protein, HSP90, is one of the most abundant proteins and is conserved throughout all species (1). Classically, HSP90 was known to associate with several steroid hormone receptors (2) and the Src family of transforming tyrosine kinases (3). HSP90 is now established to be a general molecular chaperone that supports the functionality of a wide variety of signal transducing proteins (4–6). Besides steroid hormone receptors, growing numbers of protein kinases have been shown to associate with HSP90. The list of HSP90-associating kinases begins with Src (3), then casein kinase II (CK2) (7), and now includes many kinases such as Raf (8), sevenless (9), and Cdk4 (10). HSP90 protects CK2 from aggregation and inactivation (7, 11), and functions of several kinases are assisted by HSP90 (12, 13).
Several molecular chaperone-related proteins have been reported to function in concert with HSP90 (5). For example, steroid hormone receptor complexes contain one of the HSP90-associating immunosuppressant-binding proteins, immunophilins (14). These include several FK506-binding proteins such as FKBP52 (15) and a cyclosporin-binding cyclophilin, CyP40 (16). Both of them were recently shown to possess chaperone-like activity in vitro by themselves (17–19).
FKBP52 (also called FKBP59, p59, HBI, or HSP56) was originally identified as the 59-kDa component, with unknown properties, of several steroid hormone receptors (15). Subsequently, this protein was reported to be identical with a high molecular weight FK506-binding protein (20–22). In contrast to FKBP12 complexed with FK506, FK506–FKBP52 complexes do not have calcineurin-inhibiting activity (23, 24). Like other immunophilins, FKBP52 also exhibits an FK506-inhibitable peptidyl prolyl isomerase (rotamase) activity (25). FKBP52 binds ATP/GTP (26), calmodulin (27), and HSP90 (15, 20, 28). Tetratricopeptide repeat (TPR) sequences localized in the C-terminal part of FKBP52 are responsible for the association with HSP90 (28). FKBP52 also interacts, by its FK506-binding domain, with the newly discovered FAP48 (29). FKBP52 has recently been reported to possess rotamase-independent in vitro chaperone-like activity (18), suggesting a role of FKBP52 in protein folding. However, the physiological roles and the functional regulation of FKBP52 in vivo remain unknown. Sequence examination has revealed that FKBP52 contains a well conserved putative phosphorylation consensus sequence for CK2 within the hinge I region (25), suggesting the possibility that CK2 phosphorylates FKBP52.
CK2 is a ubiquitously distributed Ser/Thr kinase and phosphorylates a large number of proteins (30). CK2 is suggested to function as a connection between growth signals and gene expressions; however, its precise physiological roles and regulatory mechanisms remain unknown. CK2 is one of the protein kinases tightly associated with HSP90 (7). Thus, it is conceivable that certain HSP90-binding proteins such as FKBP52 may be phosphorylated by CK2.
In this report, we describe in vivo and in vitro phosphorylation of FKBP52 by CK2, and we have determined one of the phosphorylation sites within the hinge I region. Most importantly, CK2-phosphorylated FKBP52 did not bind to HSP90. We propose that phosphorylation of HSP90-associated proteins by CK2 may be one of the mechanisms that determine the molecular composition of HSP90-containing complexes.
MATERIALS AND METHODS
Buffers.
Lysis buffer is 50 mM Tris⋅HCl/10% (vol/vol) glycerol/100 mM NaF/50 mM NaCl/2 mM EDTA/10 mM sodium pyrophosphate/1 mM DTT/1% Nonidet P-40/2 mM sodium vanadate, pH 8.0. V8 buffer is 125 mM Tris⋅HCl/1 mM EDTA/1 mM 2-mercaptoethanol/0.1% SDS/20% glycerol, pH 6.8. Phosphorylation buffer is 12 mM Tris⋅HCl/9 mM Mops/200 mM NaCl/4% glycerol/12 mM MgCl2/0.02 mM EDTA/0.02 mM DTT, pH 7.6.
Proteins, Chemicals, Peptides, and Antibodies.
CK2 was purified from porcine testis (7). HSP90 was purified from mouse L5178Y cells (31) and further purified with a Poros-heparin column (Perseptive Biosystems, Cambridge, MA). FK506 was provided by Fujisawa Pharmaceutical, Osaka, Japan. FK506 was esterified at the C32-hydroxyl group with an allyloxycarbonyl-protected amino acid, and immobilized onto Affi-Gel 10 (Bio-Rad) after removal of the protective group (32). Pept419 (Phe-135–Gly-149 of rabbit FKBP52) and Pept790 (Lys-182–Pro-201 of rabbit FKBP52) were synthesized and conjugated to keyhole limpet hemocyanin (KLH) (33). Rabbit antibodies against FKBP52 (20) and rabbit anti-HSP90 antiserum (31) were previously described.
Expression and Purification of Recombinant FKBP52.
Full-length FKBP52 (amino acids 1–458) was expressed in Sf9 cells and purified (24). FKBP52 and its deletion mutant expression vectors were described previously (24, 26, 28, 34) except those mentioned below. The mutant FKBP52 (amino acids 240–458) was generated, first by deletion of fragment EcoRV–NdeI from the mutated FKBP52 called HBI-M3 (28). The NdeI site was filled in before ligation to the EcoRV site. The StyI–StyI fragment (base pairs 767-1307) of the resulting plasmid was replaced by that from wild-type FKBP52. Subsequently the insertion (amino acids 240–458) was excised by using EcoRI and inserted into pGEX-1λT (Pharmacia). The mutant FKBP52 (amino acids 240–400) was obtained from the mutant FKBP52 (amino acids 240–458), which was cleaved by BamHI to generate a fragment delimited by the unique BamHI site (located at amino acid residue 400) and by the BamHI site of pGEX-1λT. This fragment was then inserted at the BamHI site of pGEX-1λT. All of them are expressed as glutathione S-transferase (GST) fusion proteins in Escherichia coli and purified (34).
