Abstract
Aneuploidy or chromosome imbalance is the most massive genetic abnormality of cancer cells. It used to be considered the cause of cancer when it was discovered more than 100 years ago. Since the discovery of the gene, the aneuploidy hypothesis has lost ground to the hypothesis that mutation of cellular genes causes cancer. According to this hypothesis, cancers are diploid and aneuploidy is secondary or nonessential. Here we reexamine the aneuploidy hypothesis in view of the fact that nearly all solid cancers are aneuploid, that many carcinogens are nongenotoxic, and that mutated genes from cancer cells do not transform diploid human or animal cells. By regrouping the gene pool—as in speciation—aneuploidy inevitably will alter many genetic programs. This genetic revolution can explain the numerous unique properties of cancer cells, such as invasiveness, dedifferentiation, distinct morphology, and specific surface antigens, much better than gene mutation, which is limited by the conservation of the existing chromosome structure. To determine whether aneuploidy is a cause or a consequence of transformation, we have analyzed the chromosomes of Chinese hamster embryo (CHE) cells transformed in vitro. This system allows (i) detection of transformation within 2 months and thus about 5 months sooner than carcinogenesis and (ii) the generation of many more transformants per cost than carcinogenesis. To minimize mutation of cellular genes, we have used nongenotoxic carcinogens. It was found that 44 out of 44 colonies of CHE cells transformed by benz[a]pyrene, methylcholanthrene, dimethylbenzanthracene, and colcemid, or spontaneously were between 50 and 100% aneuploid. Thus, aneuploidy originated with transformation. Two of two chemically transformed colonies tested were tumorigenic 2 months after inoculation into hamsters. The cells of transformed colonies were heterogeneous in chromosome number, consistent with the hypothesis that aneuploidy can perpetually destabilize the chromosome number because it unbalances the elements of the mitotic apparatus. Considering that all 44 transformed colonies analyzed were aneuploid, and the early association between aneuploidy, transformation, and tumorigenicity, we conclude that aneuploidy is the cause rather than a consequence of transformation.
Aneuploidy is by far the most massive genetic abnormality of cancer cells and was the first to be discovered more than 100 years ago (1). It is defined as an abnormal complement of chromosomes. In view of this, D. von Hansemann proposed in 1890 and T. Boveri in 1914 that aneuploidy is the cause of cancer (1, 2). Since then nearly all of more than 20,000 solid human cancers that have been analyzed to date proved to be aneuploid (3–5). In the 1930s and 1950s it was discovered that the aneuploidy of a given cancer typically includes clonal and nonclonal elements. The clonal elements, termed the “stemline,” lent further support to the hypothesis that cancer originated with and thus, may have been caused by, aneuploidy (4, 6–9, 100).
Aneuploidy vs. Gene Mutation Hypothesis of Cancer.
Despite near-perfect correlations, the aneuploidy–cancer hypothesis has gradually lost ground to the somatic gene mutation hypothesis, which holds that mutation converts certain cellular genes, termed protooncogenes, to dominant oncogenes, which cause cancer either alone or after mutational inactivation of tumor suppressor genes (10–14). The rise of the mutation hypothesis is based on four kinds of discoveries:
(i) The “confusing plethora” (6) of aneuploidy patterns found in every histological type of cancer soon discouraged faith in the aneuploidy hypothesis: no cancer-specific aneuploidy was ever found (3, 4). Even the cells of a given “clonal” cancer proved to be heterogeneous with regard to chromosome numbers (4, 6, 9, 100). Moreover, several investigators have claimed diploid cancers (15, 17, 93, 100). In view of this, aneuploidy has been interpreted either as a “Folgeerscheinung” (consequence) (15) that is not essential for carcinogensis (16–19), or as the cause of tumor progression (8, 16, 92, 101). In the words of Mitelman et al., aneuploidy is “secondary” because, first, “practically every numerical change may be found—even as the sole anomaly—in almost every neoplastic disorder; second, because the pathogenetic significance of such abnormalities is totally unknown; and, third, because no chromosomal breakpoints are involved” (5)—although a cancer-specific or consistent aneuploidy is not postulated by Boveri’s hypothesis.
(ii) Soon after the discovery of genes, some classical carcinogens were shown to be mutagenic, such as x-rays in 1927 (22) and alkylating agents in the 1940s (23). As more and more carcinogens were found to be mutagenic with ever more sensitive mutagenicity tests (17, 24–27, 102), the gene mutation hypothesis reached a high point in 1973 with the premise that “carcinogens are mutagens” (28).
(iii) Since the Chronic Myelocytic Leukemia (CML)-specific Philadelphia chromosome was first shown to be the product of a balanced translocation in 1973 (29), 215 “recurrent,” balanced translocations were found in certain cancers (5). Because the genes at the breakpoints are mutated by rearrangements, they are considered “primary” causes of cancer (16).
(iv) Point-mutated genes from rare cancer cells, particularly ras genes, act as dominant transforming genes in transfected, aneuploid mouse cell lines (30–32). This observation is currently interpreted as functional support for the hypothesis that mutation converts protooncogenes to dominant cancer- or oncogenes (10–12), although mutant ras does not transform diploid human or animal cells (33–35).
As a result of these developments, aneuploidy now is not even discussed as a cause of cancer in text books of molecular biology (10–12). In the meantime, however, the gene mutation hypothesis has not succeeded in a number of ways.
(i) It has not succeeded to identify a dominant cellular oncogene or a combination of genes that transform normal diploid, human, or animal cells to cancer cells (26, 34, 36). Indeed the first putative cellular oncogene, mutated ras, was just reinterpreted by one of its discoverers as a senescence gene (37). Dominant transforming function of the mutated ras genes proved to be a transfection artifact, which elevates expression in transformed mouse cells 100- to 1,000-fold compared with the cancers from which it was isolated (34, 35).
(ii) It has not succeeded to prove that a balanced translocation, such as the Philadelphia chromosome, is sufficient for cancer. Instead, the Philadelphia chromosome seems to induce the clonal hyperplasia of terminally differentiating myelocytes, termed CML, from which a clonal cancer typically emerges at a low but predictable probability. This transition is called blast crisis and is associated with aneuploidy (4, 38, 39).
(iii) It has not succeeded to reconcile the relatively high probability of protooncogene mutation with the much lower probability of cancer. The probability that a human cell becomes a cancer cell is only about 1 in 3 × 1016, because a human lifespan corresponds to about 1016 cells (25, 33), because no more than one in three humans develops cancer in a lifetime (25, 33, 40, 41), and because cancer is clonal (10, 25). By contrast, the probability that a human cell contains a mutated protooncogene is 1 in 109, because the spontaneous mutation rate is 1 in 109 nucleotides per human and animal DNA per mitosis (12, 34, 42, 43) and because human DNA contains 109 nucleotides.
