Abstract
When SIN1 (MAPKAP1) was used as the bait in a two-hybrid screen of a human bone marrow cDNA library, its most frequent partner was poly(rC) binding protein 2 (PCBP2/hnRNP-E2), which associates with the N-terminal domain of SIN1 and can be coimmunoprecipitated with SIN1 and the cytoplasmic domain of the IFN receptor IFNAR2 from HeLa cells. SIN1, but not PCBP2, also associates with the receptors that bind TNFα. PCBP2 is known to bind pyrimidine-rich repeats within the 3′ UTR of mRNAs and has been implicated in control of RNA stability and translation and selective cap-independent transcription. RNAi silencing of either SIN1 or PCBP2 renders cells sensitive to basal and stress-induced apoptosis. Stress in the form of TNFα and H2O2 treatments rapidly raises the cell content of SIN1 and PCBP2, an effect reversible by inhibiting MAPK14. A meta analysis of human microarray information with an algorithm that discerns similarities in gene-regulatory profiles shows that SIN1 and PCBP2 are generally coregulated with large numbers of genes implicated in both cell survival and death and in cellular stress responses, including RNA translation and processing. We predict that SIN1 is a scaffold protein that organizes antiapoptotic responses in stressed cells, whereas PCBP2, its binding partner, provides for the selective expression of cell survival factors through posttranslational events.
Keywords: apoptosis, interferon, stress kinase-interacting protein 1, yeast two-hybrid assay
When faced with environmental stressors such as reactive oxygen species, inflammatory shock, or viral infection, cells may react by either boosting protective mechanisms or undergoing programmed cell death. A delicate balance is maintained between pathways that promote survival or death, and this balance can be affected by a number of cytokines, including members of the IL, IFN, and TNF families (1). The type 1 IFN in particular have the ability to promote (2, 3) or prevent (4) cell death depending on the circumstances. Exposure of cells to stress induces compensatory activation of multiple intracellular signaling pathways, among which is the one controlled by p38 MAPK (MAPK14), also known as stress-activated protein kinase (5).
We previously used yeast two-hybrid genetic screens to identify stress kinase-interacting protein 1 (SIN1; MAPKAP1) as a factor that associates with the IFN receptor subunit IFNAR2 (6). Cells in which the SIN1 gene has been ablated or where gene expression had been knocked down by RNAi silencing are susceptible to stress (7, 8). SIN1 is represented as a single gene in all metazoan species and fungi so far examined, is highly conserved in vertebrates, and is expressed in most, if not all, tissues of the mouse (6). Despite this ubiquity, it has until recently been poorly studied, and its mechanisms of action remain unclear. It was originally described as a gene product that modulated RAS function in Saccharomyces cerevisiae (9). Later, a yeast two-hybrid screen of a Schizosaccharomyces pombe cDNA library identified an apparent ortholog of the Avo1 gene that binds the StyI/Spc1, stress-activated MAP kinase (SAPK) (10). SIN1 forms complexes with the stress-associated kinases JNK/MAPK8 (11) and MEKK2/MAP3K2 (12), as well as MAPK14 and the transcription factor ATF2 (13). It is also an essential, stabilizing component of mammalian target of rapamycin complex 2 (TORC2) (7). A fission yeast strain lacking Avo1 was sterile and sensitive to multiple types of stress, including heat shock, and had delayed cell cycles compared with a parental strain (10). The present study, which identifies an RNA binding protein, PCBP2, as a functional partner for SIN1, further implicates SIN1 as central to cellular responses to stress and control of apoptosis in eukaryotic cells through involvement in control of RNA stability and translation.
Results
Identification of Poly(rC) Binding Protein 2 (PCBP2) as a Partner for SIN1.
Yeast library screening with full-length SIN1 as bait yielded 85 positive colonies harboring both the activation domain (AD) and DNA-binding domain vectors, of which 49 had ORFs without frame shifts [supporting information (SI) Table S1]. The gene products included ribosomal protein S20, whose interaction with the bait may be nonspecific, hemoglobin α2 (HBA2), defensin α1 (DEFA1), TGFβ-induced apoptosis protein 1 (TAIP2/FAM130A2), WAS protein family member 2 (WAVE2/WASF2), and cardiac troponin I-interacting protein kinase (TNNI3K). Two frequent cDNAs encoded polyglutamine binding protein 1 (PQBP1) and poly(rC) binding protein 2 (PCBP2, also known as hnRNP E2 or αCP-2). Here we have concentrated on PCBP2, in part because its mRNA is an activator of 2′,5′ oligoadenylate synthetase (OAS1) (14), a gene induced by type 1 IFN, and because PCBP2 also plays a role in controlling mRNA stability (15), suggesting a mechanism whereby SIN1 may promote cell survival in response to stress.
Interaction of SIN1 with PCBP2 in Vitro and in Vivo.