In Vitro Phosphorylation of FKBP52 by CK2.
Purified FKBP52 (1 μg) was incubated with CK2 (40 ng) in the phosphorylation buffer. Radiolabeled ATP (40 μM, 0.2 MBq per assay) was added and the mixture was incubated at 30°C for 30 min. The reaction was stopped by boiling 5 min in the SDS sample buffer.
In Vivo Phosphorylation of FKBP52.
For expression of FKBP52 in mammalian cells, cDNA encoding rabbit FKBP52 (amino acids 6–458) was inserted into the EcoRI site of pSVK3 (Pharmacia). COS7 cells were transfected by electroporation (230 V, 960 μF, 0.4 cm gap) with the expression plasmid. Three days later, cells were labeled for 6 h in phosphate-free Dulbecco’s modified Eagle’s medium (DMEM)/10% FCS supplemented with 4 MBq per dish (100 mm) of [32P]orthophosphate. Cells were solubilized in 400 μl per dish of the lysis buffer supplemented with protease inhibitors. The cell lysates were centrifuged at 15,000 × g for 60 min at 2°C and precleared with Affi-Gel 10. Then the lysates were mixed with FK506–Affi-Gel 10 for 6 h at 4°C. The FK506-beads were washed twice with the lysis buffer, three times with the lysis buffer supplemented with 0.65 M NaCl, and twice with 50 mM Tris⋅HCl, pH 7.4. The FK506-bound proteins were eluted from the gel by boiling with the SDS sample buffer.
V8-Peptide Mapping.
The bands corresponding to the in vivo- and in vitro-phosphorylated FKBP52 were cut out from the gel, crushed in the V8 buffer, and applied onto a discontinuous 15% polyacrylamide gel. Then 10 μl of 0.1 mg/ml V8 protease was overlaid, and the products were electrophoresed.
Gel Electrophoresis.
SDS/PAGE was performed using discontinuous 10% or 5–20% acrylamide gradient gels. For the detection of smaller deletion mutants of FKBP52, 15% acrylamide gels were used. For the detection of KLH-peptide conjugates, 6% acrylamide gels were used.
HSP90-Binding Assay.
HSP90-binding activity of FKBP52 was assayed as described (28) with slight modification. FKBP52 was mixed with HSP90 in a buffer containing 12.5% glycerol, 25 mM Hepes, 50 mM KCl, 15 mM Tris, 30 mM NaCl, 2.5 mM MgCl2, 1 mM EDTA, 10 mM 2-mercaptoethanol, and 0.01% Tween 20, pH 7.6, and incubated 60 min at 30°C. The mixtures were analyzed by native PAGE (4–15% polyacrylamide). The bands were cut out from the gel, homogenized in the V8 buffer, and then subjected to SDS/PAGE.
RESULTS
CK2 Phosphorylated Purified FKBP52 in Vitro.
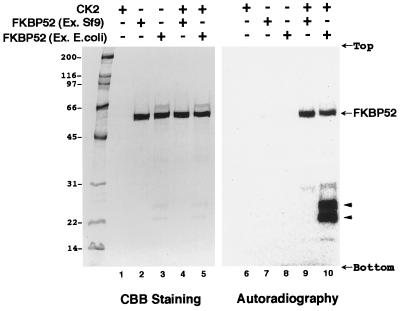
Recombinant full-length rabbit FKBP52 was expressed in Sf9 cells and purified. The FKBP52 obtained was more than 95% pure (Fig. 1, lane 2). FKBP52 was incubated with purified CK2 in the presence of [γ-32P]ATP. Clearly, FKBP52 was phosphorylated in vitro by CK2 (lane 9). No phosphorylation was observed when CK2 was omitted (lane 7), indicating that this phosphorylation is not a result of autophosphorylation or phosphorylation by contaminating kinases. The amount of CK2 (40 ng) was too low to be visualized.
Figure 1.
Phosphorylation of purified FKBP52 by CK2. Recombinant rabbit FKBP52 expressed in Sf9 cells (lanes 2, 4, 7, 9) or expressed in E. coli (lanes 3, 5, 8, 10) was incubated with (lanes 4, 5, 9, 10) or without (lanes 2, 3, 7, 8) CK2 in the presence of [γ-32P]ATP and analyzed by SDS/PAGE. Coomassie brilliant blue (CBB) staining (lanes 1–5) and autoradiography (lanes 6–10) are shown. As a control, CK2 was incubated without FKBP52 (lanes 1 and 6). The molecular masses (kDa) of markers are indicated on the left, and the position of FKBP52 is on the right. The positions of proteolytic fragments of FKBP52 are shown by arrowheads.
The same experiments were performed with bacterially expressed FKBP52. The FKBP52 obtained was more than 90% pure (Fig. 1, lane 3). A small amount of a protein with a slightly higher molecular weight than FKBP52 and faint degradation products around 22–25 kDa could be observed. Bacterially expressed FKBP52 was also a good substrate for CK2 (lane 10, indicated by an arrow). Again, this phosphorylation was not observed when CK2 was omitted (lane 8).