This means that humans should contain 105 (1014: 109) cancer cells, because they are made up of 1014 cells (25, 33). Because at least 60 putative protooncogenes have been postulated (10); and not just one but a multiplicity of mutations are thought to convert a given protooncogene to an oncogene (44); the number of cancer cells at any given time should be even several orders of magnitude higher.
In view of this, a rapidly growing number of tumor suppressor genes is now postulated that must also be mutationally inactivated for oncogenes to cause cancer (14, 36, 45–47). However, the hypothesis that multiple mutations are necessary to generate a cancer cell (e.g., “at least seven genetic events” for colon cancer) created a new paradox (46): Based on normal mutation rates, 1 in 109, colon cancer would practically never occur, because only 1 in 1063 [=(109)7] colon cells would ever become a cancer cell. Therefore, it is now postulated that prospective tumor cells have intrinsically high mutation rates, a “mutator phenotype” (48). But the gene mutation rates of most cancers are normal; only those of a few are moderately elevated (42, 46, 103, 104).
Moreover, it is unlikely that the most frequently inactivated tumor suppressor gene, p53 (47), can offer much protection against cancer, because each of 280 point mutations are now thought to inactivate this gene (49). Contrary to one reviewer, it therefore is not “surprising … that mice develop normally without the p53 gene” (105). Another hypothetical tumor suppressor gene, previously thought to prevent colon cancer, has just been reinterpreted as a developmental gene (50).
(iv) The hypothesis has not succeeded to explain the numerous unique functions and structures of cancer cells, such as invasiveness, dedifferentiation, distinct morphology, high “genetic instability” (8, 25, 92) despite normal gene mutation rates, and specific surface antigens in terms of [even 7 (46)] gene mutations (8, 11, 25). According to J. Cairns, “one of the problems is that most mutations lead to loss of function, rather than creation of new function” (25).
(v) It has not succeeded to explain the paradox of why hereditary mismatch repair deficiencies, such as Xeroderma and hereditary disposition for colorectal cancer, give rise to very specific cancers (e.g., skin and colon cancer) but not to others (46, 51–53).
(vi) It has not succeeded to explain the paradox of the growing list of nongenotoxic carcinogens (e.g., those that are not mutating DNA), such as asbestos, aromatic hydrocarbons, mineral oil, hormones, or mitotic spindle blockers including colcemid (25, 26, 54, 55).
(vii) It has not succeeded to explain why hereditary disposition to chromosome imbalance, such as Bloom’s syndrome, increases the cancer risk more than 100-fold (51, 56, 57).
(viii) It has not succeeded to explain why aneuploidy is “the rule rather than the exception” in cancer (8), although the gene mutation hypothesis predicts cancers, which are diploid, caused by “dominant” cellular oncogenes and mutant suppressor genes (10). Indeed diploidy is apparently not found among solid cancers (4, 6), except in the rare cancers caused by dominant retroviral oncogenes (58–61, 100, 106).
(ix) The 1,000-fold increase of the cancer risk with age is thought to reflect a stepwise accumulation of “mutations” (13, 25). However, because most, if not all, gene mutations of cancer cells, such as ras, myc (33–35), rb, and p53 (33, 36, 105), are heritable, cancer should be a disease of young age, because all but one of a set of mutantations could be inherited. But, if the “mutations” were aneuploidies, they would have to be accumulated somatically, because aneuploidy is not heritable (62, 99).
In view of these shortcomings and paradoxa of the mutation hypothesis, we have reexamined the aneuploidy hypothesis.
Mechanism of Carcinogenesis According to the Aneuploidy Hypothesis.
By regrouping thousands of genes encoded in chromosomes—as in speciation—aneuploidy inevitably will alter many genetic programs. We propose that this genetic revolution explains the numerous unique functions and structures of cancer cells much better than gene mutation, which is limited by the conservation of the existing chromosome structure. The consequences of the two types of mutation can also be understood by comparing the cell with an orchestra. The sound of an orchestra is changed much more readily by randomizing the number of instruments than by mutating individual players.
The aneuploidy hypothesis also predicts that many different, numerical chromosome imbalances could cause the same kind of cancer—just like many different random deletions or additions of instruments could destroy the same orchestra, the combinations being limited only by aneuploidies that are lethal or insufficient for carcinogenesis (David Rasnick, University of California, Berkeley, personal communication).
Indeed, low levels of aneuploidy occur somatically in noncancerous cells of humans and animals (96–99) and in cells maintained in vitro (8, 11, 19, 73, 80). The incidence increases with the age of the cell in the body or in the culture (8, 19, 94, 96–99). According to this view, “initiation” of carcinogenesis by hydrocarbons (25) induces aneuploidy insufficient for carcinogenesis. Conversely, the loss of the transformed phenotype of CHE cells has been correlated with a partial loss of aneuploidy (95). Moreover, low levels of aneuploidy occur congenitally (4, 56, 62). Down syndrome, which results from trisomy of the smallest human chromosome, 21, demonstrates the dramatic consequences of even minimal aneuploidy (62).
As a mechanism, we propose that chemical carcinogens, particularly the nongenotoxic ones, cause aneuploidy by interfering with mitotic chromosome disjunction (54). For example, the hydrophobic, polycyclic hydrocarbons could interfere with mitosis by physically and even covalently binding proteins (63–65) of the spindle apparatus. Alternatively, physical interference with chromosome disjunction by asbestos fibers could produce asymmetric mitosis (54). Thus, the aneuploidy hypothesis offers a rational explanation for carcinogenesis by the growing list of nongenotoxic carcinogens (26, 55).
Experimental Distinction Between Aneuploidy and Gene Mutation.
To distinguish experimentally between the hypotheses that aneuploidy is either a cause or a consequence of cancer, we decided to investigate (i) transformation as early as possible, to distinguish cause from consequence, and (ii) as many cases of transformation as possible to maximize the likelihood of finding a primary transformant without a “secondary” aneuploidy.
To meet these criteria we studied the chromosomes of Chinese hamster embryo (CHE) cells transformed in vitro, because transformation in vitro can be detected early, within 2 months after treatment (20, 66) and thus about 5 months sooner than chemical carcinogenesis (67). In addition, many more primary transformants can be generated and analyzed per cost in vitro than in animals. Several studies have documented that stable, morphological transformation of CHE cells in vitro by chemical carcinogens correlates well with tumorigenicity (20, 66, 68, 69).
To minimize transformation by gene mutation, we have used the aromatic polycyclic hydrocarbons benzo[a]pyrene (BP), methylcholanthrene (MCA), dimethylbenzanthracene (DMBA), which are nongenotoxic in CHE cells unless oxidized by liver enzymes (21, 25, 70) and colcemid (54).
The CHE in vitro system also allows a distinction between chemical and spontaneous transformation, because spontaneous transformation occurs at a much lower frequency and, on average, only many more cell divisions later (19, 71–73) than chemical transformation (20, 66, 68). We have also studied the chromosomes of spontaneously transformed CHE cells.
Finally, CHE cells were chosen to facilitate the diagnosis of aneuploidy because this animal contains only 22 chromosomes compared with the 40 of mice, the 42 of rats, the 44 of Syrian hamsters, and the 46 of humans (71).
