Abstract
Organometallic complexes offer chemistry that is not accessible to purely organic molecules and, hence, potentially new mechanisms of drug action. We show here that the presence of both an iodido ligand and a σ-donor/π-acceptor phenylazopyridine ligand confers remarkable inertness toward ligand substitution on the half-sandwich “piano-stool” ruthenium arene complexes [(η6-arene)Ru(azpy)I]+ (where arene = p-cymene or biphenyl, and azpy = N,N-dimethylphenyl- or hydroxyphenyl-azopyridine) in aqueous solution. Surprisingly, despite this inertness, these complexes are highly cytotoxic to human ovarian A2780 and human lung A549 cancer cells. Fluorescence-trapping experiments in A549 cells suggest that the cytotoxicity arises from an increase in reactive oxygen species. Redox activity of these azopyridine RuII complexes was confirmed by electrochemical measurements. The first one-electron reduction step (half-wave potential −0.2 to −0.4 V) is assignable to reduction of the azo group of the ligand. In contrast, the unbound azopyridine ligands are not readily reduced. Intriguingly the ruthenium complex acted as a catalyst in reactions with the tripeptide glutathione (γ-l-Glu-l-Cys-Gly), a strong reducing agent present in cells at millimolar concentrations; millimolar amounts of glutathione were oxidized to glutathione disulfide in the presence of micromolar ruthenium concentrations. A redox cycle involving glutathione attack on the azo bond of coordinated azopyridine is proposed. Such ligand-based redox reactions provide new concepts for the design of catalytic drugs.
Keywords: arenes, cytotoxicity, glutathione, redox reactions, ruthenium complexes
Organometallic complexes offer potential for the development of new materials with a wide range of applications (1, 2). Organometallic complexes can exhibit chemical reactivity not possessed by either the metal or organic ligands alone and this reactivity can be fine-tuned by subtle changes in the electronic and steric properties of the bound ligands, or by variation of the metal and its oxidation state. These features provide a versatile platform for drug design that is now being exploited in several areas. For example, the redox activity of the organometallic ferrocenylphenol substitutents enhances the anticancer activity of tamoxifen derivatives by conferring an ability to generate reactive oxygen species in cells (3). Here we consider the anticancer activity of ruthenium(II) arene complexes. Such complexes are well known as catalysts in industry, for example, for hydrogenation reactions (4). Although organometallic complexes can catalyze hydrogenation of intracellular biomolecules (5), metal-centered catalysts might be readily poisoned and be ineffective in biological media.
Organometallic half-sandwich RuII arenes of the type [(η6-arene)Ru(YZ)(X)]+ where YZ is typically a chelating diamine ligand (e.g., ethylenediamine, en) and X is a halide (e.g., Cl) exhibit anticancer activity in vitro and in vivo (6, 7). As with many metal-based anticancer drugs, the primary cellular target is thought to be DNA. The Ru–Cl bond is labile, but hydrolysis is suppressed extracellularly because of the high chloride concentration, ≈104 mM (8). The intracellular chloride concentration is much lower, and hydrolysis can generate a reactive site on ruthenium (Ru-OH2) for subsequent DNA binding. DNA binding leads to further “downstream” effects, ultimately resulting in cell death (9, 10). Changing the chelating ligand has dramatic effects on cytotoxicity, which is also influenced by the nature of the arene ligand (11). In this series of diamine complexes, changing the monodentate ligand X from chloride to iodide has little effect on activity; both are good leaving groups (12). This contrasts dramatically with the chlorido and iodido ruthenium(II) arene complexes reported here, which contain σ-donor/π-acceptor 2-phenylazopyridines as N,N-chelating (YZ) ligands. Unlike their en analogues, these complexes exhibit different solution chemistry, and a different mechanism of cancer cell cytotoxicity, involving catalytic redox reactions.
Results
Complexes.
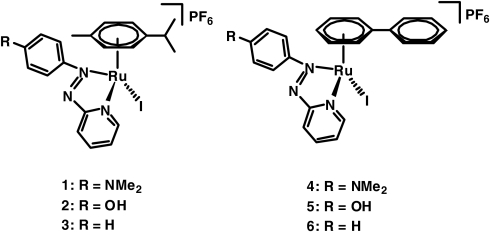
We synthesized and characterized RuII complexes containing either p-cymene (complexes 1–3) or biphenyl (complexes 4–6) as the π-bonded arene, a bidentate phenylazopyridine ligand having either NMe2, OH, or H as para substituent on the phenyl ring, and I− as the monodentate ligand (Fig. 1). The molecular structure of complex 4·MeOH was determined by x-ray crystallography [see supporting information (SI) Fig. S1]. Significant bond lengths and angles are reported in the SI Text (Table S1). The Ru–N bond lengths are comparable to analogous RuII arene phenylazopyridine chlorido complexes (13), as are the ruthenium-arene carbon distances (13). The Ru–I bond length [2.6984 (5) Å] is slightly shorter than in ruthenium(II) arene structures containing ethylenediamine as the chelating ligand {2.7116 (7) Å for [(η6-hexamethylbenzene)Ru(en)I]+ and 2.7166 (7) Å for [(η6-indan)Ru(en)I]+ (12)}. These phenylazopyridine complexes possess “3-legged piano-stool” structures and are intensely colored as a result of RuII-to-ligand (4d6-π*) charge-transfer and intraligand π–π* transitions (13).
Fig. 1.
Structures of complexes synthesized in this work.
Cytotoxicity.
Complexes 1, 2, 4, and 5 containing p-cymene or biphenyl as arene and electron-donating groups (OH, NMe2) on the phenylazopyridine ligand were highly cytotoxic to A2780 human ovarian and the A549 human lung cancer cell lines with IC50 values of 2–6 μM, whereas complexes 3 (IC50 >100 μM) and 6 (IC50 ≈40–50 μM), containing an unsubstituted phenylazopyridine ligand were much less cytotoxic (Table 1). Replacement of iodide by chloride led to a dramatic decrease in cytotoxicity (13), and the 2-phenylazopyridine ligands alone were relatively nontoxic, with the exception of Azpy-NMe2 in the A549 cell line (Table 1).