When bacterially expressed FKBP52 was used, prominent phosphorylation bands around 22–25 kDa were observed (Fig. 1, lane 10, arrowheads). These fragments were observed only when the GST-FKBP52 fusion protein was cleaved by thrombin, and the fragments bound specifically to FK506-beads (data not shown). These results indicate that these fragments were thrombin-cleavage by-products derived from the FK506-binding region of FKBP52.
In Vivo Phosphorylation of FKBP52.
The in vitro phosphorylation of FKBP52 by CK2 led us to test if FKBP52 is a phosphoprotein in vivo. COS7 cells were transfected with an FKBP52 expression plasmid, and the expression of exogenous FKBP52 was confirmed by Western blotting (data not shown). The transfected cells were labeled with [32P]orthophosphate, and FK506-binding proteins were isolated with FK506–Affi-Gel 10.
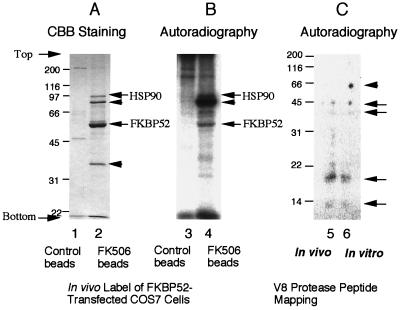
CBB staining revealed that several proteins were specifically isolated by FK506–Affi-Gel 10 (Fig. 2A, lane 2). Among them the most prominent band was the 59-kDa FKBP52 (shown by an arrow). The identity of this 59-kDa band as FKBP52 was confirmed by Western blotting (data not shown). The autoradiography of the same gel as in Fig. 2A is shown in Fig. 2B. FKBP52 was significantly phosphorylated, demonstrating that FKBP52 is a phosphoprotein in vivo.
Figure 2.
Phosphorylation of FKBP52 in vivo. COS7 cells were transfected with an expression plasmid of FKBP52 (amino acids 6–458) and labeled with [32P]orthophosphate. (A) CBB staining of proteins isolated with FK506–Affi-Gel 10 (lane 2) or with control–Affi-Gel 10 (lane 1). The positions of FKBP52 and HSP90 are indicated by arrows. Two additional associated proteins are indicated by arrowheads. (B) Autoradiography of the same gel as A. In vivo phosphorylated FKBP52 and HSP90 are indicated by arrows. (C) V8 phosphopeptide maps of in vivo- (lane 5) and in vitro- (lane 6) phosphorylated FKBP52 are shown by autoradiography. The position of original FKBP52 is indicated by an arrowhead, and phosphopeptides common to in vivo- and in vitro-phosphorylated FKBP52 are indicated by arrows.
Next, the in vivo-phosphorylation site(s) of FKBP52 was compared with that of the in vitro CK2-phosphorylated site(s) by V8-peptide mapping (Fig. 2C). Several major V8-cleavage phosphopeptides (45, 40, 18, and 12 kDa as indicated by arrows) were identical in their molecular mass for the in vivo- (lane 5) and in vitro- (lane 6) phosphorylated FKBP52. This result suggests that FKBP52 is phosphorylated on the same site(s) both in vivo and in vitro by CK2.
Several other proteins, with molecular masses of 90, 80, and 36 kDa, were isolated along with FKBP52 by FK506–Affi-Gel 10 (Fig. 2A, lane 2). The 90-kDa protein was a phosphoprotein and the 80-kDa protein was heavily phosphorylated in vivo (Fig. 2B, lane 4). The 90-kDa protein was identified by Western blotting as HSP90 (data not shown). Other proteins were not characterized further here.
Phosphorylation of FKBP52 Mutants by CK2.
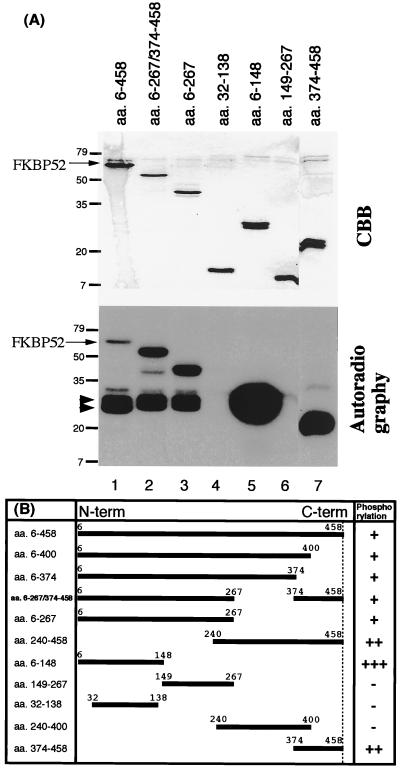
We constructed deletion mutants of FKBP52 and examined if these mutants are phosphorylated by CK2 in vitro. The purified proteins were incubated with CK2 in the presence of [γ-32P]ATP (Fig. 3A). The upper panel shows the CBB staining, and the lower panel shows the autoradiography. FKBP52 (amino acids 6–458), an internal deletion mutant (amino acids 6–267/374–458), and a C-terminal deletion mutant (amino acids 6–267) were phosphorylated by CK2 (lanes 1–3). The degradation fragments (shown by arrowheads) were phosphorylated as observed in Fig. 1. A fragment corresponding to the core region of the FK506-binding domain (amino acids 32–138, lane 4) and a fragment containing the ATP/GTP-binding domain (amino acids 149–267, lane 6) were not at all phosphorylated. In contrast, an N-terminal fragment (amino acids 6–148, lane 5) and a C-terminal fragment (amino acids 374–458, lane 7) were substrates for CK2.