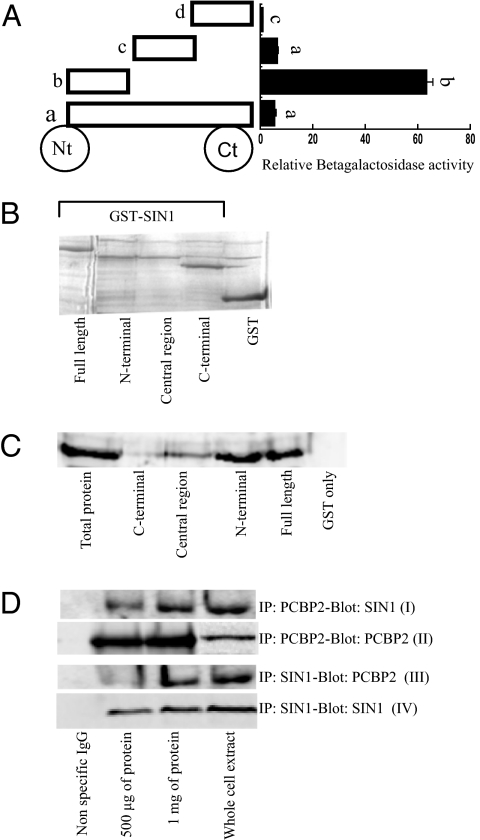
Yeast two-hybrid assays were used to identify the region of SIN1 that binds PCBP2. The strength of interaction was assessed by the intensity of α-galactosidase hydrolysis on the plate (MEL1) and by measuring liquid β-galactosidase activity (LacZ). The 180-aa N terminus provided the highest activities [Fig. 1A and Fig. S1 A and B]. Full-length SIN1 and the 190-aa central region provided modest activities, whereas the 152-aa C terminus was inactive, suggesting that the N terminus of SIN1 is the primary site of interaction with PCBP2.
Fig. 1.
Analysis of the interaction between SIN1 and PCBP2. (A) Yeast two-hybrid analysis of strength of interaction between SIN1 and PCBP2. β-Galactosidase activity (means ± SEM) from yeast strains expressing the different SIN1 constructs (full-length, a; N-terminal 180 aa, b; mid-region of 190 aa, c; C-terminal 152 aa, d). Where letters above bars differ, values differ (P < 0.05). (B) SDS/PAGE of 10 μg of each purified GST-SIN1 construct stained by Gelcode Blue. (C) GST pull-down assay. Each of the SIN1–GST fusion proteins (lanes 2–5) was incubated with 150 μg of HeLa cell extract and collected on glutathione beads. The complexes were analyzed by Western blotting with anti-PCBP2 antibody. Lane 1, total protein (50 μg of whole cell lysate); lane 6, GST-only control. (D) Coimmunoprecipitation of SIN1 and PCBP2 from cell extracts. Lanes 2 and 3, HeLa cell lysates (500 μg and 1 mg) were incubated with either anti-PCBP2 (I and II) or anti-SIN1 (III and IV). Immunocomplexes were analyzed by Western blotting with either anti-SIN1 (I and IV) or anti-PCBP2 (II and III). In lane 1 a nonspecific IgG was used for precipitation; in lane 4, 50 μg of whole-cell lysate was analyzed.
The results of the two-hybrid assay were confirmed by using in vitro GST pull-down and coimmunoprecipitation assays. GST fusion proteins were coupled to Sepharose beads (Fig. 1B). Both full-length SIN1 and its N-terminal fragment bound PCBP2 present in HeLa cell extracts, whereas the central and C-terminal regions were far less efficient in trapping the protein (Fig. 1C). Endogenous PCBP2 was then collected from HeLa cell extracts with anti-PCBP2, and immune complexes were analyzed by SDS/PAGE and Western blotting with anti-SIN1 antibody. This and the reverse experiment, in which SIN1 antibody was used to bind endogenous protein and anti-PCBP2 used for Western blotting, were consistent with the conclusion that SIN1 and PCBP2 associate in vivo (Fig. 1D, first and third blots).
We also used coimmunoprecipitation to determine whether SIN1 and PCBP2 are coimplicated in responses to two stress-related cytokines, type I IFN and TNFα. SIN1 had previously been discovered to interact with IFNAR2 in yeast two-hybrid assays (6). Here we observed that the IFNAR2 immune complex from HeLa cells contained PCBP2 as well as SIN1 (Fig. S2 A and B). IFN treatment appeared to elevate the amount of PCBP2 and SIN1 bound to IFNAR2, but not dramatically. Coimmunoprecipitation with antibodies directed against SIN1, PCBP2, and TNF receptors showed that SIN1 but not PCBP2 interacted with both TNFR1 and TNFR2 (TNFRSF1A and TNFRSF1B) (Fig. S2 C–E). As with IFNα, TNFα treatment appeared to cause a slight increase in the amount of SIN1 bound to these receptor proteins (Fig. S2 A, C, and D). The absence of PCBP2 in the complex with SIN1 and TNF receptors could be due either to steric effects excluding PCBP2 or to the relative instability of the immune complex during its isolation.
Silencing of SIN1 and PCBP2 and Its Effects on Sensitivity to Environmental Stress.