Table 1.
IC50 values (μM) for [(η6-arene)Ru(azpy-R)I]PF6 complexes, their corresponding chloride analogues [(η6-arene)Ru(azpy-R)Cl]PF6 and free phenylazopyridine ligands for A2780 ovarian and A549 lung cancer cell lines, and the first and second (where observed) electrochemical reduction potentials (DMF vs. Ag/AgCl)
| Complex* | IC50, μM |
Ered, V | |
|---|---|---|---|
| A2780 | A549 | ||
| 1 (1-Cl) | 4 (>100) | 3 (>100) | −0.40, −1.00 |
| 4 (4-Cl) | 3 (44) | 2 (49) | −0.36 |
| Azpy-NMe2 | >100 | 14 | −1.28 |
| 2 (2-Cl) | 4 (58) | 4 (>100) | −0.33, −0.77 |
| 5 (5-Cl) | 5 (18) | 6 (56) | −0.26, −0.72 |
| Azpy-OH | >100 | >100 | nd† |
| 3 (3-Cl) | >100 (>100) | >100 (>100) | −0.22, −0.74 |
| 6 (6-Cl) | 39 (>100) | 51 (>100) | −0.18, −0.67 |
| Azpy | >100 | >100 | −1.31 (vs. SCE)‡ |
*Iodido complexes 1–6 (and their chlorido analogues 1-Cl to 6-Cl in parentheses).
†nd, not determined.
‡Ref. 14.
Solution Chemistry.
We investigated whether the complexes could undergo activation by hydrolysis. We used conditions that mimic the concentration, pH, exposure time, and temperature used in biological cell tests, and thus monitored 100 μM solutions of the complexes in 10 mM phosphate buffer (containing 95% D2O/5% MeOD, pH 7.3) and at 310 K over 24 h by 1H NMR. For iodido complexes 1, 2, 4, and 5 containing phenylazopyridine ligands with OH or NMe2 substitutents, no changes in the spectra occurred (e.g., Fig. S2 for complex 4) suggesting that no hydrolysis took place. This was confirmed by ESI-MS studies of the NMR sample where only the intact cation was detected [1, m/z found 588.75 (M+), calcd 589.04; 2, m/z found 561.70 (M+), calcd 561.99; 4, m/z found 608.71 (M+), calcd 609.03; 5, m/z found 581.65 (M+), calcd 581.98]. However, the NMR spectra of the unsubstituted phenylazopyridine complexes 3 and 6 changed over the 24-h period (Figs. S3 and S4, respectively). For 6, all peaks for the intact cation disappeared and the only assignable peaks in the spectrum were due to free biphenyl. For 3, the peaks for the intact cation decreased in intensity and peaks for free p-cymene were observed. The slow decomposition of 3 and 6 (50 μM, 10 mM phosphate buffer, 310 K, 24 h) was confirmed by UV-Vis spectroscopy (Fig. S5).
Importantly, with 104 mM NaCl present (close to extracellular [Cl−]) no changes in the UV-Vis spectrum of complex 4 were observed over 24 h at 310 K (see Fig. S6), suggesting that no substitution of iodide by chloride takes place.
Electrochemical Reductions.
In general, the complexes exhibited two electrochemical reductions (in dimethylformamide), the first at ≈ −0.2 to −0.4 V and a second at ≈ −0.7 to −1 V (Table 1). Reductions were also observed near to the solvent cutoff (≈ −2 V), but were not considered further.
In all cases the first reduction (confirmed as a one-electron reduction for complex 3, n = 0.98, Fig. S7) was essentially irreversible (no reverse scan peak at 0.1 Vs−1, Fig. S8). The reduction potential becomes more positive as the arene is changed from p-cymene to biphenyl and also as the chelating azo ligand is changed from azpy-NMe2, to azpy-OH to azpy (Table 1). Based on literature assignments for other ruthenium–phenylazopyridine complexes (14, 15), this reduction can be assigned to addition of an electron into the π* orbital centered on the azo group of the phenylazopyridine ligands to form the azo anion radical (–N=N– + e− →{–N–N–}−·). The relative order therefore reflects the decreasing π-acceptor capability of the substituted azpy ligands with the addition of electron-donating groups onto the phenyl ring (16). The irreversible second reduction step is assigned to conversion to the dianionic species ({–N–N–}2−). Azo groups usually give rise to two separate electrochemical reductions in polar aprotic solvents (17). In aqueous media the two-electron reduction of azo groups is accompanied by proton transfer to give hydrazo groups (NH–NH) in a single step (17).
Reactions with Glutathione.
In cells, the major reducing agent is the tripeptide glutathione (GSH), a thiol present at millimolar concentrations (18). We were thus interested to follow the reactions of the ruthenium complexes with GSH. HPLC and electrospray ionization (ESI)-MS studies showed that the initial step in the reaction of excess GSH with [(η6-biphenyl)Ru(azpy-NMe2)I]PF6, complex 4, and the corresponding chlorido complex involves complete substitution of halide by GS−, to give [(η6-biphenyl)Ru(azpy-NMe2)GS]+ (m/z obs 788.0, calcd 789.2; Fig. S9). Half-lives for this substitution were determined as 1.3 min (kobs = 8.73 × 10−3 s−1) for the chlorido complex 4-Cl, and 17.5 min for the iodido complex 4 (kobs = 6.58 × 10−4 s−1; 298 K by UV-Vis spectroscopy). Thus iodide substitution by GS− occurs 14 times more slowly than chloride substitution. During 1H NMR studies of reactions of [(η6-biphenyl)Ru(azpy-NMe2)I]PF6, 4, and [(η6-biphenyl)Ru(azpy-OH)I]PF6, 5, with glutathione (carried out in deoxygenated solutions under N2 or Ar to avoid autoxidation), it became apparent that these complexes can act as catalysts for the oxidation of GSH to GSSG.