Figure 3.
Phosphorylation of various deletion mutants of FKBP52 by CK2. (A) Various deletion mutants of FKBP52 were phosphorylated in vitro by CK2. CBB staining is shown in the upper panel and autoradiography in the lower panel. Mutants are designated by their amino acid residue numbers on the top of lanes. The positions of wild-type FKBP52 and molecular mass markers (kDa) are shown on the left. The low molecular mass proteolytic fragments of FKBP52 are indicated by arrowheads. (B) Schematic representation of the phosphorylation of FKBP52 mutants by CK2. Mutants are designated by their amino acid residue numbers, and the degrees of the phosphorylation by CK2 are indicated on the right as follows: −, not at all phosphorylated; +, moderately phosphorylated; ++, strongly phosphorylated; and +++, very strongly phosphorylated.
The phosphorylation experiments were performed with several other deletion mutants, and all of the results are summarized in Fig. 3B. On the basis of these results, we concluded that the N-terminal fragment (amino acids 6–148) and the C-terminal fragment (amino acids 374–458) contain phosphorylation site(s), whereas the residual regions of FKBP52 have no CK2-phosphorylation site.
The CK2-Phosphorylation Site Within the N-Terminal Region of FKBP52.
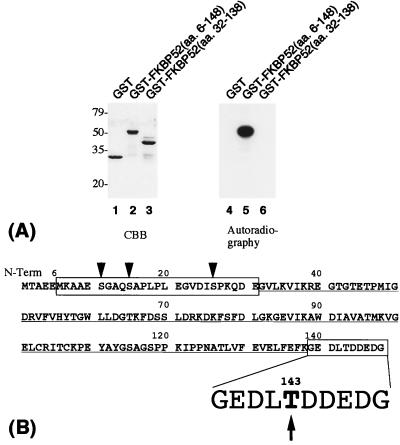
To further narrow down the phosphorylation site(s) within FKBP52 (amino acids 6–148), we used GST-fusion proteins without thrombin cleavage to avoid the interference of the phosphorylation of the proteolytic fragments that possess almost the same mobility as FKBP52 (amino acids 6–148) (see Fig. 3A). Only GST-FKBP52 (amino acids 6–148), but neither GST-FKBP52 (amino acids 32–138) nor GST, was phosphorylated by CK2 (Fig. 4A). The result clearly indicated that amino acids 6–31 and/or amino acids 139–148 are responsible for the phosphorylation by CK2.
Figure 4.
Phosphorylation sites of FKBP52 by CK2. (A) GST (lanes 1 and 4), GST-FKBP52 (amino acids 6–148) (lanes 2 and 5), and GST-FKBP52 (amino acids 32–138) (lanes 3 and 6) were phosphorylated by CK2 in vitro without thrombin cleavage. CBB staining (lanes 1–3) and autoradiography (lanes 4–6) are shown. (B) The amino acid sequence of the corresponding region of rabbit FKBP52. The result in A indicates that the boxed regions should contain the CK2-phosphorylation site(s), whereas the underlined region should not. Nonconsensus serines are indicated by arrowheads, and Thr-143, which agrees well with the CK2-phosphorylatable motif, is in bold type and indicated by an arrow.
The amino acid sequence of the corresponding region of rabbit FKBP52 is shown in Fig. 4B. The amino acid sequence 6–31 contains three serine residues, and amino acid sequence 139–148 contains one threonine residue. It is established that CK2 phosphorylates Ser/Thr residues surrounded by negatively charged amino acids, especially at the C-terminal side of the phosphorylatable Ser/Thr (35). An acidic residue at position +3 is shown to be essential for the phosphorylation (35). None of the serines within amino acids 6–31 matches the criteria. On the other hand, Thr-143 is surrounded by two acidic amino acids in its N-terminal side and a stretch of four acidic amino acids in its C-terminal side, and thus is a very good consensus sequence for CK2-phosphorylation. These sequence examinations strongly suggest that Thr-143 is a phosphorylation site for CK2.
Effect of a Plausible Phosphorylation Site Peptide on the CK2-Phosphorylation.
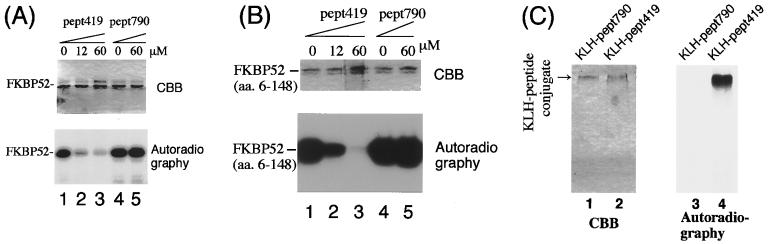
A fixed amount of FKBP52 (amino acids 6–458) was subjected to phosphorylation by CK2 in the presence of synthetic peptides Pept419 (amino acids 135–149, containing Thr-143) or Pept790 (amino acids 182–201, as a control). The phosphorylation of FKBP52 was markedly inhibited by Pept419 in a dose-dependent manner (Fig. 5A, lanes 1–3), whereas Pept790 did not inhibit the phosphorylation even at the highest concentration (lane 5). The phosphorylation of the FKBP52 (amino acids 6–148) was also inhibited by Pept419 (Fig. 5B, lanes 1–3), but not by Pept790 (lanes 4–5). Pept419 also inhibited the phosphorylation by CK2 of HSP90 (a substrate for CK2) and that of the CK2-specific substrate peptide (RRREEETEEE) at almost the same concentrations (data not shown). These results indicate that Pept419 inhibits the phosphorylation by CK2 in a competitive manner, suggesting that Pept419 itself is phosphorylated by CK2.