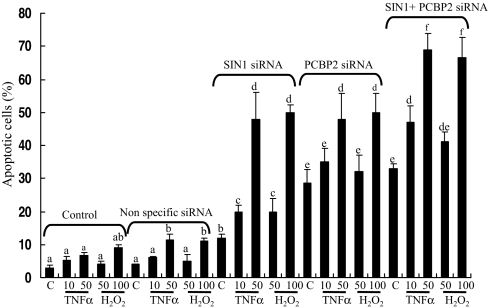
siRNAs targeted to SIN1 (Ambion ID 130550) and PCBP2 (ID 143967) but not a negative control siRNA (Ambion control 1) efficiently knocked down both SIN1 and PCBP2 in HeLa and NIH 3T3 cells at ≈40 nM concentration (Table S2 and Fig. S3 A and B). We then examined whether such silencing led to increased rates of apoptosis and influenced responses to environmental stress induced by treatment with either TNFα or H2O2. The nonspecific siRNA provided a slightly higher rate of background apoptotic cell death than was observed in the nontransfected cells. The siRNA control cells also became slightly more sensitive to cell death induced by TNFα and H2O2, presumably as the outcome of cell injury caused by transfection. Silencing of SIN1 approximately doubled apoptosis to ≈12% in HeLa cells (Fig. S4 A and B) and to 10% in 3T3 cells (Fig. S4 C and D), even in the absence of any stressor. At the highest concentrations TNFα and H2O2 treatment killed about half the cells after SIN1 silencing. Knockdown of PCBP2 expression was about as effective at increasing both background cell death and apoptosis in response to TNFα and H2O2 as knockdown of SIN1. A combination of SIN1 and PCBP2 siRNA increased apoptosis above that from the single treatments, with almost 70% of the cells dying in response to 50 ng/ml TNFα and 100 μM H2O2 (Fig. 2). We conclude that SIN1 and PCBP2 participate in cell survival pathways that protect cells from apoptosis.
Fig. 2.
Apoptosis in response to cellular stress after siRNA silencing of SIN1 and PCBP2 in HeLa cells. HeLa cells were not transfected (control) or were transfected with a nonspecific siRNA, SIN1 siRNA, PCBP2 siRNA (each 40 nM), or SIN1 plus PCBP2 siRNAs (each 20 nM). After 72 h, cells were left untreated (C) or were exposed to TNFα (10 or 50 ng/ml) or H2O2 (50 or 100 μM) for 6 h. Cells were TUNEL-stained and counterstained with propidium iodide. Results are shown as mean percentage of apoptotic cells ± SEM from five experiments, with a minimum of three fields of >300 cells counted in each. Where letters differ above bars within individual treatments, values are different (P < 0.05).
Expression of SIN1 and PCBP2 Protein Under Environmental Stress.
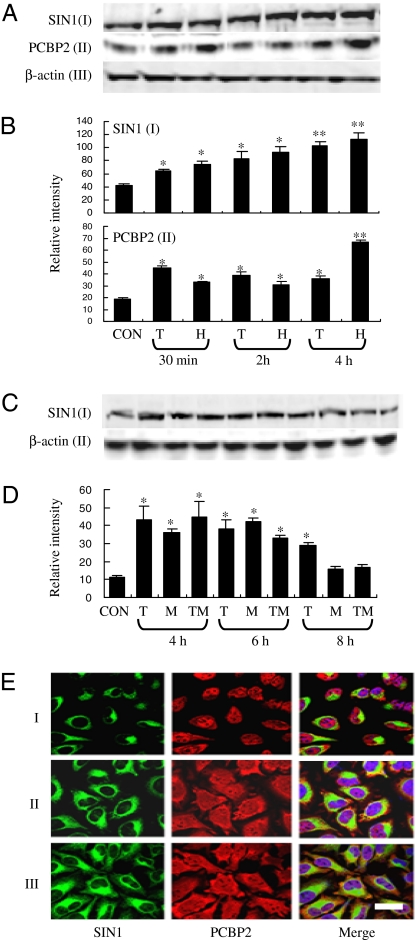
The concentration of SIN1 and PCBP2 in HeLa cells rose significantly (P < 0.05) within 30 min of exposure to TNFα and H2O2 relative to control cells and continued to climb over the first 4 h of treatment, when concentrations had increased almost 3-fold for each protein (Fig. 3 A and B). Treatment of control cells with the proteasome inhibitor MG132 in absence of TNFα and H2O2 also led to an approximate tripling of SIN1 within 4 h (Fig. 3 C and D), suggesting that SIN1 is normally turned over relatively rapidly through proteasome activity. These levels were sustained over 6 h but declined by 8 h, although no such dropoff was noted in either untreated control cells (data not shown) or cells exposed to TNFα alone. The decline in SIN1 between 6 h and 8 h after MG132 treatment was not due to the toxicity of the compound, because MG132 did not alter the percentage of viable cells as determined by either trypan blue exclusion (data not shown) or TUNEL staining (Fig. S5A). An effect similar to that observed with MG132 was noted with a second proteasomal inhibitor, lactacystin (Fig. S5B), strongly suggesting that the rise in SIN1 concentrations accompanying cellular stress may be due to reduced proteasomal activity.