The NMR spectra showed that incubation of 10 mM GSH with 100 μM 4 or 5 (in 10 mM phosphate, pH 7.2, 85% H2O, 10% D2O, 5% acetone-d6, 310 K, followed over 24 h) led to a steady oxidation of 3.7 mM (turnover frequency = 0.30 h−1, R2 = 0.98) and 4.6 mM, (turnover frequency = 0.37 h−1, R2 = 0.99), respectively, of GSH to GSSG (Fig. S10), as evidenced by the appearance of new peaks at δ 3.30 ppm corresponding to the β-CH2 of GSSG (Fig. S10 Inset). The formation of GSSG was further confirmed by ESI-MS spectra of these solutions after 24 h incubation, which showed peaks consistent with the presence of glutathione (m/z obs 305.2, calcd for GS− 305.1) and glutathione disulfide (m/z obs 611.1, calcd for GSSG 611.2).
LC-MS attempts to detect intermediates in the catalytic cycle were unsuccessful, probably because they are transient. Interestingly, bubbles formed in the reaction tubes during the course of the reaction, suggesting that a gas had been produced. A variety of methods were used in attempts to detect H2 during the course of the reaction (colorimetric assays, NMR, and gas chromatography). However, none of these led to H2 detection (see SI Text). The GC analysis showed that small amounts of O2 were present in the head space of reactions despite thorough initial purging with N2 or Ar. No H2O2 was detected in the reaction mixtures by using peroxide test sticks.
Under similar conditions, neither of the free ligands Azpy-NMe2 nor Azpy-OH caused any oxidation of GSH to GSSG. Hence, the redox activity of 2-phenylazopyridine ligands depends on being coordinated to ruthenium.
Detection of Reactive Oxygen Species (ROS) in A549 Cancer Cells.
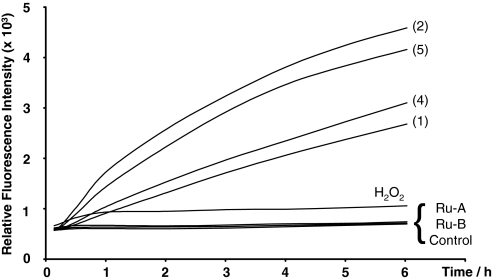
The molecular probe 2′,7′-dichlorodihydrofluorescein-diacetate (DCFH-DA) is taken up by live cells, is hydrolyzed to 2′,7′-dichlorodihydrofluorescein (DCFH), and in the presence of ROS is oxidized to highly fluorescent 2′,7′-dichlorofluorescein (DCF) (19). This probe measures general oxidative stress (20). Fig. 2 shows the increase in fluorescence detected over time after A549 cancer cells were exposed to ruthenium compounds preloaded with DCFH-DA. Compounds 1, 2, 4, and 5 all caused an increase in DCF fluorescence with time, indicating that they all increase the levels of ROS inside A549 cancer cells.
Fig. 2.
Relative increase in DCF fluorescence detected over time after exposure to 25 μM Ru compounds (1, 2, 4, and 5), Ru-A ([(η6-bip)Ru(en)Cl]PF6), Ru-B ([(η6-tha)Ru(en)Cl]PF6), and H2O2. For each compound the fluorescence was averaged over 12 wells (n = 12).
In contrast, the ethylenediamine chelates [(η6-biphenyl)Ru(en)Cl]PF6 or [(η6-tetrahydroanthracene)Ru(en)Cl]PF6 (Ru-A and Ru-B, respectively, in Fig. 2) caused no increase in DCF fluorescence, showing that these two compounds do not generate ROS.
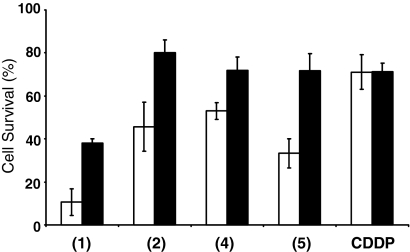
The thiol levels in A549 cells were increased by preincubation with 5 mM N-acetyl-l-cysteine (NAC). The cells were washed to remove excess NAC before treatment with the complexes. Fig. 3 shows that after treatment with complexes 1, 2, 4, or 5 for 24 h incubation followed by 96 h recovery time, there is greater cell survival for the cells that have increased thiol levels, whereas increased thiol levels had little effect on survival of cells treated with cisplatin.
Fig. 3.
Percentage cell survival after 24 h exposure to ruthenium compounds at 1 μM (4), or 5 μM (1, 2, and 5), and cisplatin (CDDP) as a control (5 μM) and 96 h recovery for A549 lung cancer cells (lighter) and A549 cells pretreated with 5 mM N-acetylcysteine for 2 h before drug addition to increase intracellular thiol levels (darker). The error bars are standard deviations from an average of three wells.
Discussion
Cytoxicity Without Hydrolysis.
Many examples are known of metal anticancer complexes containing metal–halide bonds that are activated in cells by hydrolysis and subsequently bind to DNA as a target, for example, PtII diam(m)ine complexes, including the drug cisplatin (21), and diamino RuII arene complexes (22). In contrast, the organometallic iodido RuII arene azopyridine complexes studied in this article are inert toward hydrolysis and yet are still cytotoxic.
For example, complexes 1, 2, 4, and 5 are all resistant to hydrolysis over 24 h at 310 K and at micromolar concentrations and physiological pH. Complexes containing unsubstituted azpy (3 and 6) were not as cytotoxic toward the two cancer cell lines. This low activity can be correlated with their instability in phosphate-buffered solutions (Figs. S3–S5). High cytotoxicity is therefore associated with iodido azopyridine complexes that are highly stable in aqueous media. The corresponding chlorido azpy complexes are much less stable in water, with slow hydrolysis of both Ru–Cl and Ru–arene bonds occurring [t1/2 9–21 h (13)], and these complexes are much less cytotoxic (Table 1). Iodide is less electronegative than chloride, behaves more like a simple two-electron donor, and forms stronger bonds to RuII. The increased electron density at the ruthenium center is therefore available for backdonation onto the arene, which also strengthens the RuII–arene bonds. Hydrolysis rates of RuII arenes decrease when a π-acceptor-chelating ligand is incorporated into the complex, as we have discussed elsewhere (13).