Figure 5.
Phosphorylation of the Thr-143-containing peptide by CK2. Recombinant FKBP52 (amino acids 6–458) (A) or FKBP52 (amino acids 6–148) (B) was phosphorylated by CK2 in the presence of Pept419 or Pept790. The phosphorylation mixtures were analyzed by CBB staining (Upper) or by autoradiography (Lower). (C) Phosphorylation of Pept419 by CK2. Pept419-KLH or Pept790-KLH was phosphorylated in vitro by CK2. CBB staining (lanes 1 and 2) and autoradiography (lanes 3 and 4) are shown. The position of the peptide-conjugates is indicated by an arrow.
Phosphorylation of Pept419 by CK2.
Pept419 and Pept790 are too small to see the phosphorylation by SDS/PAGE, thus we conjugated these peptides to KLH (33) and examined the phosphorylation of the conjugates. Only KLH-Pept419 (Fig. 5C, lane 4), but not KLH-Pept790 (lane 3), was phosphorylated by CK2. This result indicated that Pept419 is phosphorylated by CK2, determining that Thr-143 (the only phosphorylatable residue within Pept419) is a phosphorylation site of FKBP52 by CK2.
CK2-Phosphorylated FKBP52 Does Not Bind HSP90.
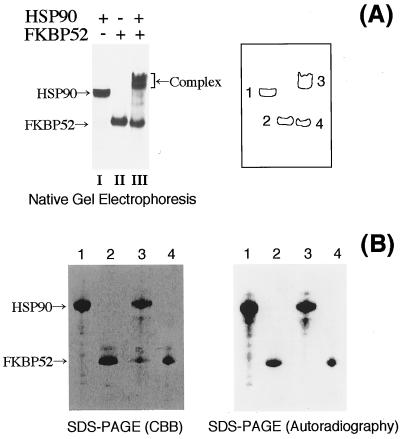
Importantly, we found that CK2-phosphorylated FKBP52 does not bind HSP90. Bacterially expressed FKBP52 (2 μg) was incubated with CK2 (40 ng) and [γ-32P]ATP, and then further incubated with or without HSP90 (3 μg). The mixtures were subjected to native PAGE to detect the HSP90-FKBP52 complexes. The CBB staining pattern is shown in Fig. 6A (lanes I–III), and each band was designated as illustrated on the right. Both HSP90 (band 1) and FKBP52 (band 2) migrated as single bands. When HSP90 and FKBP52 were mixed, the HSP90 band significantly shifted upward (band 3), with a concomitant decrease of the FKBP52 band (band 4 as compared with band 2), indicating the HSP90–FKBP52 association.
Figure 6.
CK2-phosphorylated FKBP52 does not bind HSP90. (A) Purified recombinant FKBP52 was phosphorylated by CK2 in the presence of [γ-32P]ATP in vitro. HSP90-binding activity of FKBP52 was examined by native PAGE and CBB staining is shown. Lane I, HSP90 alone (3 μg); lane II, FKBP52 alone (2 μg); or lane III, FKBP52 (2 μg) was mixed with HSP90 (3 μg). The positions of HSP90, FKBP52, and FKBP52–HSP90 complexes are indicated by arrows. Outlines of protein bands are illustrated on the right with the numbers 1–4. (B) Bands 1–4 of A were excised and analyzed by re-electrophoresis on an SDS/polyacrylamide gel. CBB staining (Left) and corresponding autoradiography (Right) are shown. The positions of HSP90 and FKBP52 are indicated by arrows on the left.
To analyze the molecular components of the complexes and their phosphorylation states, we cut out protein bands (bands 1–4) from the native gel, and re-electrophoresed them in the presence of SDS. The CBB staining pattern of the re-electrophoresis is shown in Fig. 6B Left. The lane numbers correspond to the band numbers of Fig. 6A. As expected, HSP90 was recovered from band 1 and band 3 (indicated by an arrow), and FKBP52 was recovered from bands 2, 3, and 4 (shown by an arrow). This result confirms that band 3 contains both HSP90 and FKBP52 as components. The autoradiography of the same gel is shown in Fig. 6B Right. As expected, phosphorylated HSP90 was recovered from bands 1 and 3, because HSP90 was phosphorylated by CK2 during the second incubation period. Phosphorylated FKBP52 was observed in bands 2 and 4, indicating that a part of FKBP52 was phosphorylated by CK2. Importantly, band 3 contained no phosphorylated FKBP52, whereas FKBP52 was clearly visible by CBB staining. Longer exposure of the same gel failed to detect any phosphorylated FKBP52 in band 3 (data not shown). This result clearly indicates that CK2-phosphorylated FKBP52 was not included in the HSP90–FKBP52 complexes. In other words, the HSP90-binding activity of FKBP52 was lost by CK2 phosphorylation. The phosphorylation of HSP90 had no effect on the association with FKBP52, as phosphorylated HSP90 was recovered from the up-shifted complexes (Fig. 6B, lane 3).