A preliminary report of this work was presented at the Nineteenth International Symposium of the International Association for Comparative Leukemia Research and Related Diseases in Mannheim, Germany, in July 1997 (74).
MATERIALS AND METHODS
Subconfluent cultures of CHE cells, derived from a single male, 18-day embryo (71), that had been kept growing for one or two consecutive passages in culture were incubated for 2–3 hr with 0.1 μg/ml of colcemid (GIBCO/BRL). The cells were then rinsed with phosphate-buffered physiological saline, dissociated with trypsin at 37°C, mixed with 1 ml complete culture medium, and centrifuged for 6 min at 500 rpm at room temperature. Subsequently, the cells were incubated at room temperature for 12–20 min in 75 mM KCl (GIBCO/BRL) and then fixed with ethanol/acetic acid (3:1; vol/vol) exactly as described by GIBCO/BRL. About 15- to 20-μl aliquots of cells suspended in fixative were dripped with an Eppendorf pipette onto microscope slides, and metaphase chromosomes were directly counted with a Leitz Phase contrast microscope at ×200 and ×400 magnification. The chromosome numbers of metaphases were counted by one of us (e.g., P.D., A.K., C.R., or A.W.), photographed with a Polaroid camera attached to the microscope, and verified by one or two others.
RESULTS
Transformation of Chinese Hamster Embryo Cells with Nongenotoxic Carcinogens.
CHE cells were seeded at a density of one million per 10-cm culture dish in DMEM supplemented with 5% fetal calf serum, 5% calf serum, 0.5% dimethyl sulfoxide, and either BP at 3 μM, or DMBA at 1 μM, or MCA at 1 μM until they reached confluency. Medium with carcinogen was changed every 2 to 3 days. After another passage of 4-fold growth in the presence of carcinogen, cells were maintained as confluent cultures without dimethyl sulfoxide and carcinogen in two parallel dishes. If no transformed colonies appeared after incubation for 2 weeks, one of these was passaged after 4-fold dilution and the other was further incubated without dilution. The rationale of this strategy was to permit only transformed cells that are able to overgrow confluent and contact-inhibited normal cells to form colonies—a situation similar to the origin of a cancer in an animal or human. Control cultures of CHE cells were maintained likewise but without carcinogen.
About 3 weeks after the beginning of carcinogen treatment, approximately 300 foci of refractile cells appeared in BP- and MCA-treated cultures. Six weeks after the first DMBA treatment about 100 foci appeared per 10-cm dish. Because of the known cytotoxicity of DMBA (75), cultures needed extra time in carcinogen-free medium to reach confluence. No such foci appeared in the control cultures. Within the following month most of these initial BP, MCA, and DMBA foci either disappeared or formed a homogenous background. The induction of apparently reversible hyperplasia by polycyclic hydrocarbons has been described previously for cells in culture (75–77, 107) and also for cells in animals (78).
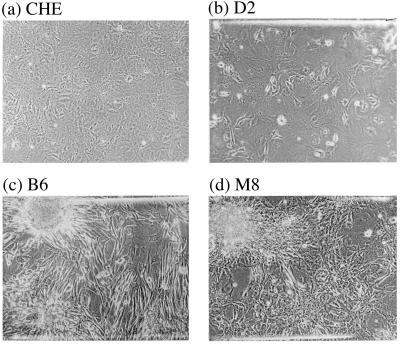
In the second month after carcinogen treatment and without further cell transfer, initially five to six and subsequently more multilayered, spindle-shaped, or epithelial colonies appeared. These colonies continued to grow over the monolayer of nongrowing cells and have not reversed their transformed phenotype to this date. Ten to 15 of these stably transformed colonies were picked from each carcinogen-treated culture and grown up into clonal cultures for chromosome analysis. Fig. 1 shows the morphology of normal CHE cells from a control culture (a), of a colony of DMBA-transformed CHE cells, termed D2, (b), of a colony of BP-transformed CHE cells, termed B6, (c), and of a colony of MCA-transformed CHE cells, termed M8 (d).
Figure 1.
The morphology of Chinese Hamster embryo (CHE) cells (a), a colony of dimethylbenzanthracene-transformed CHE cells, termed D2 (b), a colony of methylcholanthrene-transformed CHE cells, termed B6 (c), and a colony of benzo[a]pyrene-transformed CHE cells, termed M8 (d). Note that the colony D2 is a mixture of transformed and untransformed CHE cells, whereas colonies M8 and B6 consist almost only of transformed cells.
All Colonies of Chemically Transformed Chinese Hamster Cells Are Aneuploid.
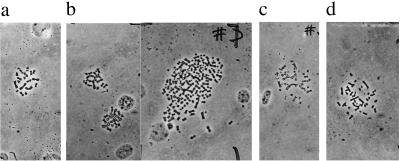
It is shown in Tables 1, 2, 3 that all 10 colonies of BP- , 15 colonies of DMBA-, and 13 colonies of MCA-transformed CHE cells analyzed were between 50 and 100% aneuploid. The chromosome numbers ranged from far below 22, to far above 22, e.g., up to and above tetraploidy as shown by a few examples in Fig. 2. By contrast, 86% of the control CHE cells had the normal karyotype of 22 chromosomes (Table 1). The difference between this percentage of diploidy and the expected 100% appears to reflect technical shortcomings of the method (71, 72, 79, 80).
Table 1.
Chromosome numbers of colonies of Chinese hamster cells transformed with benzopyrene (B) in vitro and untreated controls
| Cell type | Metaphases, n | Number of chromosomes per cell | Frequency, %
|
||
|---|---|---|---|---|---|
| <22 | 22, n22 | >22 | |||
| B1 | 12 | 29, 35, 35, 35, 36, 38, 38, 39, 39, 40, 42, 42 | 0 | 0 | 100 |
| B2 | 8 | 21, 22, 22, 22, 37, 37, 37, 41 | 12 | 38 | 50 |
| B3 | 12 | 16, 18, 19, 21, 25, 30, 31, 32, 33, 36, 37, 37 | 33 | 0 | 67 |
| B6 | 13 | 22, 25, 25, 33, 34, 35, 37, 37, 41, 41, 41, 45, 68 | 0 | 8 | 92 |
| B8 | 18 | 13, 14, 16, 18, 20, 21, 21, 21, 22, 23, 24, 24, 24, 24, 25, 26, 26, 34 | 44 | 6 | 50 |
| B10 | 11 | 16, 18, 19, 20, 20, 21, 36, 36, 36, 36, 41 | 54 | 0 | 46 |
| B11 | 13 | 18, 25, 32, 33, 36, 38, 40, 41, 42, 44, 62, 62, 79 | 8 | 8 | 84 |
| B12 | 18 | 16, 16, 21, 21, 22, 22, 23, 23, 33, 35, 35, 36, 38, 39, 39, 40, 44, 45 | 22 | 17 | 61 |
| B14 | 24 | 11, 14, 17, 17, 17, 18, 18, 18, 21, 22, 22, 22, 22, 22, 22, 22, 23, 23, 23, 23, 24, 33, 38, 45 | 38 | 29 | 33 |
| B15 | 5 | 13, 17, 18, 23, 27 | 60 | 0 | 40 |
| CHE | 35 | 17, 17, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 23, 31, 44, 46 | 6 | 86 | 9 |
Table 2.