The dramatic increase in cytotoxicity of iodido complexes compared with chlorido complexes observed here is not seen for RuII arene complexes containing the σ-donor ethylenediamine as the chelating ligand; compare, for example, [(η6-benzene)Ru(en)Cl]PF6, IC50 = 17 μM, [(η6-benzene)Ru(en)I]PF6, IC50 = 20 μM (12). In the case of ethylenediamine complexes, the Ru–I bonds are converted to Ru–Cl bonds during the cytotoxicity assays because of the large excess of NaCl in the cell culture medium. In contrast, the iodido phenylazopyridine complexes do not appear to react with excess NaCl (see Fig. S5 for 4).
The iodido complexes also react more slowly with GSH than the chlorido complexes. In cells, GSH exerts a major protective role as a detoxifier of electrophilic substances (E), such as metals, forming thioether conjugates GS-E (18). These conjugates are eliminated via an ATP-dependent GS-E pump (23). GS-conjugation is the major deactivation mechanism for platinum complexes such as cisplatin (24). The greater resistance of the Ru–I bond to conjugation may further enhance their potency compared with chlorido complexes.
Ligand-Based Redox Reactions.
The coordination of phenylazopyridine ligands to arene-ruthenium centers shifts their reduction potentials to biologically accessible values and results in cytotoxic complexes. Phenylazopyridine ligands can accept two electrons in one electrochemically accessible, lowest unoccupied molecular orbital, which is primarily azo in character (25). Coordination of these ligands to the dipositive ruthenium center allows ligand-based reduction to occur at a much more positive potential; the first electrochemical reduction for the free ligand azpy-NMe2, for example, occurs at −1.28 V, compared with −0.40 V for complex 1 and −0.36 V for complex 4 (Table 1).
Catalytic Activity Toward Glutathione.
Ruthenium complexes 4 and 5 oxidize GSH to GSSG catalytically, unlike the free ligands azpy-NMe2 and azpy-OH. Here we have shown that under physiologically relevant conditions micromolar concentrations of organometallic azpy complexes can deplete millimolar amounts of GSH over 24 h by catalytic oxidation to GSSG.
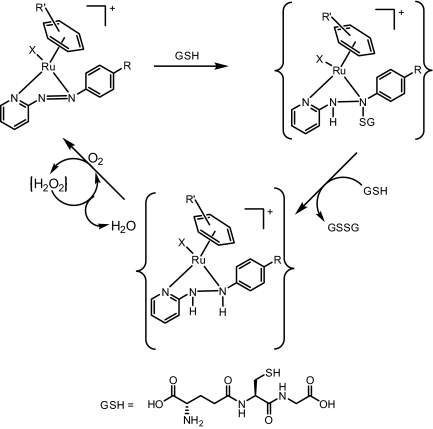
A possible catalytic cycle for the conversion of GSH into GSSG is shown in Fig. 4. The initial step involves the conjugation of GSH across the azo double bond and subsequent reduction of the azo group to an hydrazo group with GSH oxidation. Such a process is reported to occur for the azo ester C6H5N=NCOOCH3 (26) and for the diamide (CH3)2NCON=NCON(CH3)2 (27). However for these reported reactions, a stoichiometric amount of complex is required, that is, the compounds are not catalytic. Because no H2 was detected during the course of the reaction, it seems likely that the second step in which the azo N=N group is regenerated, and the catalytic cycled is completed, proceeds via concomitant hydrogenation of dissolved O2 to generate H2O2. Given that no H2O2 was detected in the reaction mixture, it is reasonable to propose that iodide (displaced from Ru by the initial substitution reaction with GSH) causes its decomposition (I− + H2O2 → IO− + H2O; IO− + H2O2 → I− + H2O + O2). Such catalytic behavior of iodide toward H2O2 is well known (28) and explains the observed gas bubbles (O2).
Fig. 4.
Proposed cycle for the catalytic oxidation of GSH to its corresponding disulfide GSSG by ruthenium(II) arene phenylazopyridine complexes. Initially X is iodide, which is displaced by GS− during the early stages. Released iodide can catalyze the decomposition of H2O2. Bound GSH at pH 7 has deprotonated thiol and carboxyl groups, and a protonated amino group.
Examples in the literature of azo groups facilitating the oxidation of GSH include the Hodgkins and non-Hodgkins lymphoma drug procarbazine [CH3NH–NHCH2C6H4CONHCH(CH3)2], an hydrazo compound that is easily oxidized to the corresponding azo compound and re-reduced via GSH oxidation to GSSG (29). The reoxidation of this hydrazo group is facilitated by O2 inside cells, which is hydrogenated to form H2O2.
Oxidation of GSH to GSSG was not induced by the free ligands. This is perhaps not surprising because electron-donating groups attached to azo groups (here OH or NMe2) decrease their ability to oxidize GSH (17). Coordination to RuII is necessary to make the azo nitrogen atoms positive enough for the redox reaction to occur.
Detection of Reactive Oxygen Species.
The dichlorofluorescein fluorescence experiments showed that the iodido RuII arene N,N-dimethyl- and hydroxy-azopyridine complexes 1, 2, 4, and 5 cause a buildup of reactive oxygen species in A549 lung cancer cells (Fig. 2). Reactive Oxygen Species (ROS) present a higher reactivity than molecular oxygen (30), and encompass not only free radicals such as the superoxide (O2•−), hydroperoxyl radical (HO2•) and the hydroxyl radical (HO•), but also hydrogen peroxide (H2O2) and singlet oxygen (1O2). ROS are continuously generated in small quantities during normal cellular processes and are involved in several biological functions, such as cell growth and signaling (30). Excessive ROS generation inside cells is harmful, causing oxidation of biomolecules such as lipids, cell membranes, proteins, and DNA, and giving rise to oxidative stress (30). Importantly, no DCF fluorescence was detected for ruthenium arene complexes containing ethylenediamine (en) as chelating ligand, illustrating a fundamental difference in the mechanism of action of these two classes (azpy/en) of cytotoxic ruthenium arene complexes.