Phosphorylation of FKBP52 by CK2 Does Not Affect the FK506-Binding Activity of FKBP52.
Finally, the FK506-binding activity of FKBP52 was assayed by retention on FK506–Affi-Gel 10. Recombinant FKBP52 bound strongly to FK506–Affi-Gel 10, irrespective of the presence or absence of ATP (data not shown). CK2-phosphorylated FKBP52 was still able to bind to FK506–Affi-Gel 10 (data not shown). Thus, it was concluded that phosphorylation by CK2 does not inhibit the FK506-binding activity of FKBP52. The isolation of in vivo phosphorylated FKBP52 by FK506–Affi-Gel 10 (Fig. 2) also supported the conclusion that phosphorylated FKBP52 binds FK506.
DISCUSSION
Immunophilins possess peptidyl prolyl cis–trans isomerase activity (36). Recently, many immunophilins have been reported to function as molecular chaperones for certain substrates (17–19). Although the role of the isomerase activity in the chaperone activity of immunophilins is still ambiguous, immunophilins are now believed to maintain the structure of many target proteins in concert with other chaperones. In fact, several immunophilins have been reported to bind HSP90 by means of their TPR domains (16, 28, 37) and were recovered in steroid hormone receptor complexes (14, 15).
The function of immunophilins must be regulated by certain mechanisms inside cells. Our results show that the HSP90-binding activity of FKBP52 is lost upon phosphorylation by CK2. It should be noted that not all of FKBP52 was phosphorylated in vivo (as revealed by a stoichiometric analysis of Fig. 2); thus, a large amount of unphosphorylated FKBP52 that associates with HSP90 remains within cells. In fact, we could co-isolate HSP90 with FKBP52 from cell lysates (see Fig. 2). Although several immunophilins have been reported to be phosphorylated in vivo or in vitro (see, for example, refs. 32 and 38), this is, to our knowledge, the first evidence of the functional regulation of immunophilins by phosphorylation. Another prominent property of FKBP52 is to bind FK506 (25). The FK506-binding activity of FKBP52 was not affected by the CK2 phosphorylation. Conversely, addition of up to 50 μM FK506 had no effect on the phosphorylation of FKBP52 by CK2 (data not shown). It seems that the relationship between the CK2-mediated phosphorylation and the FK506-binding ability of FKBP52 is irrelevant.
We demonstrated that CK2 phosphorylates FKBP52 both in vivo and in vitro, and we determined that one of the phosphorylation sites is Thr-143. Thr-143 locates to a short stretch of sequence where acidic amino acids are clustered, just after the FK506-binding domain. This stretch has been estimated to be a hinge region, which connects the FK506-binding domain with other regions of the molecule (39). Therefore, the CK2-induced phosphorylation could affect the steric interaction between different domains of FKBP52. Such an intramolecular conformational change could also explain why the phosphorylation of Thr-143 prevented HSP90 binding, which is mapped to the TPR-containing C terminus of FKBP52 (28). Czar et al. (40) reported that this hinge region might serve as a nuclear localization signal recognition sequence, consistent with the possibility that FKBP52 is involved in glucocorticoid receptor trafficking to the nucleus.
Interestingly, truncated FKBP52 (amino acids 6–148) was a much better substrate for CK2 than was full-length FKBP52 (see Fig. 3). Addition of fragments comprising other regions of FKBP52 in any combination tested (e.g., three fragments, amino acids 149–267, amino acids 240–400, and amino acids 374–458, added at the same time) did not inhibit the phosphorylation of FKBP52 (amino acids 6–148) by CK2 (data not shown). Thus, the enhancement of the phosphorylation after the removal of amino acids 149–458 was due not to the relief of CK2 inhibition by this region but rather to the removal of steric hindrance at the phosphorylation site, Thr-143. As only 5 amino acids are left at the C-terminal side of Thr-143 in FKBP52 (amino acids 6–148), it would be plausible that the region is highly flexible, extending from the globular FK506-binding domain, and very easily accessible to CK2. The phosphorylation site may be more hindered within the overall three-dimensional structure of full-length FKBP52 and less accessible for CK2.
Our results do not exclude the possibility that other sites of FKBP52 were also phosphorylated. FKBP52 (amino acids 374–458) was also phosphorylated by CK2 in vitro (see Fig. 3), indicating that at least one additional CK2-phosphorylation site is located in this part of the molecule. Indeed, the rabbit FKBP52 sequence stops with amino acids 450SQSQVETEA458, in which Thr-456 may be a candidate site for CK2 phosphorylation. The prevention of HSP90-binding activity of FKBP52 by the CK2 phosphorylation could be explained by the fact that the TPR region resides in the C-terminal part of FKBP52 (28). In addition, several minor phosphopeptides were observed in addition to the CK2-phosphorylation sites when in vivo phosphorylated FKBP52 was analyzed (see Fig. 2C, lane 5), suggesting that certain kinases other than CK2 also phosphorylate FKBP52 in vivo, as implied by sequence analysis (20, 25).