Chromosome numbers of colonies of Chinese hamster cells transformed with dimethylbenzanthracene (D) in vitro
| Cell type | Metaphases, n | Number of chromosomes per cell | Frequency, %
|
||
|---|---|---|---|---|---|
| <22 | 22, n22 | >22 | |||
| D1 | 22 | 16, 20, 20, 21, 22, 22, 22, 29, 32, 32, 33, 36, 37, 38, 38, 38, 38, 39, 39, 40, 42, 70 | 18 | 14 | 68 |
| D2 | 32 | 16, 17, 17, 18, 20, 20, 21, 21, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 23, 28, 29, 34, 38, 41, 42, 44, 54, 61, 80 | 25 | 44 | 31 |
| D3 | 11 | 14, 15, 18, 18, 21, 24, 24, 25, 32, 33, 39 | 45 | 0 | 55 |
| D4 | 21 | 16, 18, 18, 18, 18, 19, 19, 20, 20, 20, 21, 21, 21, 22, 22, 22, 22, 22, 22, 22, 36 | 62 | 33 | 5 |
| D42 | 21 | 11, 17, 17, 17, 19, 20, 21, 21, 21, 22, 23, 23, 23, 23, 23, 23, 23, 24, 28, 28, 46 | 47 | 1 | 52 |
| D43 | 15 | 15, 19, 19, 20, 20, 20, 20, 21, 22, 22, 22, 22, 22, 24, 24 | 54 | 33 | 13 |
| D5 | 7 | 34, 36, 36, 38, 38, 129, >100 | 0 | 0 | 100 |
| D6-M | 28 | 15, 15, 15, 16, 16, 17, 18, 18, 18, 18, 20, 20, 21, 21, 21, 21, 22, 22, 22, 22, 22, 27, 27, 27, 27, 30, 41, 44 | 57 | 21 | 21 |
| D8 | 16 | 15, 15, 16, 17, 20, 20, 21, 21, 21, 21, 22, 22, 22, 22, 22, 24 | 63 | 31 | 6 |
| D9 | 13 | 21, 21, 21, 22, 22, 23, 23, 23, 23, 23, 24, 47, 48 | 23 | 15 | 62 |
| D10 | 37 | 13, 14, 14, 14, 16, 17, 18, 19, 21, 21, 21, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 23, 23, 23, 24, 24, 24, 24, 24, 26, 31, 36, 46, | 30 | 38 | 32 |
| D11 | 10 | 19, 20, 20, 21, 21, 21, 22, 22, 24, 26 | 60 | 20 | 20 |
| D12 | 25 | 16, 18, 19, 20, 21, 21, 21, 21, 21, 22, 22, 22, 22, 22, 22, 23, 23, 23, 23, 23, 23, 23, 23, 31, 36 | 36 | 24 | 40 |
| D14 | 40 | 13, 16, 17, 18, 19, 19, 21, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 23, 23, 23, 23, 23, 24, 24, 24, 26, 39, 39, 41, 44 | 18 | 52 | 30 |
| D15 | 16 | 18, 18, 18, 19, 20, 20, 20, 20, 22, 24, 30, 30, 32, 41, 49, 58 | 50 | 6 | 44 |
Table 3.
Chromosome numbers of colonies of Chinese hamster cells transformed with methylcholanthrene (M) in vitro
| Cell type | Metaphases, n | Number of chromosomes per cell | Frequency, %
|
||
|---|---|---|---|---|---|
| <22 | 22, n22 | >22 | |||
| M1 | 9 | 20, 24, 27, 29, 44, 46, 48, 60, 88 | 11 | 22 | 67 |
| M2 | 9 | 18, 18, 23, 30, 31, 38, 40, 41, 47 | 22 | 0 | 78 |
| M3 | 29 | 13, 19, 19, 20, 20, 21, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 22, 23, 23, 23, 35, 42, 43, 60, 76 | 21 | 52 | 27 |
| M34 | 12 | 16, 17, 18, 19, 20, 20, 22, 23, 23, 24, 24, 24 | 50 | 8 | 42 |
| M4 | 18 | 20, 21, 21, 22, 22, 22, 22, 22, 22, 23, 23, 23, 23, 24, 27, 29, 30, 42 | 17 | 33 | 50 |
| M5 | 11 | 29, 33, 33, 37, 38, 38, 38, 39, 52, 80, 80 | 0 | 0 | 100 |
| M6 | 19 | 16, 18, 19, 21, 21, 21, 22, 23, 24, 24, 24, 24, 25, 29, 30, 30, 30, 31, >60 | 32 | 5 | 63 |
| M7 | 22 | 16, 19, 19, 20, 21, 21, 21, 22, 22, 22, 22, 22, 22, 22, 22, 22, 23, 23, 24, 24, 24, 26 | 32 | 41 | 27 |
| M8 | 19 | 16, 20, 20, 20, 22, 22, 25, 30, 31, 33, 36, 37, 38, 39, 40, 40, 41, 41, 42 | 21 | 11 | 68 |
| M9 | 10 | 16, 19, 20, 20, 20, 21, 21, 22, 35, 44 | 70 | 20 | 10 |
| M10 | 21 | 16, 16, 20, 20, 20, 21, 21, 22, 22, 22, 22, 22, 22, 22, 22, 24, 30, 33, 34, 35, 56, | 33 | 38 | 21 |
| M11 | 10 | 24, 28, 28, 34, 35, 38, 38, 57, 60, 91 | 10 | 0 | 90 |
| M12 | 8 | 20, 21, 23, 34, 36, 38, 40, 51 | 25 | 0 | 75 |
Figure 2.
(a) The 22 chromosomes of a normal CHE cell. (b) The 19, 20, and more than 100 chromosomes of cells from the dimethylbenzanthracene-transformed colony D42. (c) The 34 chromosomes of a cell from the methylcholanthrene-transformed colony M34. (d) The 41 chromosomes of a cell from the benzo[a]pyrene-transformed colony B6.
Two lines of evidence suggest that the diploid cells in some transformed colonies are contaminating normal cells rather than diploid-transformed cells. First, the percentage of diploid cells in a given colony was directly proportional to the percentage of untransformed cells. For example, about half of the cells of colony D2 appeared morphologically untransformed (Fig. 1b), and about 44% of the cells of this colony were diploid (Table 2). By contrast, practically all cells of colonies M8 and B6 appeared transformed (Fig. 1 c and d), and 89% (Table 3) and 92% (Table 1) of their cells, respectively, were aneuploid. Second, preliminary subcloning (i.e., selection of a focal area of highly transformed cells from cultures with mixed phenotypes) increased the percentage of aneuploidy in two out of three cases. Subcolony D42 from colony D4 (Table 2) and subcolony M34 from colony M3 (Table 3) were 99% and 92% aneuploid compared with the 67% (D4) and 48% (M3) of their progenitors. By contrast, subcolony D43 had the same percentage of aneuploidy, 67%, as the progenitor D4 (Table 2).