Fig. 3.
Effects of TNFα and H2O2 on SIN1 and PCBP2 expression (A, B, and E) and of MG132 (C and D) on SIN1 protein expression in HeLa cells. (A) HeLa cells were left untreated (CON) or were exposed to either 50 ng/ml TNFα (T) or 100 μM H2O2 (H) for 30 min, 2 h, or 4 h in serum-free medium. Total cell lysates (100 μg) were analyzed by Western blotting with anti-SIN1 (I) or anti-PCBP2 (II). III shows β-actin as loading control. The white “gaps” between some of the images indicate where irrelevant lanes have been omitted to construct the figure. (B) Relative band intensities from three experiments identical to that performed in A plotted as means ± SEM (*, differs from control, P < 0.05; **, P < 0.01). (C) HeLa cells were left untreated (CON) or were exposed to 50 ng/ml TNFα (T), 10 μM MG132 (M), or TNFα and MG132 together (TM) for 4 h, 6 h, and 8 h in serum-free medium. Cell lysates (100 μg of protein) were analyzed by Western blotting with anti-SIN1 (I) or anti-β-actin (II). (D) Relative band intensities from three experiments identical to that performed in C plotted as means ± SEM. (E) Localization of SIN1 (green) and PCBP2 (red) in HeLa cells (DAPI, blue). Cells were untreated (I) or were treated with 50 ng/ml TNFα (II) or 100 μM H2O2 (III) for 6 h. Cells were immunostained for either SIN1 or PCBP2. (Scale bar: 20 μm.) A more complete time course is provided in Fig. S6.
After TNFα and H2O2 addition, HeLa cells became more flattened, but, despite the shape change, SIN1 distribution did not change greatly and remained cytoplasmic with a strong, perinuclear localization (Fig. 3E and Fig. S6). As anticipated, SIN1 fluorescence in the treated cells was enhanced with increased time of exposure to TNFα and H2O2. PCBP2, which in untreated cells was more strongly concentrated in nuclei than in cytoplasm, showed the reverse pattern after treatment with either stressor. Presumably this change in location provides PCBP2 an opportunity to interact with SIN1, although the localization patterns of the two antigens were clearly not identical after stress, as PCBP2 became concentrated most strongly at the cell margins, especially at projections that resemble focal adhesions.
Possible Link Between the MAPK14 Pathway and TNFα Up-Regulation of SIN1.
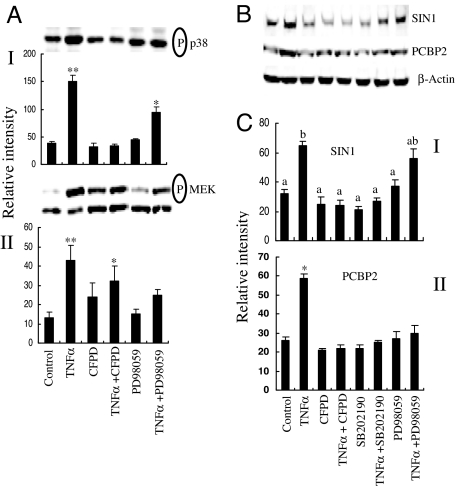
TNFα markedly increased the phosphorylation of p38MAPK (MAPK14) in HeLa cells (Fig. 4AI), and this effect, as expected, was blocked by inhibitors of the MAPK14 pathway but not by the MEK1/2 (MAP2K1/2) inhibitor PD98059. TNFα treatment also augmented phosphorylation of MAP2K1/2, a process that could be largely blocked by PD98059 and to a lesser extent by the MAPK14 pathway inhibitors (Fig. 4AII).
Fig. 4.
Effects of treatment with TNFα and MAP kinase inhibitors on concentrations of phosphorylated MAP kinases, SIN1, and PCBP2 in HeLa cells. (A) Western blot analysis and scanning quantitation by using antibodies to phosphorylated p38 MAP kinase (MAPK14) (I) and phosphorylated MEK1/2 (MAP2K1/2) (II) after culture for 4 h in serum-free medium in the presence of TNFα (50 ng/ml), the p38 MAPK inhibitor CFPD (10 μM), and MEK inhibitor PD98059 (10 μM). Values are means ± SEM of densitometric scans for three independent experiments (*, differs from control, P < 0.05; **, P < 0.01). (B and C) Western blot analysis and scanning quantitation of SIN1 (I) and PCBP2 (II) after culture for 4 h in the presence of TNFα (50 ng/ml), CFPD (10 μM), SB2022190, and PD98059 (10 μM). Values are means ± SEM of scans for three independent experiments. In C, values are significantly different (P < 0.05) where letters above bars differ (I) or are marked by an asterisk (II).
We then examined whether MAPK pathway inhibitors had the ability to interfere with the up-regulation of SIN1 expression by TNFα in HeLa cells. After 4 h of exposure to TNFα, the concentrations of SIN1 and PCBP2 in cell extracts were approximately double that in controls (Fig. 4 B and C), with the increase largely reversed by the presence of either MAPK14 inhibitor. Concentrations did not fall significantly in response to the inhibitors in cells not exposed to TNFα. PD98059 had a tendency to reverse the TNFα-induced increase in SIN1 concentrations, but the values were not significant.