To assess the impact of ROS inside A549 lung cancer cells, intracellular thiol levels were increased by adding the antioxidant N-acetyl-l-cysteine, which is believed to act by raising intracellular concentrations of cysteine and hence GSH, and also by direct reaction of its thiol group with ROS (31). Treatment of cells with N-acetyl-l-cysteine increased the survival of cells treated with RuII arene phenylazopyridine compounds (Fig. 3) providing evidence that ROS are implicated in cell death.
Oxidative Stress Causes Cell Death.
Oxidative stress is caused by a serious imbalance between the levels of ROS in a cell and its antioxidant defenses in favor of the former (20). On the basis that GSH is the primary cellular antioxidant, it seems likely that the depletion of antioxidants is the primary mechanism by which ROS levels are increased and by which oxidative stress occurs in the systems studied here. Because cancer cells (mainly multidrug-resistant cells) are more sensitive to the additional exposure to ROS than normal cells (32), the proposal has been made that ROS can be used therapeutically to treat cancer (27, 31). Several established anticancer drugs are known to generate ROS or other radicals, including adriamycin (33), arsenic trioxide (34), and cisplatin (35).
Conclusions
Organometallic complexes present versatile platforms for drug design. Appropriate choice of the metal, its oxidation state, and the ligands can allow fine-tuning of the steric and electronic properties of the complex and hence its biological activity and mechanism of action. In the present case we have assembled RuII complexes containing a π-bonded arene ligand together with a strongly chelated σ-donor/π-acceptor azopyridine ligand and iodide as a strong monodentate ligand to produce complexes that are relatively inert toward activation by hydrolysis and yet potently cytotoxic toward cancer cells.
These azopyridine complexes undergo activation by reduction. Whereas azopyridine ligands alone are difficult to reduce, the reduction potentials are biologically accessible when the azopyridine is coordinated to RuII. The complexes can induce redox reactions inside cancer cells leading to an increase in the accumulation of reactive oxygen species. Intriguingly iodido RuII arene azopyridine complexes can act as catalysts for the oxidation of the major intracellular reducing agent glutathione to glutathione disulfide, in a cycle that appears to involve ligand-centered reduction of the azo bond by glutathione forming an hydrazo intermediate, followed by formation of glutathione disulfide and regeneration of the azo bond. Catalytic drugs with ligand-centered mechanisms might have the advantage of not being so readily poisoned as metal-centered catalysts and therefore have greater potential for biological activity.
Materials and Methods
Materials.
4-(2-Pyridylazo)-N,N-dimethylaniline (azpy-NMe2), KI, NH4PF6, dichlorodihydrofluorescein diacetate, glutathione, and N-acetyl-l-cysteine were purchased from Sigma-Aldrich and ascorbic acid from Alfa Aesar. Methanol was anhydrous (Sigma-Aldrich) or dried over Mg/I2, and anhydrous DMF (Aldrich) was used for electrochemical experiments. The starting materials [(η6-arene)RuCl2]2 (arene = p-cymene, biphenyl) were prepared by reaction of RuCl3·xH2O with the reduced form of the arene (11), and the corresponding iodido dimers [(η6-arene)RuI2]2 from the reaction of [(η6-arene)RuCl2]2 with excess KI. The syntheses of 2-phenylazopyridine (azpy) and 4-(2-pyridylazo)phenol (azpy-OH) have been reported previously (13). Full details of the synthesis and characterization of complexes are in the SI Text.
NMR Spectroscopy.
NMR spectra were recorded on either a Bruker Avance 600 MHz equipped with either a TXI [1H, 13C, 15N] x,y,z-gradient probe or a TXI [1H, 13C, 15N] z-gradient cryoprobe, a Bruker Avance 800 MHz equipped with a TCI [1H, 13C, 15N] z-gradient cryoprobe, or a Bruker DPX 360 MHz spectrometer equipped with a QNP [1H, 13C, 15N] probe. 1H NMR signals were referenced to the residual solvent, δ 7.27 (CDCl3), 2.52 (DMSO-d6), and for aqueous solutions (containing D2O or H2O/D2O mixtures) dioxane was added as an internal reference (δ 3.75). The water resonance was suppressed by using a 1D double-pulsed field gradient spin echo (DPFGSE) experiment (36) or presaturation method. All spectra were recorded at 298 or 310 K and data were processed by using XwinNMR (Version 3.6, Bruker).
X-Ray Crystallography.
Diffraction data were collected at 150 K by using Mo Kα radiation (λ = 0.71073 Å) on a Bruker Smart Apex CCD diffractometer. The crystal structure of 4 was solved by using Patterson methods (DIRDIF) (37) and refined against F2 by using SHELXL-97 (38). The H atoms that are part of methyl and hydroxyl groups were located in a difference map, and the groups treated as rotating rigid bodies; other H atoms were placed in calculated positions.
Electrospray Mass Spectrometry.
ESI-MS was obtained on a Micromass Platform II mass spectrometer and solutions were infused directly. The capillary voltage was 3.5 V and the cone voltage was typically varied between 10 and 45 V. The source temperature depended on the solvent used. Both positive and negative ion modes were used (as appropriate).
UV-Vis Spectroscopy.
A Perkin-Elmer Lambda-16 UV/Vis spectrophotometer was used with 1-cm path length quartz cuvettes (0.5 ml) and a PTP1 Peltier temperature controller. Spectra were recorded at 310 K and were processed by using UV-Winlab software for Windows 95.
pH Measurements.
The pH values of aqueous solutions were measured at ≈298 K by using a Corning pH meter 240 equipped with a Thermo micro combination KCl or KNO3 electrode calibrated with pH 4, pH 7, and pH 10 buffer solutions (Sigma-Aldrich). The pH meter readings for D2O solutions were recorded without correction for the effect of deuterium on the glass electrode, and are termed pH*.
Cyclic Voltametry and Coulometry.