Transformed Colonies of Colcemid-Treated Cells Are Aneuploid.
It has been shown that colcemid causes aneuploidy in CHE cells (79) and also transforms Syrian Hamster cells in culture (81). Following these procedures, we have treated CHE cells with 0.02 μg/ml colcemid thrice for 2 days in consecutive transfers. Several stable foci appeared between 2 and 3 months later, of which two, C1 and C3, were subjected to chromosome analysis. As can be seen in Table 4, C1 was 54% and C3 was 83% aneuploid. Their chromosome numbers deviated less from 22 than those of hydrocarbon-transformed colonies (Table 4).
Table 4.
Chromosome numbers of colonies of Chinese hamster cells transformed spontaneously (K) and with colcemid (C) in vitro
| Cell type | Metaphases, n | Number of chromosomes per cell | Frequency, %
|
||
|---|---|---|---|---|---|
| <22 | 22, n22 | >22 | |||
| K2 | 13 | 16, 18, 20, 20, 20, 22, 22, 22, 22, 22, 23, 40, 44 | 38 | 38 | 23 |
| K3 | 12 | 21, 21, 21, 21, 22, 22, 22, 22, 22, 22, 22, 33 | 33 | 58 | 8 |
| K6 | 15 | 12, 14, 14, 19, 19, 19, 20, 20, 20, 22, 23, 24, 24, 24, 24 | 60 | 7 | 33 |
| K7 | 13 | 12, 16, 18, 20, 21, 21, 21, 22, 22, 23, 23, 23, 24 | 54 | 15 | 31 |
| C1 | 11 | 13, 16, 21, 22, 22, 22, 22, 22, 23, 34, 42 | 27 | 46 | 27 |
| C3 | 24 | 14, 22, 22, 22, 23, 23, 24, 24, 24, 24, 24, 24, 24, 24, 24, 24, 24, 24, 24, 40, 40, 42, 44, 65 | 4 | 17 | 79 |
Spontaneous Transformants Are also Aneuploid.
During the 4 months in which all the above colonies of transformed cells were generated from chemically treated cultures, no colonies of transformed cells had appeared in eight 10-cm dishes of control cells kept in the same incubator with two weekly medium changes and monthly transfers at 4-fold dilutions. By the end of the fourth month two colonies had appeared in two separate dishes, termed K1 and K2, and five more, K3 to K7, appeared in the fifth month. Of these, only four colonies, K2, K3, K6, and K7, grew progressively into mass cultures. Each of these colonies was aneuploid (Table 4). But their chromosome numbers deviated little from 22 and 44, respectively.
Tumorigenicity of Chemically Transformed Cells.
Tumors were observed in two out of two 4-month-old, inbred Chinese hamsters (71) inoculated with 300,000 MCA-transformed cells of colony M8 (Fig. 1, Table 3), and in two out of two hamsters inoculated with 300,000 BP-transformed cells of colony B6 (Fig. 1, Table 1) 2–3 months after inoculation. The diameters of the M8 tumors were 1.5 and 0.4 cm and those of the B6 tumors were 1 and 0.3 cm. No tumor was observed in a control hamster inoculated with 300,000 CHE cells.
DISCUSSION
One Hundred Percent Correlation Between Aneuploidy and Transformation.
We have observed that 38 out of 38 transformed colonies and subcolonies of carcinogen-treated CHE cells, 2 out of 2 transformed colonies of colcemid-treated CHE cells, and 4 out of 4 spontaneously transformed colonies of CHE cells were 50 to 100% aneuploid. Because 39 of these 44 transformed colonies contained more than 50% of aneuploid cells, the aneuploidy must have originated in the same cell from which the transformed colony originated.
In the remaining five colonies aneuploidy could have originated in the second mitosis after transformation, because about 50% of their cells were diploid. However, this seems unlikely because the diploid cells of these colonies appeared to be untransformed contaminants that were lost after subcloning for the transformed phenotype. Moreover, it has been observed previously that even numerically diploid cells from MCA-induced cancers of Chinese hamsters have abnormal chromosome compositions, e.g., they are pseudodiploid (82). Thus, even CHE transformants with 22 chromosomes may be aneuploid. Nevertheless, single-cell cloning is necessary to determine whether any diploid transformants can be isolated from our colonies.
Considering that each 1 of 44 transformed colonies was aneuploid and the early association between aneuploidy, transformation, and tumorigenicity, we conclude that aneuploidy is the cause rather than a consequence of transformation. It remains to be determined whether transforming aneuploidy was achieved in one or several steps.
The aneuploidy hypothesis offers rational solutions for the shortcomings and paradoxa of the gene mutation hypothesis itemized above.
In contrast to our conclusion, it has been argued that in some CHE cells passaged in culture, “the neoplastic process proceeded … without any detectable cytogenetic abnormality” (19). However, even 20 generations before the carcinogenicity test, the respective CHE cells were already 44% aneuploid (19). Others have argued that aneuploidy is a consequence of transformation (16). However, cancers and in vitro transformants of the Chinese hamster generated by the dominant oncogene of Rous sarcoma virus, rather than by aneuploidy, are frequently diploid (58, 61, 72).
Aneuploidy vs. Gene Mutation.
Because there is no functional test for oncogenicity of mutated cellular genes (see above) or tumor suppressor genes (105), we cannot determine whether aneuploidy depends on gene mutation for transformation.
However, aromatic polycyclic hydrocarbons are not genotoxic in most test systems (24, 25), including Chinese hamster cells in vitro—unless oxidized by liver enzymes (21, 70). Outside the liver they persist even in animals metabolically unchanged for weeks and months (83–85). Nevertheless, others have found mutated ras genes in some but not all Syrian hamster embryo cells transformed by BP (108), but these genes do not transform hamster or any other embryo cells (33, 35). Thus, there is no proof that the low degree of genotoxicity of metabolically oxidized polycyclic hydrocarbons is in fact essential for carcinogenicity (8, 25, 26). On the contrary, polycyclic hydrocarbons in which metabolic oxidation sites are blocked still register in carcinogenicity tests (86) and cause tumors in animals without mutating ras genes (109). There is also no evidence that colcemid-induced (54) and spontaneous transformation is based on gene mutation.
In view of this and of the near-perfect correlation between aneuploidy and nonviral, solid cancers (4, 6), the burden is now on the gene mutation hypothesis to prove that mutated cellular genes are sufficient for carcinogenesis.
Is Aneuploidy the Cause of Chromosome Number Instability?