Several cochaperones have been revealed to function by physically and functionally interacting with HSP90 (5, 6). These cochaperones include FKBP52, CyP40, p23, DnaJ, and Cdc37. Recently, it was reported that Cdc37 is homologous to pp50, which was identified as a component of HSP90–pp60v-Src complexes (3, 10). More recently, Cdc37 was reported to function as a molecular chaperone (41) assisting activities of many protein kinases such as Cdc28, Cdk4, CK2, Raf, and v-Src (42). Cochaperones are thought to play an important role in certain protein complexes such as pp60v-src and steroid hormone receptors, which contain HSP90 as a common component. However, depending on the complexes, additional different protein components are included in these heteromeric structures. Immunophilins are frequently recovered as components of steroid hormone receptors complexes (14, 15), whereas Cdc37 is not. Conversely, Cdc37 is a component of many HSP90–kinase complexes (42), whereas immunophilins are not. Thus, certain regulatory mechanisms that determine the molecular composition of chaperone-containing complexes probably exist. Because FKBP52 loses its HSP90-binding activity upon phosphorylation by CK2 as reported here, the immunophilin cannot be a component of active CK2–HSP90 complexes. We propose that this kind of phosphorylation-regulated association–dissociation of chaperones might be a possible mechanism that determines the molecular composition of chaperone–target complexes. As FKBP52 is suggested to play a role in glucocorticoid receptor trafficking, we also propose that phosphorylation events might be involved in the molecular assembly of multiple chaperones with steroid hormone receptors, hence modulating steroid hormone receptor-mediated signal transduction and immunosuppression.
Acknowledgments
We thank the Fujisawa Pharmaceutical Company for providing us FK506. We are very grateful to Drs. E. Nishida and Y. Hashimoto for encouragement, and colleagues in the Institut National de la Santé et de la Recherche Médicale Unité 33 for helpful discussions. Y.M. was supported by a fellowship of the Institut National de la Santé et de la Recherche Médicale. This work was supported in part by grants-in-aid from the Ministry of Education, Science and Culture of Japan.
ABBREVIATIONS
- HSP
heat shock protein
- FKBP
FK506-binding protein
- CK2
casein kinase II
- GST
glutathione S-transferase
- KLH
keyhole limpet hemocyanin
- TPR
tetratricopeptide repeat
- CBB
Coomassie brilliant blue
References
- 1.Parsell D A, Lindquist S. Annu Rev Genet. 1993;27:437–496. doi: 10.1146/annurev.ge.27.120193.002253. [DOI] [PubMed] [Google Scholar]
- 2.Joab I, Radanyi C, Renoir M, Buchou T, Catelli M G, Binart N, Mester J, Baulieu E-E. Nature (London) 1984;308:850–853. doi: 10.1038/308850a0. [DOI] [PubMed] [Google Scholar]
- 3.Brugge J S, Erickson E, Erickson R L. Cell. 1981;25:363–372. doi: 10.1016/0092-8674(81)90055-6. [DOI] [PubMed] [Google Scholar]
- 4.Rutherford S L, Zuker C S. Cell. 1994;79:1129–1132. doi: 10.1016/0092-8674(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 5.Buchner J. FASEB J. 1996;10:10–19. [PubMed] [Google Scholar]
- 6.Yahara, I., Minami, Y. & Miyata, Y. (1998) Ann. New York Acad. Sci., in press. [DOI] [PubMed]
- 7.Miyata Y, Yahara I. J Biol Chem. 1992;267:7042–7047. [PubMed] [Google Scholar]
- 8.Stancato L F, Chow Y-H, Hutchison K A, Perdew G H, Jove R, Pratt W B. J Biol Chem. 1993;268:21711–21716. [PubMed] [Google Scholar]
- 9.Cutforth T, Rubin G M. Cell. 1994;77:1027–1036. doi: 10.1016/0092-8674(94)90442-1. [DOI] [PubMed] [Google Scholar]
- 10.Stepanova L, Leng X, Parker S B, Harper J W. Genes Dev. 1996;10:1491–1502. doi: 10.1101/gad.10.12.1491. [DOI] [PubMed] [Google Scholar]
- 11.Miyata Y, Yahara I. Biochemistry. 1995;34:8123–8129. doi: 10.1021/bi00025a019. [DOI] [PubMed] [Google Scholar]
- 12.Schulte T W, Blagosklonny M V, Ingui C, Neckers L. J Biol Chem. 1995;270:24585–24588. doi: 10.1074/jbc.270.41.24585. [DOI] [PubMed] [Google Scholar]
- 13.Xu Y, Lindquist S. Proc Natl Acad Sci USA. 1993;90:7074–7078. doi: 10.1073/pnas.90.15.7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baulieu E-E. Ann New York Acad Sci. 1995;761:50–55. doi: 10.1111/j.1749-6632.1995.tb31368.x. [DOI] [PubMed] [Google Scholar]
- 15.Renoir J-M, Radanyi C, Faber L E, Baulieu E-E. J Biol Chem. 1990;265:10740–10745. [PubMed] [Google Scholar]