A Bioinformatics Analysis for Genes That Are Coexpressed with SIN1 and PCBP2.
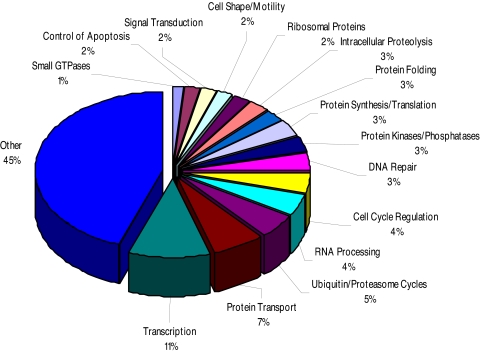
Our experiments show that a reduction in SIN1 and PCBP2 expression exacerbates cell death in response to stress and that TNFα increases expression of these proteins in part via MAPK14 signaling. We therefore used a bioinformatics approach to identify gene networks linked to these gene products and to determine whether they too were associated with cellular response to stress. In particular, we sought genes that demonstrated expression profiles similar to those of SIN1 and PCBP2 across a broad range of cell types and physiological situations (Table S3). To this end, we queried 13 human datasets (Table S3), each based on >50 Affymetrix U133 Arrays, and selected those genes, so-called “profile neighbors,” that showed the strongest (P < 0.01) association with SIN1 and PCBP2 expression patterns, respectively (Dataset S1, 1,864 genes; Dataset S2, 1,337 genes). Of the coregulated genes, 984 (Dataset S3) are profile neighbors to both SIN1 and PCBP2, i.e., are common to Dataset S1 and Dataset S2. Gene ontologies were used to group these 984 genes into functional classes (Fig. 5). The categories with the largest numbers of coregulated genes were in transcription and its control (101 genes), DNA repair (34 genes), and regulation of the cell cycle (41 genes). Another large class of 44 genes had functions relating to RNA processing and included several RNA binding proteins. A large number of mitochondrial ribosome proteins were also represented. Additional groupings included genes directly implicated in control of apoptosis, small GTPases, protein kinases (including MAPK1, MAP2K2, MAPKAPK3, MAP3K11, RAF1, and AKT1), protein phosphatases, and genes with roles in cell motility and shape change.
Fig. 5.
Classes of annotated genes that demonstrate expression profiles similar to SIN1 and PCBP2. Meta analyses of microarray data were used to identify genes that are significantly coregulated with both SIN1 and PCBP2 (Dataset S3). Gene ontologies were used to identify functional classes of the 984 annotated coregulated genes.
PCBP1 is prominent in the neighbor list (Dataset S3), suggesting that its expression is closely tied to that of SIN1 and PCBP2. Among other genes whose expression is most highly connected to that of SIN1 and PCBP2 are APH1A, an essential component of the γ-secretase complex; PGD (phosphogluconate dehydrogenase), a rate-limiting step of the pentose phosphate pathway whose activity generates NADPH; TIMM23, a protein translocase found on the inner mitochondrial membrane; NDUF, a subunit of NADH-ubiquinone oxidoreductase; and DDOST (a dolichol oligosaccharide transferase), which could be involved in responses within the endoplasmic reticulum to stress (16).
Discussion
The study provides further evidence that SIN1 plays a critical role in one or more pathways that modulate stress responses in mammalian cells. SIN1 had been poorly studied until recently, although it is currently commanding considerable attention (17). SIN1 was first identified as a component of stress pathways in yeast species (9). It forms a component of the TORC2 complex in S. cerevisiae (18), Dictyostelium (19), and mammalian cells (7, 8). Importantly, TORC2, with SIN1 playing an essential directive role, regulates responses to nutrient stressors through its ability to phosphorylate and control the activity of the cell survival kinase AKT1 (protein kinase B/rac protein kinase), whose gene expression profile significantly correlates with SIN1 and PCBP2 in microarray analyses (Dataset S3). In addition to its presence in the TORC2 complex, SIN1 interacts with various stress-associated kinases (11–13) and small GTPases (20), which are also highly represented in the list of SIN1/PCBP2 expression profile neighbors. Because there is no evidence that the rather featureless SIN1 polypeptide has any catalytic properties (20, 21), it may provide a platform for organizing responses to a variety of external cell stressors. The data presented here, for example, that SIN1 expression is necessary to protect mouse and human cells from apoptosis and that its expression is up-regulated when cells are treated with either TNFα or H2O2, are entirely consistent with such a role.
Because PCBP2 and SIN1 interact directly, and siRNA silencing of either PCBP2 or SIN1 expression had almost identical effects on apoptosis, it seems reasonable to conclude that the two proteins are part of a pathway that protects against stress-induced cell death. This inference, which is discussed further below, is further supported by the meta analysis finding that PCBP2 and SIN1 genes are coregulated with at least 900 genes, the majority of which have roles in cell survival and cell death after stress.