All chemically transformed CHE colonies were highly heterogeneous with regard to chromosome number, with a minority being hypodiploid (<22) and a majority being hyperdiploid (>22). This result was unexpected in view of the presumably clonal origin of these colonies from single, chemically transformed cells.
However, others have found the same heterogeneity of chromosome numbers in colonies of chemically transformed CHE cells (20, 68, 87), in chemically induced cancers of Chinese hamsters (67, 82), and in chemically induced cancers of other animals, including rats (88–90), mice (9), Syrian hamsters (54, 91), and in spontaneous cancers of man (6, 92, 93, 100).
We propose that this karyotypic heterogeneity is the direct consequence of aneuploidy, because aneuploidy can perpetually destabilize the chromosome number.
This could be achieved by the synthesis of abnormal concentrations of spindle proteins or histones or of abnormal numbers of centrosomes encoded by abnormal numbers of chromosomes (110–112). The resulting asymmetric and multipolar mitoses have been observed in cancer cells since 1890 (1, 2, 9, 93, 100). An alternative explanation suggests that mutation of a gene involved in chromosome segregation destabilizes the chromosome number (101). However, such a specific mutation seems unlikely in our case because all 44 of our colonies were transformed by nongenotoxic carcinogens.
This proposal resolves the apparent contradiction between the “genetic instability” of cancer cells (92) and their genetic stability defined by specific stem lines (7, 94) and by the conservation of normal frequencies of gene mutation (42, 103, 104): The causative aneuploidy and those chromosomes that control gene mutations are conserved, whereas the balance of chromosomes not essential for these functions would be subject to perpetual variation. Further work analyzing the chromosome numbers of clonal cultures derived from single transformed cells is necessary to test this proposal.
Cancer Prevention.
If cancer is caused by aneuploidy, cancer prevention could be improved by eliminating from food and drugs substances that cause aneuploidy.
Acknowledgments
We thank David Rasnick (University of California at Berkeley) and Harvey Bialy (Nature Biotechnology, New York) for critical reviews of the manuscript. Most of this work was carried out and supported by the III Medizinische Klinik in Mannheim of the University of Heidelberg. P.D. was the recipient of a guest professorship from the University of Heidelberg and support from the Forschungsfond der Fakultaet Fuer Klinische Medizin Mannheim and from Friedel and Prof. Mathias Hafner (Mannheim) and from Siggi and Max Duesberg while working at the III Medizinische Klinik during a sabbatical leave from University of California at Berkeley. All work in Berkeley and some of that in Mannheim was supported by Robert Leppo (philanthropist, San Francisco), the Abraham J. and Phyllis Katz Foundation (New York), the Nathan Cummings Foundation (San Francisco), and additional donations from Carol J. Wilhelmy (philanthropist, San Mateo), the A. Robbins Research International (San Diego), John Chen (Microsoft, Seattle), and other private sources.
ABBREVIATIONS
- CHE cells
Chinese hamster embryo cells
- MCA
methylcholanthrene
- DMBA
dimethylbenzanthracene
- BP
benzo[a]pyrene
References
- 1.von Hansemann D. Virchows Arch Pathol Anat. 1890;119:299–326. [Google Scholar]
- 2.Boveri T. Zur Frage der Enstehung Maligner Tumoren. Jena, Germany: Gustav Fischer Verlag; 1914. [Google Scholar]
- 3.Mitelman F. Catalogue of Chromosome Aberrations in Cancer. New York: Wiley–Liss; 1994. [DOI] [PubMed] [Google Scholar]
- 4.Sandberg A A. The Chromosomes in Human Cancer and Leukemia. New York: Elsevier Science; 1990. [Google Scholar]
- 5.Mitelman, F., Mertens, F. & Johansson, B. (1997) Nat. Genet.15, Suppl., 417–474. [DOI] [PubMed]
- 6.Heim S, Mitelman F. Cancer Cytogenetics. New York: Liss; 1987. [Google Scholar]
- 7.Heim S, Mandahl N, Mitelman F. Cancer Res. 1988;48:5911–1916. [PubMed] [Google Scholar]
- 8.Pitot H C. Fundamentals of Oncology. New York: Dekker; 1986. [Google Scholar]
- 9.Winge O. Z Zellforsch Mikrosk Anat. 1930;10:683–735. [Google Scholar]
- 10.Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson J D. Molecular Biology of the Cell. New York: Garland; 1994. [Google Scholar]
- 11.Lodish H, Baltimore D, Berk A, Zipursky S L, Matsudaira P, Darnell J. Molecular Cell Biology. by Freeman, New York and Oxford, U.K.: Scientific American Books; 1995. [Google Scholar]
- 12.Lewin B. Genes V. Oxford, U.K.: Oxford Univ. Press; 1994. [Google Scholar]
- 13.Cairns J. Matters of Life and Death: Perspectives on Public Health, Molecular Biology, Cancer, and the Prospects for the Human Race. Princeton, NJ: Princeton Univ. Press; 1997. [Google Scholar]
- 14.Levine A. Annu Rev Biochem. 1993;62:623–651. doi: 10.1146/annurev.bi.62.070193.003203. [DOI] [PubMed] [Google Scholar]
- 15.Andres A H. Z Zellforsch Mikrosk Anat. 1932;16:88–122. [Google Scholar]
- 16.Johansson B, Mertens F, Mitelman F. Genes Chromosomes Cancer. 1996;16:155–163. doi: 10.1002/(SICI)1098-2264(199607)16:3<155::AID-GCC1>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 17.Bauer K-H. Das Krebsproblem. Berlin: Springer; 1963. [Google Scholar]
- 18.DiPaolo J A, Popescu N C. Am J Pathol. 1976;85:709–738. [PMC free article] [PubMed] [Google Scholar]
- 19.Kraemer P, Ray A, Bartholdi M F, Cram S L. Cancer Genet Cytogenet. 1987;27:273–287. doi: 10.1016/0165-4608(87)90010-0. [DOI] [PubMed] [Google Scholar]