- 16.Ratajczak T, Carrello A, Mark P J, Warner B J, Simpson R J, Moritz R L, House A K. J Biol Chem. 1993;268:13187–13192. [PubMed] [Google Scholar]
- 17.Duina A A, Chang H-C J, Marsh J A, Lindquist S, Gaber R F. Science. 1996;274:1713–1715. doi: 10.1126/science.274.5293.1713. [DOI] [PubMed] [Google Scholar]
- 18.Bose S, Weikl T, Bugl H, Buchner J. Science. 1996;274:1715–1717. doi: 10.1126/science.274.5293.1715. [DOI] [PubMed] [Google Scholar]
- 19.Freeman B C, Toft D O, Morimoto R I. Science. 1996;274:1718–1720. doi: 10.1126/science.274.5293.1718. [DOI] [PubMed] [Google Scholar]
- 20.Lebeau M-C, Massol N, Herrick J, Faber L E, Renoir J-M, Radanyi C, Baulieu E-E. J Biol Chem. 1992;267:4281–4284. [PubMed] [Google Scholar]
- 21.Yem A W, Tomasselli A G, Heinrikson R L, Zurcher-Neely H, Ruff V A, Johnson R A, Deibel M R., Jr J Biol Chem. 1992;267:2868–2871. [PubMed] [Google Scholar]
- 22.Tai P-K K, Albers M W, Chang H, Faber L E, Schreiber S L. Science. 1992;256:1315–1318. doi: 10.1126/science.1376003. [DOI] [PubMed] [Google Scholar]
- 23.Wiederrecht G, Hung S, Chan H K, Marcy A, Martin M, Calaycay J, Boulton D, Sigal N, Kincaid R L, Siekierka J J. J Biol Chem. 1992;267:21753–21760. [PubMed] [Google Scholar]
- 24.Lebeau M-C, Myagkikh I, Rouviere-Fourmy N, Baulieu E-E, Klee C B. Biochem Biophys Res Commun. 1994;203:750–755. doi: 10.1006/bbrc.1994.2246. [DOI] [PubMed] [Google Scholar]
- 25.Peattie D A, Harding M W, Fleming M A, DeCenzo M T, Lippke J A, Livingston D J, Benasutti M. Proc Natl Acad Sci USA. 1992;89:10974–10978. doi: 10.1073/pnas.89.22.10974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le Bihan S, Renoir J-M, Radanyi C, Chambraud B, Joulin V, Catelli M-G, Baulieu E-E. Biochem Biophys Res Commun. 1993;195:600–607. doi: 10.1006/bbrc.1993.2088. [DOI] [PubMed] [Google Scholar]
- 27.Massol N, Lebeau M-C, Renoir J-M, Faber L E, Baulieu E-E. Biochem Biophys Res Commun. 1992;187:1330–1335. doi: 10.1016/0006-291x(92)90448-t. [DOI] [PubMed] [Google Scholar]
- 28.Radanyi C, Chambraud B, Baulieu E-E. Proc Natl Acad Sci USA. 1994;91:11197–11201. doi: 10.1073/pnas.91.23.11197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chambraud B, Radanyi C, Camonis J H, Shazand K, Rajkowski K, Baulieu E-E. J Biol Chem. 1996;271:32923–32929. doi: 10.1074/jbc.271.51.32923. [DOI] [PubMed] [Google Scholar]
- 30.Allende J E, Allende C C. FASEB J. 1995;9:313–323. doi: 10.1096/fasebj.9.5.7896000. [DOI] [PubMed] [Google Scholar]
- 31.Koyasu S, Nishida E, Kadowaki T, Matsuzaki F, Iida K, Harada F, Kasuga M, Sakai H, Yahara I. Proc Natl Acad Sci USA. 1986;83:8054–8058. doi: 10.1073/pnas.83.21.8054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fretz H, Albers M W, Galat A, Standaert R F, Lane W S, Burakoff S J, Bierer B E, Schreiber S L. J Am Chem Soc. 1991;113:1409–1411. [Google Scholar]
- 33.Renoir J M, Pahl A, Keller U, Baulieu E-E. C R Acad Sci Paris. 1993;316:1410–1416. [PubMed] [Google Scholar]
- 34.Chambraud B, Rouviere-Fourmy N, Radanyi C, Hsiao K, Peattie D A, Livingston D J, Baulieu E-E. Biochem Biophys Res Commun. 1993;196:160–166. doi: 10.1006/bbrc.1993.2229. [DOI] [PubMed] [Google Scholar]
- 35.Meggio F, Marin O, Pinna L A. Cell Mol Biol Res. 1994;40:401–409. [PubMed] [Google Scholar]
- 36.Galat A. Eur J Biochem. 1993;216:689–707. doi: 10.1111/j.1432-1033.1993.tb18189.x. [DOI] [PubMed] [Google Scholar]
- 37.Ratajczak T, Carrello A. J Biol Chem. 1996;271:2961–2965. doi: 10.1074/jbc.271.6.2961. [DOI] [PubMed] [Google Scholar]
- 38.Jin Y J, Burakoff S J. Proc Natl Acad Sci USA. 1993;90:7769–7773. doi: 10.1073/pnas.90.16.7769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Callebaut I, Renoir J-M, Lebeau M-C, Massol N, Burny A, Baulieu E-E, Mornon J-P. Proc Natl Acad Sci USA. 1992;89:6270–6274. doi: 10.1073/pnas.89.14.6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Czar M J, Lyons R H, Welsh M J, Renoir J-M, Pratt W B. Mol Endocrinol. 1995;9:1549–1560. doi: 10.1210/mend.9.11.8584032. [DOI] [PubMed] [Google Scholar]
- 41.Kimura Y, Rutherford S L, Miyata Y, Yahara I, Freeman B C, Yue L, Morimoto R I, Lindquist S. Genes Dev. 1997;11:1775–1785. doi: 10.1101/gad.11.14.1775. [DOI] [PubMed] [Google Scholar]
- 42.Hunter T, Poon Y C. Trends Cell Biol. 1997;7:157–161. doi: 10.1016/S0962-8924(97)01027-1. [DOI] [PubMed] [Google Scholar]