PCBP2 and its close relative PCBP1 are members of a family of widely expressed proteins that bind nucleic acids through conserved KH-box domains (15, 22). Both recognize sequences harboring tandem pyrimidine-rich elements with poly(C) stretches within the 3′ UTRs and can confer increased stability (23–25) and in some cases translational silencing (25). In addition to stabilizing mRNAs, PCBP2 has been implicated in regulating transcription (26) and in enhancing translation through a cap-independent mechanism involving an internal ribosome entry site in the 5′ UTR (IRES) (15). Several mammalian genes involved in protecting against apoptosis are believed to be translated by cap-independent processes (27), presumably as a means of evading global down-regulation of protein synthesis and the caspase-based destruction of the translational initiation factor EIF4GI (28, 29), thereby allowing selective survival genes to be preferentially expressed when the stress experienced by the cell is not cripplingly severe. Significantly, the gene for EIF4G1 is coregulated with SIN1 and PCBP2 (Dataset S3). PCBP2 and other RNA binding proteins, if appropriately directed, have the potential to exercise a variety of means either to silence or to enhance translation of select transcripts after cells confront changes in their extracellular environment (15).
The meta analysis of human microarray data supports the hypothesis that SIN1 plays a central, directive role in controlling apoptosis. With few exceptions, genes and pathways regulated in concert with SIN1 are involved in reacting to various forms of stress. SIN1 appears to occupy an important node in a network of pathways that safeguard cells against environmental affronts and subsequently allow the cells either to die or to recover from damage. PCBP2, which is as vital as SIN1 in shielding against apoptosis, is also expressed coordinately with genes that encode large numbers of cell survival as well as cell death factors.
Apoptosis is a finely tuned process. Severe stress initiates cell death whereas transient or less severe trauma may induce a prosurvival response (29). Both TNFR1 and TNFR2 are capable of triggering apoptosis (1), but this process can be modulated by factors such as MAPK8 signaling, polyamines, and silencer of death domain protein (30, 31). Similarly, type 1 IFN can cause programmed cell death (2, 32) and up-regulate genes that drive apoptosis (3, 33, 34), most likely as part of an antiviral response. However, type I IFNs are not, under most circumstances, cytotoxic and can act as survival factors (4). SIN1, which binds the C terminus of IFNAR2 and also forms complexes with TNFR1 and TNFR2 (Fig. S2), may help to control the balance between cell survival and cell death decisions after either type I IFN or TNFα binding to their receptors. The SIN1/PCBP2/IFNAR2 complex may represent the initiating step in a previously unrecognized IFN-mediated signaling pathway involved in cell survival.
In conclusion, we predict that SIN1 is intimately involved in survival decisions by the cell. We suggest that, as a potential scaffold protein, SIN1 can receive input from a variety of signaling pathways and then set in motion processes that either protect against apoptosis or drive programmed cell death, including the regulation of selective mRNA translation through the mobilization of RNA binding proteins such as PCBP2.
Materials and Methods
Yeast Two-Hybrid Screen.
A human bone marrow library, pretransformed into Y187 yeast, was mixed with a concentrated culture of AH109 pretransformed with full-length SIN1 in YPDA medium. The mixed yeast culture was maintained at 30°C at 30 rpm (Innova44 shaker incubator; New Brunswick Scientific, Edison, NJ) for 24–30 h and then plated on an SD/_Trp-Leu-His-Ade plate containing kanamycin and 15 mM 3-aminotriazole. Yeast colonies >2 mm in diameter were picked for further analysis. See SI Materials and Methods for details.
Interaction Between Truncated SIN1 Polypeptides and PCBP2.
Y187 yeast transformed with PCBP2 in an AD vector were mated with AH109 yeasts having SIN1 deletion constructs (N-terminal, middle half, and C-terminal; Fig. S1 A and B and Table S4). After selection on SD/_Trp-Leu-His-Ade plates, the relative strength of MEL1 and LacZ reporter gene expression was assessed. For full details see SI Materials and Methods.
GST Pull-Down Assay, Coimmunoprecipitation, and Western Blotting.
GST pull-down assays, coimmunoprecipitation, and blotting were performed by standard procedures (35). See SI Materials and Methods for details.
Cell Culture.
HeLa (CCL-2.2) cells (American Type Culture Collection) were cultured in Eagle's MEM with 2 mM l-glutamine and Earle's balanced salts containing 1.5 g/liter NaH2CO3, 0.1 mM nonessential amino acids, 1.0 mM Na pyruvate, and 10% FBS. NIH 3T3 cells (CRL-1658) were grown in DMEM with 4 mM l-glutamine adjusted to contain 1.5 g/liter NaH2CO3 and 10% FBS.
Transfection of siRNA.
The siRNA designed to silence SIN1 and PCBP2 gene expression and a nonspecific control were purchased from Ambion (Table S2). The transfection reagent used for all siRNAs was siPORTNeoEX. Three siRNA were tested for each gene, and the one giving optimal silencing was selected.
TUNEL Staining and Estimation of Apoptosis.