- 20.Connell J R. Br J Cancer. 1984;50:167–177. doi: 10.1038/bjc.1984.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conney A H. Cancer Res. 1982;42:4875–4917. [PubMed] [Google Scholar]
- 22.Muller H J. Science. 1927;66:84–87. doi: 10.1126/science.66.1699.84. [DOI] [PubMed] [Google Scholar]
- 23.Auerbach C, Robson J M, Carr J G. Science. 1947;105:243–247. doi: 10.1126/science.105.2723.243. [DOI] [PubMed] [Google Scholar]
- 24.Burdette W J. Cancer Res. 1955;15:201–226. [PubMed] [Google Scholar]
- 25.Cairns J. Cancer: Science and Society. San Francisco: Freeman; 1978. [Google Scholar]
- 26.Lijinsky W. Environ Mol Mutagen. 1989;14:78–84. doi: 10.1002/em.2850140615. [DOI] [PubMed] [Google Scholar]
- 27.Rous P. Nature (London) 1959;183:1357–1361. doi: 10.1038/1831357a0. [DOI] [PubMed] [Google Scholar]
- 28.Ames B, Durston W E, Yamaski E, Lee F D. Proc Natl Acad Sci USA. 1973;70:2281–2285. doi: 10.1073/pnas.70.8.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rowley J D. Nature (London) 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 30.Tabin C J, Bradley S M, Bargmann C I, Weinberg R A, Papageorge A G, Scolnick E M, Dhar R, Lowy D R, Chang E H. Nature (London) 1982;300:143–149. doi: 10.1038/300143a0. [DOI] [PubMed] [Google Scholar]
- 31.Reddy E P, Reynolds R K, Santos E, Barbacid M. Nature (London) 1982;300:149–152. doi: 10.1038/300149a0. [DOI] [PubMed] [Google Scholar]
- 32.Logan J, Cairns J. Nature (London) 1982;300:104–105. doi: 10.1038/300104a0. [DOI] [PubMed] [Google Scholar]
- 33.Duesberg P H, Schwartz J R. Prog Nucleic Acid Res Mol Biol. 1992;43:135–204. doi: 10.1016/s0079-6603(08)61047-8. [DOI] [PubMed] [Google Scholar]
- 34.Duesberg P. Science. 1995;267:1407–1408. doi: 10.1126/science.7794335. [DOI] [PubMed] [Google Scholar]
- 35.Hua V Y, Wang W, K, Duesberg P H. Proc Natl Acad Sci USA. 1997;94:9614–9619. doi: 10.1073/pnas.94.18.9614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stanbridge E J. Annu Rev Genet. 1990;24:615–657. doi: 10.1146/annurev.ge.24.120190.003151. [DOI] [PubMed] [Google Scholar]
- 37.Weinberg R A. Cell. 1997;38:573–575. doi: 10.1016/s0092-8674(00)81897-8. [DOI] [PubMed] [Google Scholar]
- 38.Koeffler H P, Golde D W. N Engl J Med. 1981;304:1201–1209. doi: 10.1056/NEJM198105143042004. [DOI] [PubMed] [Google Scholar]
- 39.Sandberg A A, Chen Z. In: Molecular Biology in Cancer Medicine. Razelle K, Talpaz M, editors. London: Martin Dunitz; 1996. [Google Scholar]
- 40.Bailar J C, Gorink H L. N Engl J Med. 1997;336:1569–1574. doi: 10.1056/NEJM199705293362206. [DOI] [PubMed] [Google Scholar]
- 41.Parker S L, Tong T, Bolden S, Wingo P A. CA Cancer J Clin. 1996;46:5–27. doi: 10.3322/canjclin.46.1.5. [DOI] [PubMed] [Google Scholar]
- 42.Strauss B S. Cancer Res. 1992;52:249–253. [PubMed] [Google Scholar]
- 43.Koshland D. Science. 1994;266:19–25. [Google Scholar]
- 44.Seeburg P H, Colby W W, Capon P J, Goeddel D V, Levinson A D. Nature (London) 1984;312:71–75. doi: 10.1038/312071a0. [DOI] [PubMed] [Google Scholar]
- 45.Pennisi E. Science. 1997;275:1876–1878. doi: 10.1126/science.275.5308.1876. [DOI] [PubMed] [Google Scholar]
- 46.Kinzler K, Vogelstein B. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 47.Culotta E, Koshland D E J. Science. 1993;262:1958–1961. doi: 10.1126/science.7903477. [DOI] [PubMed] [Google Scholar]
- 48.Loeb L A. Cancer Res. 1991;51:3075–3079. [PubMed] [Google Scholar]
- 49.Hollstein M, Sidransky D, Vogelstein B, Harris C C. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 50.Roush W. Science. 1997;276:534–535. doi: 10.1126/science.276.5312.534. [DOI] [PubMed] [Google Scholar]
- 51.Cairns J. Nature (London) 1981;289:353–357. doi: 10.1038/289353a0. [DOI] [PubMed] [Google Scholar]
- 52.Bridges B. Carcinogenesis. 1981;2:471–472. doi: 10.1093/carcin/2.5.471. [DOI] [PubMed] [Google Scholar]
- 53.Bridges B. Science. 1995;269:909. doi: 10.1126/science.7638608. (lett.). [DOI] [PubMed] [Google Scholar]
- 54.Oshimura M, Barrett J C. Environ Mutagen. 1986;8:129–159. doi: 10.1002/em.2860080112. [DOI] [PubMed] [Google Scholar]
- 55.Ashby J. In: Tumor Promoters: Biological Approaches for Mechanistic Studies and Assay Systems. Langenbach R, Elmore E, Barrett J C, editors. New York: Raven; 1988. pp. 417–430. [Google Scholar]
- 56.German J. In: Chromosomes and Cancer. German J, editor. New York: Wiley; 1974. [Google Scholar]
- 57.Pitot H C. Fundamentals of Oncology. New York: Dekker; 1978. [Google Scholar]
- 58.Kato R. Hereditas. 1968;59:63–119. doi: 10.1111/j.1601-5223.1968.tb02166.x. [DOI] [PubMed] [Google Scholar]
- 59.Mitelman F. Hereditas. 1971;69:155–186. doi: 10.1111/j.1601-5223.1971.tb02431.x. [DOI] [PubMed] [Google Scholar]
- 60.Duesberg P H. Cancer Res. 1987;47:1199–1220. [PubMed] [Google Scholar]
- 61.Mitelman F. In: Chromosomes and Cancer. German J, editor. New York: Wiley; 1974. pp. 675–693. [Google Scholar]
- 62.Hassold T J. Trends Genet. 1986;2:105–110. [Google Scholar]
- 63.Miller E C. Cancer Res. 1951;18:100–108. [PubMed] [Google Scholar]
- 64.Wiest W G, Heidelberger C. Cancer Res. 1953;13:246–261. [PubMed] [Google Scholar]
- 65.Miller E C, Miller J. Cancer Res. 1952;12:547–556. [PubMed] [Google Scholar]
- 66.Kirkland D J. Br J Cancer. 1976;34:134–144. doi: 10.1038/bjc.1976.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mitelman F, Mark J, Levan G. Hereditas. 1972;72:311–318. doi: 10.1111/j.1601-5223.1972.tb01055.x. [DOI] [PubMed] [Google Scholar]