Electrochemical studies were performed with General Purpose Electrochemical System (GPES) Version 4.5 software connected to an Autolab system containing a PSTAT20 potentiostat. All of the electrochemical techniques used a three-electrode configuration. The reference electrode was Ag/AgCl in a solution of 0.1 M [TBA][BF4] in DMF against which E1/2 for the ferrocinium/ferrocene couple was measured to be +0.55 V. The working and counter electrodes were a platinum microdisc (0.5-mm diameter) and a large surface area platinum wire, respectively. Coulometric experiments were performed in a conventional H-type cell by using large surface-area Pt working and counter electrodes. All solutions were purged with dry N2 before electrochemical study.
High Performance Liquid Chromatography (HPLC).
A Hewlett–Packard series 1100 quaternary pump and a Rheodyne sample injector with a 1.0-ml loop, an HP 1100 series UV-vis detector, and HP 1100 series Chemstation with an HP enhanced integrator were used. Analytical separations were carried out on a PLRP-S reversed-phase column (250 mm × 7.5 mm, 100 Å, 8 μm, Polymer Labs) with detection at 286 nm. The mobile phases were A, water (for HPLC application, Fisher Chemicals) containing 0.1% TFAH; and B, acetonitrile (for HPLC application, Fisher Chemicals) containing 0.1% TFAH. The flow rate was 3.5 ml·min−1. The gradient (solvent B) was 10–50% over 30 min, 50–90% to 40 min.
Gas Chromatography (GC).
A 5-Å molecular sieve column on an Agilent 7890A GC system equipped with a thermal conductivity detector was used in attempts to detect H2 with N2 as a carrier gas (for details and other methods see SI Text).
IC50 Values.
The concentration of the complexes that caused 50% inhibition of the growth (IC50) was determined by using the sulforhodamine-B (SRB) assay (39) as reported (13). For IC50 determinations on A549 cells with increased thiol levels, cells were preincubated with 5 mM N-acetylcysteine for 2 h, a concentration and time previously reported to be nontoxic for this cell line (40), and confirmed by our controls. The N-acetylcysteine was removed, cells were washed twice in PBS, fresh cell medium was added, and IC50 values were then determined.
Detection of Reactive Oxygen Species (ROS) in A549 Cancer Cells.
The detection of ROS was based on earlier experimental protocols by Wang and Joseph (19) with some modifications. The full procedure is reported in the SI Text.
Reactivity of Complexes 1–6 in Buffered Solutions.
Solutions of ruthenium complexes in MeOD were diluted in 10 mM phosphate buffer/D2O to give a final concentration of 100 μM Ru (95% D2O, 5% MeOD) and NMR spectra were recorded at 310 K after ≈15 min and after 24 h. The pH* values of the samples were 7.35 (1), 7.38 (2), 7.38 (3), 7.40 (4), 7.31 (5), and 7.32 (6), and the samples were kept in the water bath at 310 K between NMR acquisitions. After 24 h, ESIMS was obtained on samples of 1, 2, 4, and 5.
Substitution of Iodide by Chloride.
Complex 4 was dissolved in water, sonicated for 15 min to ensure dissolution, filtered, and then diluted to ≈30 μM. NaCl was added from a concentrated stock solution to give 100 mM NaCl and the corresponding UV-Vis absorbance spectrum was recorded at hourly intervals over 24 h at 310 K.
UV-Vis Spectroscopy and HPLC of Reactions of Complexes 4 and 6 with GSH.
Buffers used were filtered through Chelex-100 to remove iron and degassed by using N2/Ar before dissolving GSH. Solutions of GSH were made fresh for each experiment and the pH adjusted to ≈7 with dilute NaOH/HCl, and the solutions degassed again. Aliquots of complex 4 or 4-Cl in MeOH were added to the GSH solution (final concentrations: 50 μM Ru, 5 mM GSH, 10 mM phosphate buffer; 95% H2O, 5% MeOH); the pH values were 7.06 (4) and 7.03 (4-Cl) for UV-Vis experiments and 7.88 (4) and 7.89 (4-Cl) for HPLC separations.
The rate of substitution of Ru-Cl/I by GS− was determined by UV-Vis spectroscopy, by fitting the change in absorbance at 597 nm with time (298 K) to the appropriate equation for pseudo-first-order kinetics by using Origin version 7.5 (OriginLab Corporation), to give the observed rate constant (kobs) and the corresponding half-life (t1/2). The extent of substitution of Ru-Cl/I by GS− was followed by HPLC. The new fraction appearing over time was collected and analyzed by ESI-MS.
Detection of GSH Oxidation to GSSG.
Buffers were filtered through Chelex-100 to remove iron and degassed by using N2/Ar before dissolving GSH. After dissolution the pH of the GSH solution was adjusted to ≈7 by addition of dilute NaOH/HCl and solutions were degassed again. Solutions of GSH were made fresh for each experiment and aliquots of complexes 4 or 5 dissolved in acetone-d6 were added [final concentrations 100 μM Ru/ligand, GSH 10 mM (when Ru present)/9.5 mM (just ligand), 30 mM phosphate buffer; 10% D2O, 85% H2O, 5% acetone-d6], the measured pH values were 7.62 (4), 7.57 (5), 7.55 (Azpy-NMe2), and 7.59 (Azpy-OH). Solutions of GSH alone (10/9.5 mM, 10% D2O, 85% H2O, 5% acetone-d6) were also prepared at the same time [measured pH 7.40 (control for 4 and 5) and 7.60 (control for Azpy-NMe2 and Azpy-OH)] to confirm minimal autooxidation of GSH, and these samples were kept in a water bath at 310 K for 24 h, then the 1H NMR spectra were recorded. After acquisition of NMR spectra, ESI mass spectra of samples were obtained to confirm the presence of GSSG. The turnover frequency (TOF) was determined as the gradient of a plot of turnover number (TON) versus time. The TON was determined as follows: TON = (IGSSG/{IGSH + IGSSG})([GSH]0/[Ru]), where IGSSG/IGSH is the peak integral for oxidized/reduced glutathione, and [GSH]0 is the initial concentration of GSH. “Quantofix” Peroxide Test Sticks (Aldrich; detection range 1–100 mg H2O2 per liter) were used in attempts to detect H2O2 during reactions of 5 (100 μM, 10 mM GSH, 5% acetone-d6, 95% H2O, 24 h incubation at 310 K).