Transfected cells (40 nM siRNAs) were exposed to either 10 and 50 ng/ml TNFα or 50 and 100 μM H2O2. After 6 h, cells were fixed in 4% paraformaldehyde and apoptotic nuclei were identified by using the DeadEnd Fluorometric TUNEL System (Promega). See SI Materials and Methods for details.
Immunofluorescent Localization of Antigens.
Immunofluorescence localization of SIN1 and PCBP2 was performed by standard procedures. See SI Materials and Methods for details.
Predicting Functional Partners for SIN1 and PCBP2.
Human microarray data from the National Center for Biotechnology Information Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/) Simple Omnibus in Text Format (SOFT) were analyzed to determine the datasets in which SIN1 and PCBP2 showed a significant (up or down) change in expression level. In the second step, meta analysis was performed on these datasets to determine which genes were coexpressed with SIN1 and PCBP2. See SI Materials and Methods for details. Readers requiring further information should contact G.P.S.
Statistical Analyses.
The results from apoptotic assays and the densitometric scanning of Western blots are expressed as mean ± standard error. Statistical analyses were performed by ANOVA followed by least-squares mean t test with GraphPad Prism version 4.03. Significance was accepted at P < 0.05.
Supplementary Material
Acknowledgments.
We thank Dr. Mark Hannink for input during the study, Ms. Norma McCormack for editorial assistance, Dr. Shuzong Wang for preparing the rabbit antiserum to SIN1, and Dr. Mark Ellersieck for statistical advice.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0803182105/DCSupplemental.
References
- 1.Locksley RM, Killeen N, Lenardo MJ. The TNF and TNF receptor superfamilies: Integrating mammalian biology. Cell. 2001;104:487–501. doi: 10.1016/s0092-8674(01)00237-9. [DOI] [PubMed] [Google Scholar]
- 2.Chawla-Sarkar M, et al. Apoptosis and interferons: Role of interferon-stimulated genes as mediators of apoptosis. Apoptosis. 2003;8:237–249. doi: 10.1023/a:1023668705040. [DOI] [PubMed] [Google Scholar]
- 3.Leaman DW, et al. Novel growth and death related interferon-stimulated genes (ISGs) in melanoma: Greater potency of IFN-beta compared with IFN-alpha2. J Interferon Cytokine Res. 2003;23:745–756. doi: 10.1089/107999003772084860. [DOI] [PubMed] [Google Scholar]
- 4.Yang CH, Murti A, Pfeffer LM. Interferon induces NF-kappa B-inducing kinase/tumor necrosis factor receptor-associated factor-dependent NF-kappa B activation to promote cell survival. J Biol Chem. 2005;280:31530–31536. doi: 10.1074/jbc.M503120200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kyriakis JM, Avruch J. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol Rev. 2001;81:807–869. doi: 10.1152/physrev.2001.81.2.807. [DOI] [PubMed] [Google Scholar]
- 6.Wang SZ, Roberts RM. Interaction of stress-activated protein kinase-interacting protein-1 with the interferon receptor subunit IFNAR2 in uterine endometrium. Endocrinology. 2004;145:5820–5831. doi: 10.1210/en.2004-0991. [DOI] [PubMed] [Google Scholar]
- 7.Jacinto E, et al. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 2006;127:125–137. doi: 10.1016/j.cell.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 8.Yang Q, Inoki K, Ikenoue T, Guan KL. Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes Dev. 2006;20:2820–2832. doi: 10.1101/gad.1461206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colicelli J, et al. Expression of three mammalian cDNAs that interfere with RAS function in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1991;88:2913–2917. doi: 10.1073/pnas.88.7.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilkinson MG, et al. Sin1: An evolutionarily conserved component of the eukaryotic SAPK pathway. EMBO J. 1999;18:4210–4221. doi: 10.1093/emboj/18.15.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schroder W, Bushell G, Sculley T. The human stress-activated protein kinase-interacting 1 gene encodes JNK-binding proteins. Cell Signalling. 2005;17:761–767. doi: 10.1016/j.cellsig.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 12.Cheng J, et al. Mip1, an MEKK2-interacting protein, controls MEKK2 dimerization and activation. Mol Cell Biol. 2005;25:5955–5964. doi: 10.1128/MCB.25.14.5955-5964.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Makino C, Sano Y, Shinagawa T, Millar JB, Ishii S. Sin1 binds to both ATF-2 and p38 and enhances ATF-2-dependent transcription in an SAPK signaling pathway. Genes Cells. 2006;11:1239–1251. doi: 10.1111/j.1365-2443.2006.01016.x. [DOI] [PubMed] [Google Scholar]