- 68.Trewyn R W, Kerr S J, Lehman J M. J Natl Cancer Inst. 1979;62:633–637. doi: 10.1093/jnci/62.3.633. [DOI] [PubMed] [Google Scholar]
- 69.Bloch-Shtacher N, Sachs L. J Cell Physiol. 1977;93:205–212. doi: 10.1002/jcp.1040930206. [DOI] [PubMed] [Google Scholar]
- 70.Oberly T J, Rexroat M A, Bewsey B J, Richardson K K, Michaelis K C. Environ Mol Mutagen. 1990;16:260–271. doi: 10.1002/em.2850160408. [DOI] [PubMed] [Google Scholar]
- 71.Yerganian G, Leonard M. Science. 1961;133:1600–1601. doi: 10.1126/science.133.3464.1600. [DOI] [PubMed] [Google Scholar]
- 72.Hlozanek I, Donner L, Svoboda J. J Cell Physiol. 1966;68:221–235. [Google Scholar]
- 73.Shimizu T, Kato M V, Nikaido O, Suzuki F. Jpn J Cancer Res. 1995;86:546–554. doi: 10.1111/j.1349-7006.1995.tb02433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Duesberg P H, Li R, Kraemer A, Hehlmann R. J Mol Med. 1997;75:B248. (abstr.). [Google Scholar]
- 75.Berwald Y, Sachs L. J Nat Cancer Inst. 1965;35:641–661. [PubMed] [Google Scholar]
- 76.Mironescu S, Love R. Cancer Res. 1974;34:2562–2570. [PubMed] [Google Scholar]
- 77.Rivedal E, Sanner T. Carcinogenesis. 1983;4:817–820. doi: 10.1093/carcin/4.7.817. [DOI] [PubMed] [Google Scholar]
- 78.Kagawa M, Hakoi K, Yamamoto A, Futakuchi M, Hirose M. Jpn J Cancer Res. 1993;84:1120–1129. doi: 10.1111/j.1349-7006.1993.tb02811.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cox M. Cytogenet Cell Genet. 1973;12:165–174. doi: 10.1159/000130452. [DOI] [PubMed] [Google Scholar]
- 80.Cram L S, Bartholdi M F, Ray F A, Travis G L, Kraemer P M. Cancer Res. 1983;43:4828–4837. [PubMed] [Google Scholar]
- 81.Tsutsui T, Maizumi H, Barrett J C. Carcinogenesis. 1984;5:89–93. doi: 10.1093/carcin/5.1.89. [DOI] [PubMed] [Google Scholar]
- 82.Yerganian G. Ann N Y Acad Sci. 1956;63:789–792. [Google Scholar]
- 83.Uenseren E, Fieser L F. J Org Chem. 1962;27:1386–1389. [Google Scholar]
- 84.Heidelberger C, Weiss S M. Cancer Res. 1951;11:885–897. [PubMed] [Google Scholar]
- 85.Dauben W G, Mabee D. Cancer Res. 1951;11:216–220. [PubMed] [Google Scholar]
- 86.Kumari H L, Milo G E, Witiak D T. Teratogen Carcinogen Mutagen. 1990;10:247–262. doi: 10.1002/tcm.1770100308. [DOI] [PubMed] [Google Scholar]
- 87.Kirkland D J, Venitt S. Br J Cancer. 1976;34:145–152. doi: 10.1038/bjc.1976.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ahlstroem U. Hereditas. 1974;78:235–244. doi: 10.1111/j.1601-5223.1974.tb01444.x. [DOI] [PubMed] [Google Scholar]
- 89.Mitelman F, Mark J, Levan G, Levan A. Science. 1972;176:1340–1341. doi: 10.1126/science.176.4041.1340. [DOI] [PubMed] [Google Scholar]
- 90.Levan G, Levan A. Hereditas. 1975;79:161–198. doi: 10.1111/j.1601-5223.1975.tb01475.x. [DOI] [PubMed] [Google Scholar]
- 91.DiPaolo J A, Donovan P J. Exp Cell Res. 1967;48:361–377. doi: 10.1016/0014-4827(67)90361-8. [DOI] [PubMed] [Google Scholar]
- 92.Nowell P C. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 93.Koller P C. Ann N Y Acad Sci. 1956;63:793–817. doi: 10.1111/j.1749-6632.1956.tb50893.x. [DOI] [PubMed] [Google Scholar]
- 94.Levan A. Ann N Y Acad Sci. 1956;63:774–792. doi: 10.1111/j.1749-6632.1956.tb50892.x. [DOI] [PubMed] [Google Scholar]
- 95.Simi S, Musio A, Vatteroni L, Piras A, Rainaldi G. Cancer Genet Cytogenet. 1992;62:81–87. doi: 10.1016/0165-4608(92)90044-9. [DOI] [PubMed] [Google Scholar]
- 96.Harnden D G, Benn P A, Oxford J M, Taylor A M R, Webb T P. Somatic Cell Genet. 1976;2:55–62. doi: 10.1007/BF01539242. [DOI] [PubMed] [Google Scholar]
- 97.Terzi M, Hawkins T S C. Nature (London) 1975;253:361–362. doi: 10.1038/253361a0. [DOI] [PubMed] [Google Scholar]
- 98.Galloway S M, Buckton K E. Cytogenet Cell Genet. 1978;20:78–95. doi: 10.1159/000130842. [DOI] [PubMed] [Google Scholar]
- 99.Hook E B. In: Aneuploidy: Etiology and Mechanisms. Dellarco V L, Voytek P E, Hollaender A, editors. New York: Plenum; 1985. pp. 7–77. [Google Scholar]
- 100.Hauschka T S. Cancer Res. 1961;21:957–981. [PubMed] [Google Scholar]
- 101.Lengnauer C, Kinzler K W, Vogelstein B. Nature (London) 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 102.Miller J A, Miller E C. J Natl Cancer Inst. 1971;47:v–xiv. [PubMed] [Google Scholar]
- 103.Harris C C. Cancer Res. 1991;51:5023s–5044s. [PubMed] [Google Scholar]
- 104.Barrett J C, Tsutsui T, Tsly T, Oshimura M. In: Genetic Mechanisms in Carcinogenesis and Tumor Progression. Liotta C C H L A, editor. New York: Wiley; 1990. pp. 97–114. [Google Scholar]
- 105.Hartwell L. Cell. 1992;71:543–546. doi: 10.1016/0092-8674(92)90586-2. [DOI] [PubMed] [Google Scholar]
- 106.Rubin H, Temin H M. Fed Proc. 1958;17:994–1008. [PubMed] [Google Scholar]
- 107.Fernandez A, Mondal S, Heidelberger C. Proc Natl Acad Sci USA. 1980;77:7272–7276. doi: 10.1073/pnas.77.12.7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gilmer T M, Annab L A, Barrett J C. Mol Carcinogen. 1988;1:180–188. doi: 10.1002/mc.2940010306. [DOI] [PubMed] [Google Scholar]
- 109.Chakravarti D, Pelling J C, Cavalieri E L, Rogan E G. Proc Natl Acad Sci USA. 1995;92:10422–10426. doi: 10.1073/pnas.92.22.10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mayer V W, Aguilera A. Mut Res. 1990;231:177–186. doi: 10.1016/0027-5107(90)90024-x. [DOI] [PubMed] [Google Scholar]
- 111.Futcher B, Carbon J. Mol Cell Biol. 1986;6:2213–2222. doi: 10.1128/mcb.6.6.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Burke D, Gasdaska P, Hartwell L. Mol Cell Biol. 1989;9:1049–1059. doi: 10.1128/mcb.9.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]