For details of chemicals and biochemicals, synthesis and characterization of complexes, and methods and techniques used, see SI Text.
Supplementary Material
Acknowledgments.
We thank Emily Jones and Daniel Cole (Oncosense Ltd.) for assistance with the cytotoxicity tests; Lucy Moorcraft, Paul Murray, Neil Robertson, and Lesley Yellowlees (University of Edinburgh) for assistance with the electrochemistry; Colin Murrell and Gerald Chapman (University of Warwick) for assistance with GC; Martin Wills and David Morris (University of Warwick) for discussions on H2 detection and reaction mechanisms; and also members of COST Action D39 for stimulating discussions. This work was supported by a Biotechnology and Biological Sciences Research Council CASE Award (to S.J.D.) and Oncosense Ltd.
Footnotes
Conflict of interest statement: The University of Edinburgh has filed a patent application relating to these ruthenium complexes. This has been licensed to Oncosense Ltd.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Cambridge Structural Database, Cambridge Crystallographic Data Centre, Cambridge CB2 1EZ, United Kingdom (CSD reference no. 671-850). The x-ray crystallographic data for complex 4 can be found in SI Text.
This article contains supporting information online at www.pnas.org/cgi/content/full/0800076105/DCSupplemental.
References
- 1.Halpern J. Organometallic chemistry at the threshold of a new millennium. Retrospect and prospect. Pure Appl Chem. 2001;73:209–220. [Google Scholar]
- 2.Fish RH, Jaouen G. Bioorganometallic chemistry: structural diversity of organometallic complexes with bioligands and molecular recognition studies of several supramolecular hosts with biomolecules, alkali-metal ions, and organometallic pharmaceuticals. Organometallics. 2003;22:2166–2177. [Google Scholar]
- 3.Vessieres A, et al. Modification of the estrogenic properties of diphenols by the incorporation of ferrocene. Generation of antiproliferative effects in vitro. J Med Chem. 2005;48:3937–3940. doi: 10.1021/jm050251o. [DOI] [PubMed] [Google Scholar]
- 4.Ohkuma T, et al. The hydrogenation/transfer hydrogenation network: asymmetric hydrogenation of ketones with chiral η6-arene/N-tosylethylenediamine-ruthenium(II) catalysts. J Am Chem Soc. 2006;128:8724–8725. doi: 10.1021/ja0620989. [DOI] [PubMed] [Google Scholar]
- 5.Lo HC, Buriez O, Kerr JB, Fish RH. Biorganometallic chemistry Part 11. Regioselective reduction of NAD+ models with [Cp*Rh(bpy)H]+: structure-activity relationships and mechanistic aspects in the formation of the 1,4-NADH derivatives. Angew Chem Int Ed. 1999;38:1429–1432. doi: 10.1002/(SICI)1521-3773(19990517)38:10<1429::AID-ANIE1429>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 6.Morris RE, et al. Inhibition of cancer cell growth by ruthenium(II) arene complexes. J Med Chem. 2001;44:3616–3621. doi: 10.1021/jm010051m. [DOI] [PubMed] [Google Scholar]
- 7.Aird RE, et al. In vitro and in vivo activity and cross resistance profiles of novel ruthenium (II) organometallic arene complexes in human ovarian cancer. Br J Cancer. 2002;86:1652–1657. doi: 10.1038/sj.bjc.6600290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang F, et al. Kinetics of aquation and anation of ruthenium(II) arene anticancer complexes, acidity and X-ray structures of aqua adducts. Chem Eur J. 2003;9:5810–5820. doi: 10.1002/chem.200304724. [DOI] [PubMed] [Google Scholar]
- 9.Novakova O, et al. Correlation between cytotoxicity and DNA binding of polypyridyl ruthenium complexes. Biochemistry. 1995;34:12369–12378. doi: 10.1021/bi00038a034. [DOI] [PubMed] [Google Scholar]
- 10.Hayward RL, et al. Investigation of the role of Bax, p21/Waf1 and p53 as determinants of cellular responses in HCT116 colorectal cancer cells exposed to the novel cytotoxic ruthenium(II) organometallic agent, RM175. Cancer Chemother Pharmacol. 2005;55:577–583. doi: 10.1007/s00280-004-0932-9. [DOI] [PubMed] [Google Scholar]
- 11.Habtemariam A, et al. Structure-activity relationships for cytotoxic ruthenium(II) arene complexes containing N,N-, N,O-, and O,O-chelating ligands. J Med Chem. 2006;49:6858–6868. doi: 10.1021/jm060596m. [DOI] [PubMed] [Google Scholar]
- 12.Wang F, et al. Controlling ligand substitution reactions of organometallic complexes: tuning cancer cell cytotoxicity. Proc Natl Acad Sci USA. 2005;102:18269–18274. doi: 10.1073/pnas.0505798102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dougan SJ, Melchart M, Habtemariam A, Parsons S, Sadler PJ. Phenylazo-pyridine and phenylazo-pyrazole chlorido ruthenium(II) arene complexes: arene loss, aquation, and cancer cell cytotoxicity. Inorg Chem. 2006;45:10882–10894. doi: 10.1021/ic061460h. [DOI] [PubMed] [Google Scholar]
- 14.Goswami S, Mukherjee RN, Chakravorty A. Chemistry of ruthenium. 12. Reactions of bidentate ligands with diaquabis[2-(arylazo)pyridine]ruthenium(II) cation. Stereoretentive synthesis of tris chelates and their characterization: metal oxidation, ligand reduction, and spectroelectrochemical correlation. Inorg Chem. 1983;22:2825–2832. [Google Scholar]