- 14.Molinaro RJ, et al. Selection and cloning of poly(rC)-binding protein 2 and Raf kinase inhibitor protein RNA activators of 2′,5′-oligoadenylate synthetase from prostate cancer cells. Nucleic Acids Res. 2006;34:6684–6695. doi: 10.1093/nar/gkl968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Makeyev AV, Liebhaber SA. The poly(C)-binding proteins: A multiplicity of functions and a search for mechanisms. RNA. 2002;8:265–278. doi: 10.1017/s1355838202024627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanjay A, Fu J, Kreibich G. DAD1 is required for the function and the structural integrity of the oligosaccharyltransferase complex. J Biol Chem. 1998;273:26094–26099. doi: 10.1074/jbc.273.40.26094. [DOI] [PubMed] [Google Scholar]
- 17.Polak P, Hall MN. mTORC2 caught in a SINful Akt. Dev Cell. 2006;11:433–434. doi: 10.1016/j.devcel.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 18.Loewith R, et al. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell. 2002;10:457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 19.Lee S, et al. TOR complex 2 integrates cell movement during chemotaxis and signal relay in Dictyostelium. Mol Biol Cell. 2005;16:4572–4583. doi: 10.1091/mbc.E05-04-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schroder WA, et al. Human Sin1 contains Ras-binding and pleckstrin homology domains and suppresses Ras signalling. Cell Signalling. 2007;19:1279–1289. doi: 10.1016/j.cellsig.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 21.Wang SZ, Roberts RM. The evolution of the Sin1 gene product, a little known protein implicated in stress responses and type I interferon signaling in vertebrates. BMC Evol Biol. 2005;5:13. doi: 10.1186/1471-2148-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dejgaard K, Leffers H. Characterisation of the nucleic-acid-binding activity of KH domains. Different properties of different domains. Eur J Biochem. 1996;241:425–431. doi: 10.1111/j.1432-1033.1996.00425.x. [DOI] [PubMed] [Google Scholar]
- 23.Kong J, Sumaroka M, Eastmond DL, Liebhaber SA. Shared stabilization functions of pyrimidine-rich determinants in the erythroid 15-lipoxygenase and alpha-globin mRNAs. Mol Cell Biol. 2006;26:5603–5614. doi: 10.1128/MCB.01845-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitchell P, Tollervey D. mRNA stability in eukaryotes. Curr Opin Genet Dev. 2000;10:193–198. doi: 10.1016/s0959-437x(00)00063-0. [DOI] [PubMed] [Google Scholar]
- 25.Ostareck-Lederer A, Ostareck DH. Control of mRNA translation and stability in haematopoietic cells: The function of hnRNPs K and E1/E2. Biol Cell. 2004;96:407–411. doi: 10.1016/j.biolcel.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 26.Kim SS, et al. Poly(C) binding protein family is a transcription factor in mu-opioid receptor gene expression. Mol Pharmacol. 2005;68:729–736. doi: 10.1124/mol.105.012245. [DOI] [PubMed] [Google Scholar]
- 27.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 28.Clemens MJ, Bushell M, Jeffrey IW, Pain VM, Morley SJ. Translation initiation factor modifications and the regulation of protein synthesis in apoptotic cells. Cell Death Differ. 2000;7:603–615. doi: 10.1038/sj.cdd.4400695. [DOI] [PubMed] [Google Scholar]
- 29.Nevins TA, Harder ZM, Korneluk RG, Holcik M. Distinct regulation of internal ribosome entry site-mediated translation following cellular stress is mediated by apoptotic fragments of eIF4G translation initiation factor family members eIF4GI and p97/DAP5/NAT1. J Biol Chem. 2003;278:3572–3579. doi: 10.1074/jbc.M206781200. [DOI] [PubMed] [Google Scholar]
- 30.Jiang Y, Woronicz JD, Liu W, Goeddel DV. Prevention of constitutive TNF receptor 1 signaling by silencer of death domains. Science. 1999;283:543–546. doi: 10.1126/science.283.5401.543. [DOI] [PubMed] [Google Scholar]
- 31.Bhattacharya S, Ray RM, Johnson LR. Prevention of TNF-alpha-induced apoptosis in polyamine-depleted IEC-6 cells is mediated through the activation of ERK1/2. Am J Physiol. 2004;286:G479–G490. doi: 10.1152/ajpgi.00342.2003. [DOI] [PubMed] [Google Scholar]
- 32.Pokrovskaja K, Panaretakis T, Grander D. Alternative signaling pathways regulating type I interferon-induced apoptosis. J Interferon Cytokine Res. 2005;25:799–810. doi: 10.1089/jir.2005.25.799. [DOI] [PubMed] [Google Scholar]
- 33.Chen Y, et al. Effect of interferon-tau administration on endometrium of nonpregnant ewes: A comparison with pregnant ewes. Endocrinology. 2006;147:2127–2137. doi: 10.1210/en.2005-1310. [DOI] [PubMed] [Google Scholar]
- 34.Wang B, Xiao C, Goff AK. Progesterone-modulated induction of apoptosis by interferon-tau in cultured epithelial cells of bovine endometrium. Biol Reprod. 2003;68:673–679. doi: 10.1095/biolreprod.102.006924. [DOI] [PubMed] [Google Scholar]
- 35.Chou MT, Wang J, Fujita DJ. Src kinase becomes preferentially associated with the VEGFR, KDR/Flk-1, following VEGF stimulation of vascular endothelial cells. BMC Biochem. 2002;3:32. doi: 10.1186/1471-2091-3-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.