- 15.Goswami S, Chakravarty AR, Chakravorty A. Chemistry of ruthenium. 7. Aqua complexes of isomeric bis[(2-arylazo)pyridine]ruthenium(II) moieties and their reactions: solvolysis, protic equilibriums, and electrochemistry. Inorg Chem. 1983;22:602–609. [Google Scholar]
- 16.Krause RA, Krause K. Chemistry of bipyridyl-like ligands. 3. Complexes of ruthenium(II) with 2-((4-nitrophenyl)azo)pyridine. Inorg Chem. 1984;23:2195–2198. [Google Scholar]
- 17.Patai S, editor. The Chemistry of Hydrazo, Azo and Azoxy Groups. Bristol, UK: Wiley; 1975. [Google Scholar]
- 18.Meister A, Anderson ME. Glutathione. Annu Rev Biochem. 1983;52:711–760. doi: 10.1146/annurev.bi.52.070183.003431. [DOI] [PubMed] [Google Scholar]
- 19.Wang H, Joseph JA. Quantifying cellular oxidative stress by dichlorofluorescein assay using microplate reader. Free Radical Biol Med. 1999;27:612–616. doi: 10.1016/s0891-5849(99)00107-0. [DOI] [PubMed] [Google Scholar]
- 20.Halliwell B, Whiteman M. Measuring reactive species and oxidative damage in vivo and in cell culture: How should you do it and what do the results mean? Br J Pharmacol. 2004;142:231–255. doi: 10.1038/sj.bjp.0705776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang CX, Lippard SJ. New metal complexes as potential therapeutics. Curr Opin Chem Biol. 2003;7:481–489. doi: 10.1016/s1367-5931(03)00081-4. [DOI] [PubMed] [Google Scholar]
- 22.Yan YK, Melchart M, Habtemariam A, Peacock AFA, Sadler PJ. Catalysis of regioselective reduction of NAD+ by ruthenium(II) arene complexes under biologically relevant conditions. J Biol Inorg Chem. 2006;11:483–488. doi: 10.1007/s00775-006-0098-5. [DOI] [PubMed] [Google Scholar]
- 23.Balendiran GK, Dabur R, Fraser D. Role of glutathione in cancer. Cell Biochem Funct. 2004;22:343–352. doi: 10.1002/cbf.1149. [DOI] [PubMed] [Google Scholar]
- 24.Bose RN. Biomolecular targets for platinum antitumor drugs. Mini-Rev Med Chem. 2002;2:103–111. doi: 10.2174/1389557024605500. [DOI] [PubMed] [Google Scholar]
- 25.Santra BK, Lahiri GK. Cobalt-mediated direct and selective aromatic thiolation in the complex [CoIII(o-SC6H4N=NC5H4N)2]ClO4. Synthesis, spectroscopic characterisation and electron-transfer properties. J Chem Soc Dalton Trans. 1997:1883–1888. [Google Scholar]
- 26.Kosower EM, Kosower NS. Lest I forget thee, glutathione. Nature. 1969;224:117–120. doi: 10.1038/224117a0. [DOI] [PubMed] [Google Scholar]
- 27.Kosower EM, Miyadera T. Glutathione. 6. Probable mechanism of action of diazene antibiotics. J Med Chem. 1972;15:307–312. doi: 10.1021/jm00273a023. [DOI] [PubMed] [Google Scholar]
- 28.Hansen JC. The iodide-catalyzed decomposition of hydrogen peroxide: A simple computer-interfaced kinetics experiment for general chemistry. J Chem Educ. 1996;73:728–732. [Google Scholar]
- 29.Renschler MF. The emerging role of reactive oxygen species in cancer therapy. Eur J Cancer. 2004;40:1934–1940. doi: 10.1016/j.ejca.2004.02.031. [DOI] [PubMed] [Google Scholar]
- 30.Gomes A, Fernandes E, Lima JLFC. Fluorescence probes used for detection of reactive oxygen species. J Biochem Biophys Methods. 2005;65:45–80. doi: 10.1016/j.jbbm.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 31.Aruoma OI, Halliwell B, Hoey BM, Butler J. The antioxidant action of N-acetylcysteine: its reaction with hydrogen peroxide, hydroxyl radical, superoxide, and hypochlorous acid. Free Radical Biol Med. 1989;6:593–597. doi: 10.1016/0891-5849(89)90066-x. [DOI] [PubMed] [Google Scholar]
- 32.Agostinelli E, Seiler N. Non-irradiation-derived reactive oxygen species (ROS) and cancer: Therapeutic implications. Amino Acids. 2006;31:341–355. doi: 10.1007/s00726-005-0271-8. [DOI] [PubMed] [Google Scholar]
- 33.DeAtley SM, et al. Antioxidants protect against reactive oxygen species associated with adriamycin-treated cardiomyocytes. Cancer Lett. 1999;136:41–46. doi: 10.1016/s0304-3835(98)00306-1. [DOI] [PubMed] [Google Scholar]
- 34.Chou W-C, et al. Role of NADPH oxidase in arsenic-induced reactive oxygen species formation and cytotoxicity in myeloid leukemia cells. Proc Natl Acad Sci USA. 2004;101:4578–4583. doi: 10.1073/pnas.0306687101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siomek A, et al. Severe oxidatively damaged DNA after cisplatin treatment of cancer patients. Int J Cancer. 2006;119:2228–2230. doi: 10.1002/ijc.22088. [DOI] [PubMed] [Google Scholar]
- 36.Hwang T-L, Shaka AJ. Water suppression that works. Excitation sculpting using arbitrary wave-forms and pulsed-field gradients. J Magn Reson Ser A. 1995;112:275–279. [Google Scholar]
- 37.Beurskens PT, et al. The DIRDIF-99 program system. The Netherlands: Crystallography Laboratory, University of Nijmegen; 1999. [Google Scholar]
- 38.Sheldrick GM. SHELXL-97. Göttingen, Germany: University of Göttingen; 1997. [PubMed] [Google Scholar]
- 39.Skehan P, et al. New colorimetric cytotoxicity assay for anticancer-drug screening. J Natl Cancer Inst. 1990;82:1107–1112. doi: 10.1093/jnci/82.13.1107. [DOI] [PubMed] [Google Scholar]
- 40.Moodie FM, et al. Oxidative stress and cigarette smoke alter chromatin remodeling but differentially regulate NF-kappaB activation and proinflammatory cytokine release in alveolar epithelial cells. FASEB J. 2004;18:1897–1899. doi: 10.1096/fj.04-1506fje. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.